Changes in the Histology of Walnut (Juglans regia L.) Infected with Phomopsis capsici and Transcriptome and Metabolome Analysis
Abstract
:1. Introduction
2. Results
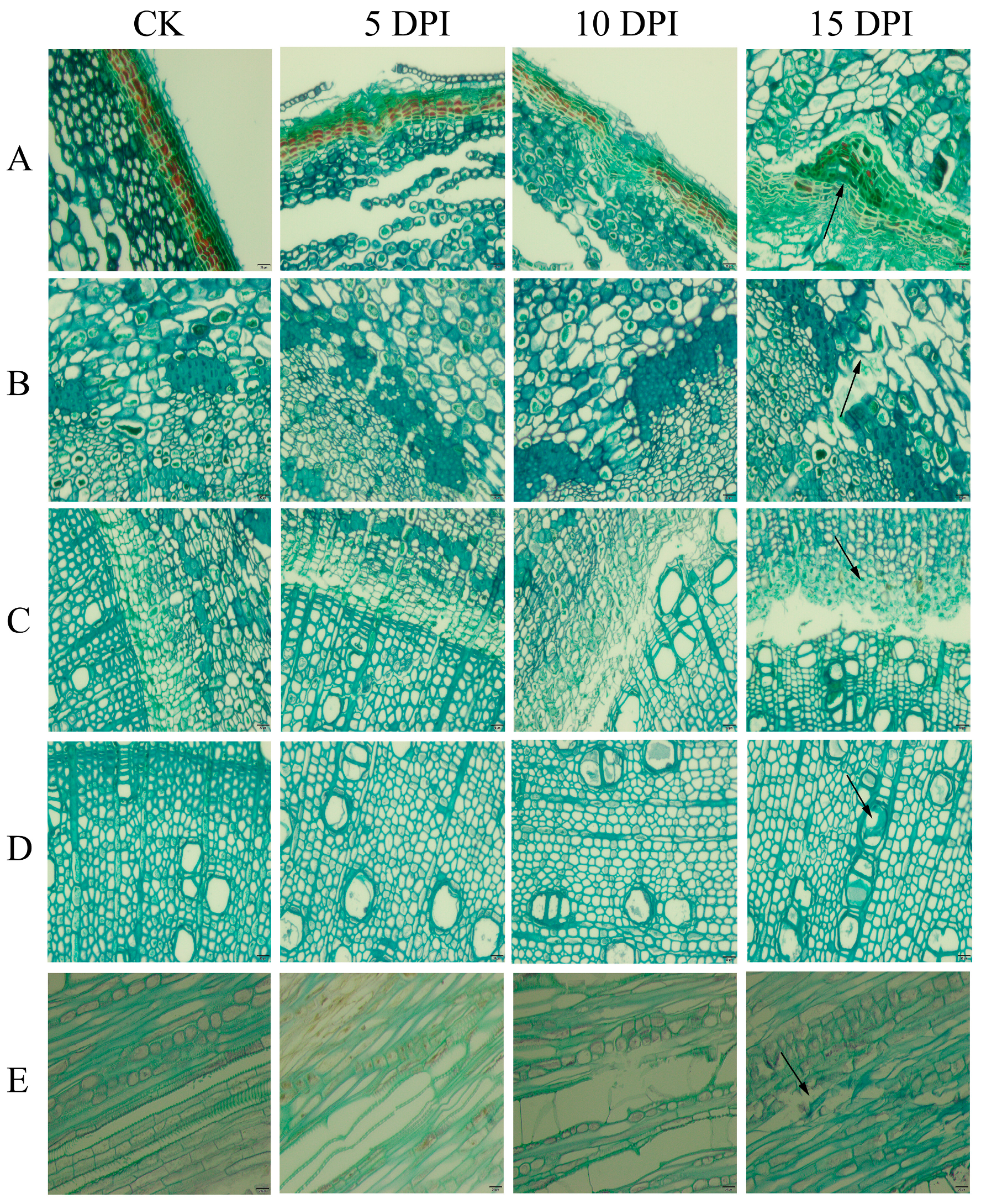
2.1. Section Observation of Walnut in Response to P. capsici Infection
2.2. Transcriptomic Results of Walnut Response to P. capsici Infection
2.2.1. RNA Quality Detection and Sequencing Results
2.2.2. Identification of Infected DEG and Functional Analysis
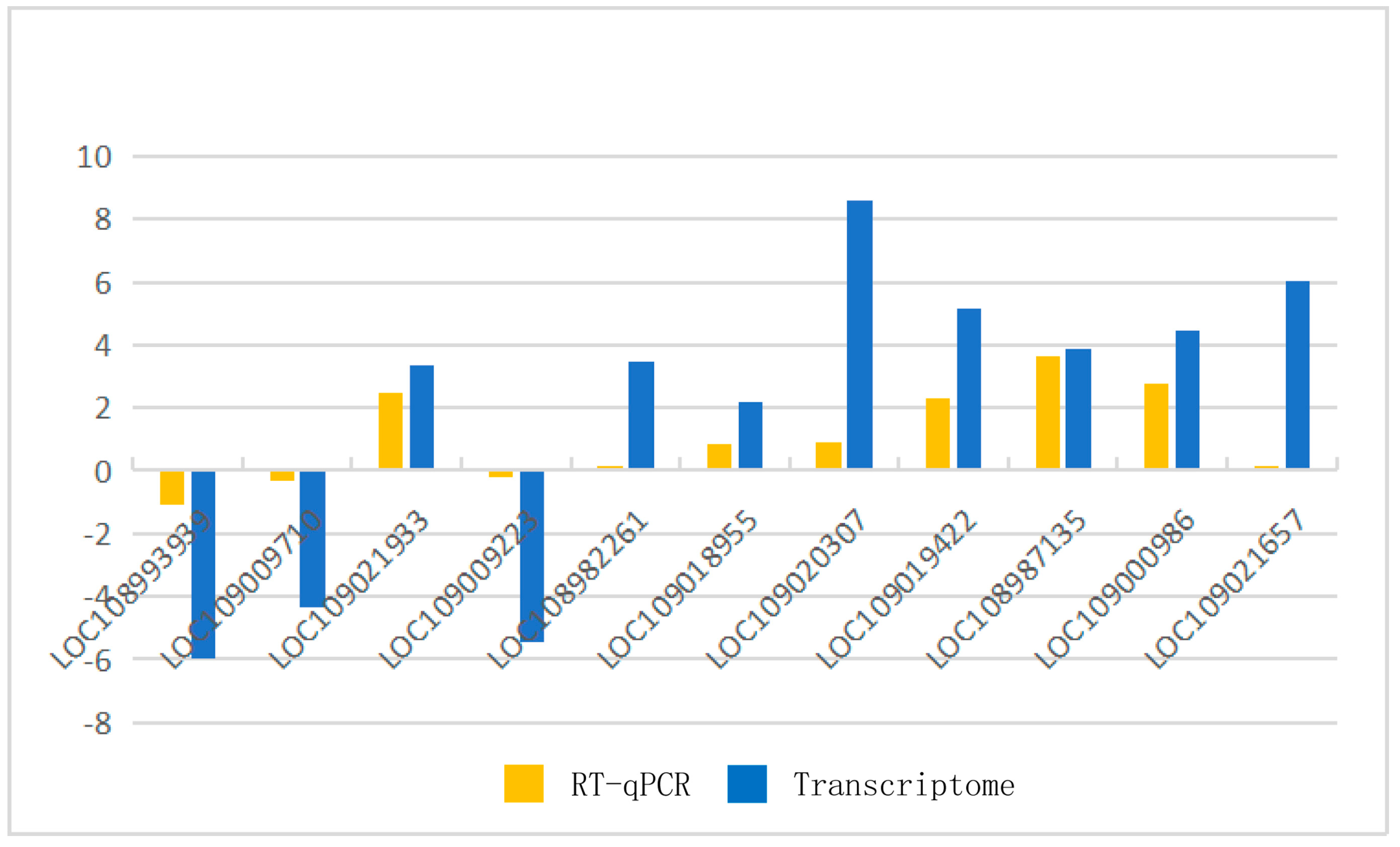
2.2.3. Verification by Real-Time Quantitative Polymerase Chain Reaction (RT-qPCR)
2.3. Metabolome Data Analysis
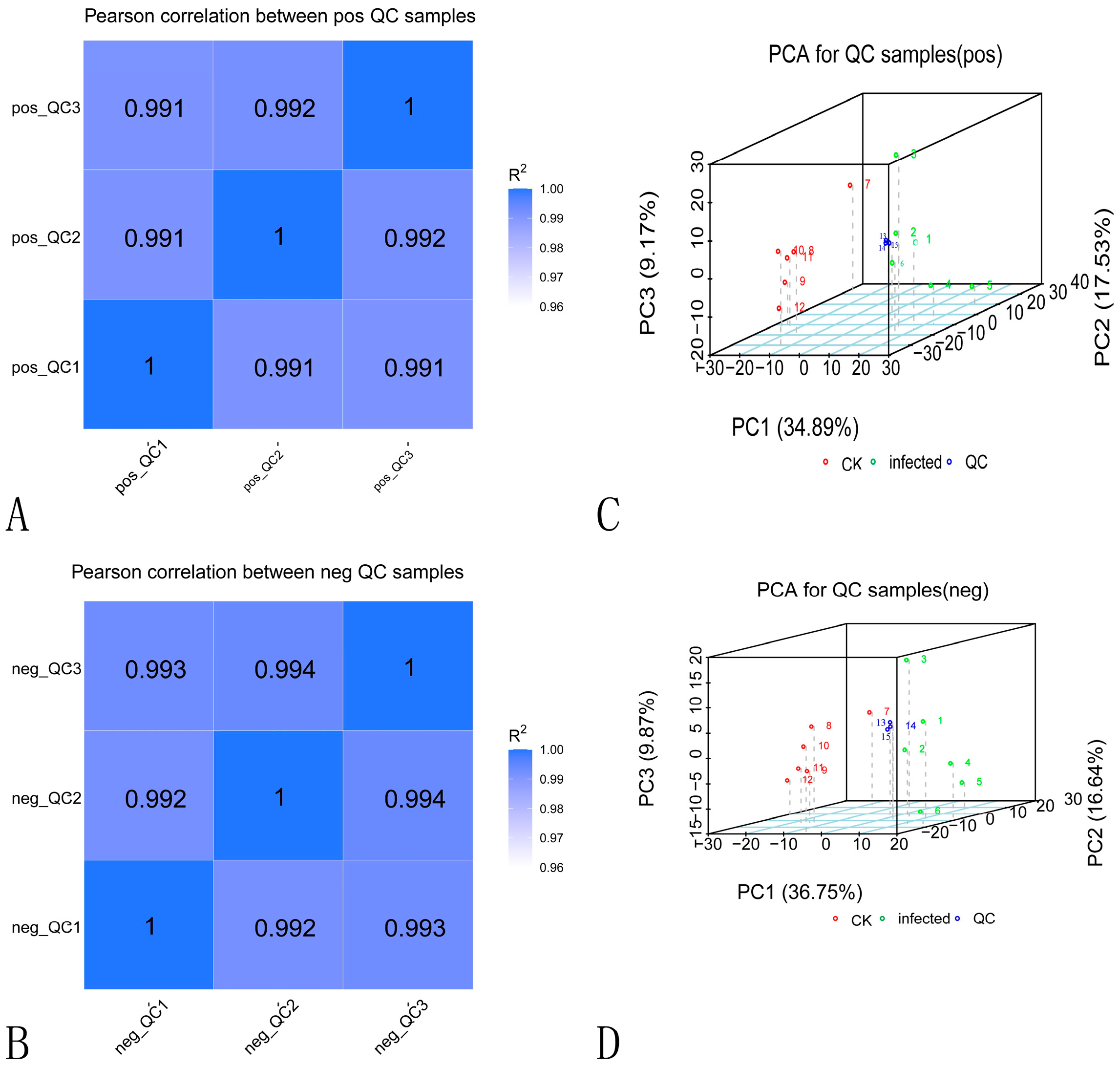
2.3.1. Data Quality Control
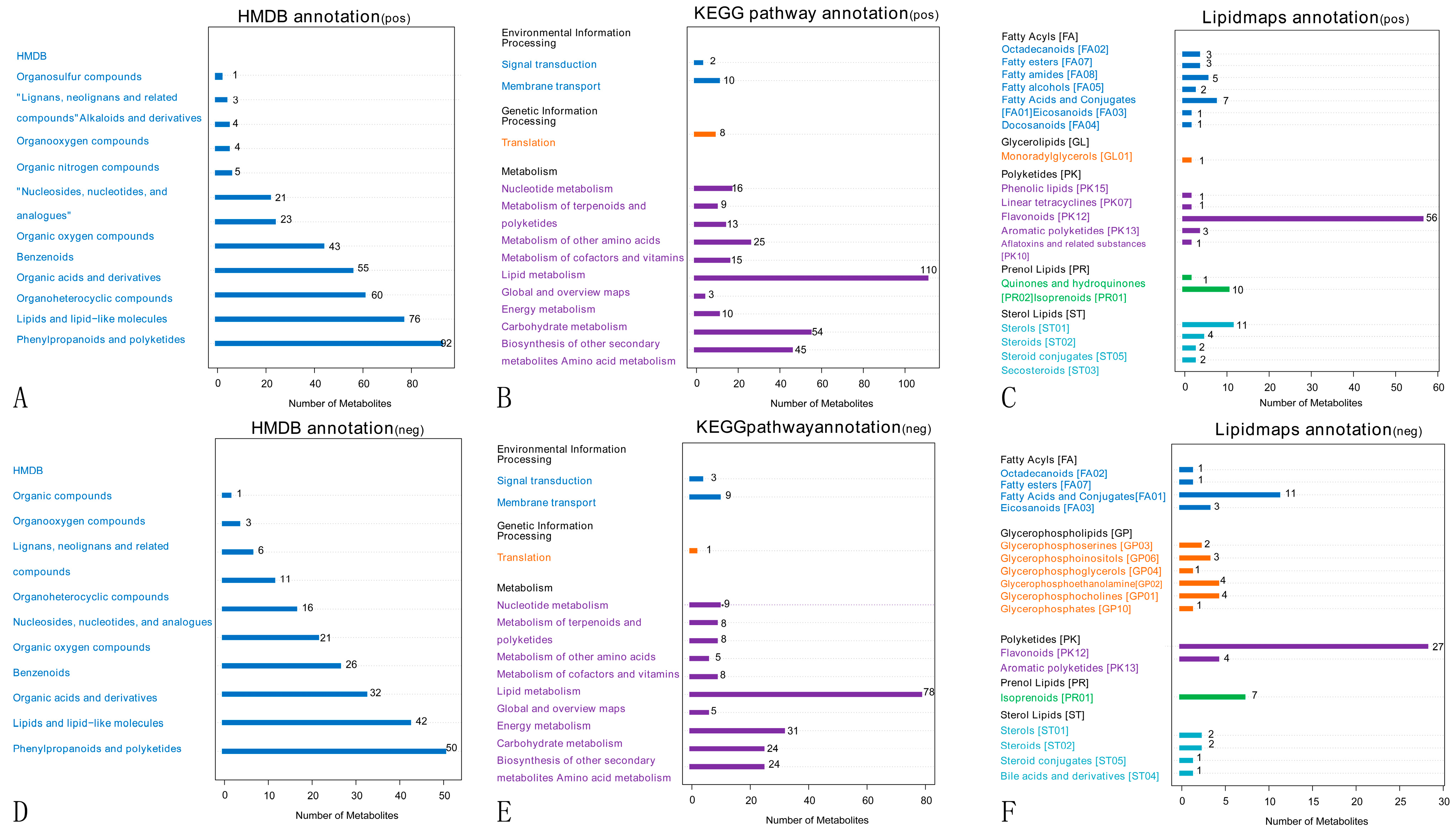
2.3.2. Metabolite Pathways and Classification Notes
2.3.3. DEM Screening and KEGG Pathway Annotation
2.3.4. Transcriptome and Metabolome Association Analysis
3. Discussion
4. Materials and Methods
4.1. Plant Materials and Pathogenic Bacterial Inoculation
4.2. Paraffin Sectioning of Walnut Branches
4.3. RNA Sequencing
4.4. Transcriptome Analysis
4.5. RT-qPCR Verification
4.6. Metabolite Extraction and Detection
4.7. Data Processing and Metabolite Identification
4.8. Data Analysis
4.9. Metabolome and Transcriptome Association Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Conflicts of Interest
References
- Zhao, B.; Liu, F.; Gong, Y.; Zhang, X.; Wang, X.; Wang, Y. The breeding and the performance of ‘Liaoning 4′ walnut cultivar. Breeding Sci. 2020, 70, 547–550. [Google Scholar] [CrossRef] [PubMed]
- Wang, F.; Zeng, Q.; Liu, C.; Xu, X.; Zhu, T.; Liu, Y.; Yang, H.; Han, S.; Yang, H.; Li, S.; et al. Branch blight of Juglans sigillata caused by Juglanconis appendiculata in China. Plant Dis. 2022, 107, 224. [Google Scholar] [CrossRef] [PubMed]
- Mihaescu, C.; Dunea, D.; Basa, A.G.; Frasin, L.N. Characteristics of Phomopsis juglandina (Sacc.) Hohn. Associated with Dieback of Walnut in the Climatic Conditions of Southern Romania. Agronomy 2021, 11, 46. [Google Scholar] [CrossRef]
- Filippi, C.V.; Corro Molas, A.; Dominguez, M.; Colombo, D.; Heinz, N.; Troglia, C.; Maringolo, C.; Quiroz, F.; Alvarez, D.; Lia, V.; et al. Genome-Wide Association Studies in Sunflower: Towards Sclerotinia sclerotiorum and Diaporthe/Phomopsis Resistance Breeding. Genes 2022, 13, 2357. [Google Scholar] [CrossRef] [PubMed]
- Santos, J.M.; Vrandecic, K.; Cosic, J.; Duvnjak, T.; Phillips, A.J.L. Resolving the Diaporthe species occurring on soybean in Croatia. Persoonia 2011, 27, 9–19. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, X.; He, P.; Zhu, T.; Pan, Z. Characterization of a new pathogen in walnut twig blight. Microbiol. China 2017, 44, 1339–1348. [Google Scholar]
- Kaluzna, M.; Fischer-Le Saux, M.; Pothier, J.F.; Jacques, M.; Obradovic, A.; Tavares, F.; Stefani, E. Xanthomonas arboricola pv. juglandis and pv. corylina: Brothers or distant relatives? Genetic clues, epidemiology, and insights for disease management. Mol. Plant Pathol. 2021, 22, 1481–1499. [Google Scholar] [CrossRef]
- Reza Pouralibaba, H.; Perez-de-Luque, A.; Rubiales, D. Histopathology of the infection on resistant and susceptible lentil accessions by two contrasting pathotypes of Fusarium oxysporum f.sp lentis. Eur. J. Plant Pathol. 2017, 148, 53–63. [Google Scholar] [CrossRef] [Green Version]
- Yang, J.; Zhang, Y.; Wang, X.; Wang, W.; Li, Z.; Wu, J.; Wang, G.; Wu, L.; Zhang, G.; Ma, Z. HyPRP1 performs a role in negatively regulating cotton resistance to V-dahliae via the thickening of cell walls and ROS accumulation. BMC Plant Biol. 2018, 18, 339. [Google Scholar] [CrossRef]
- Chowdhury, J.; Henderson, M.; Schweizer, P.; Burton, R.A.; Fincher, G.B.; Little, A. Differential accumulation of callose, arabinoxylan and cellulose in nonpenetrated versus penetrated papillae on leaves of barley infected with Blumeria graminis f. sp hordei. New Phytol. 2014, 204, 650–660. [Google Scholar] [CrossRef]
- Dangl, J.L.; Horvath, D.M.; Staskawicz, B.J. Pivoting the Plant Immune System from Dissection to Deployment. Science 2013, 341, 746–751. [Google Scholar] [CrossRef] [Green Version]
- Heese, A.; Hann, D.R.; Gimenez-Ibanez, S.; Jones, A.M.E.; He, K.; Li, J.; Schroeder, J.I.; Peck, S.C.; Rathjen, J.P. The receptor-like kinase SERK3/BAK1 is a central regulator of innate immunity in plants. Proc. Natl. Acad. Sci. USA 2007, 104, 12217–12222. [Google Scholar] [CrossRef] [Green Version]
- Khanam, S.; Bauters, L.; Singh, R.R.; Verbeek, R.; Haeck, A.; Sultan, S.M.D.; Demeestere, K.; Kyndt, T.; Gheysen, G. Mechanisms of resistance in the rice cultivar Manikpukha to the rice stem nematode Ditylenchus angustus. Mol. Plant Pathol. 2018, 19, 1391–1402. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jiang, N.; Zhang, S. Effects of Combined Application of Potassium Silicate and Salicylic Acid on the Defense Response of Hydroponically Grown Tomato Plants to Ralstonia solanacearum Infection. Sustainability 2021, 13, 3750. [Google Scholar] [CrossRef]
- Maeda, H.A. Evolutionary Diversification of Primary Metabolism and Its Contribution to Plant Chemical Diversity. Front. Plant Sci. 2019, 10, 881. [Google Scholar] [CrossRef]
- Erb, M.; Kliebenstein, D.J. Plant Secondary Metabolites as Defenses, Regulators, and Primary Metabolites: The Blurred Functional Trichotomy. Plant Physiol. 2020, 184, 39–52. [Google Scholar] [CrossRef] [PubMed]
- Lacerda, J.W.F.; Siqueira, K.A.; Vasconcelos, L.G.; Bellete, B.S.; Dall’Oglio, E.L.; Sousa Junior, P.T.; Faraggi, T.M.; Vieira, L.C.C.; Soares, M.A.; Sampaio, O.M. Metabolomic Analysis of Combretum lanceolatum Plants Interaction with Diaporthe phaseolorum and Trichoderma spirale Endophytic Fungi through H-1-NMR. Chem. Biodivers. 2021, 18, e2100350. [Google Scholar] [CrossRef] [PubMed]
- Zamljen, T.; Veberic, R.; Hudina, M.; Slatnar, A. The Brown Marmorated Stink Bug (Halyomorpha halys Stal.) Influences Pungent and Non-Pungent Capsicum Cultivars’ Pre- and Post-Harvest Quality. Agronomy 2021, 11, 2252. [Google Scholar] [CrossRef]
- Arbona, V.; Gomez-Cadenas, A. Metabolomics of Disease Resistance in Crops. Curr. Issues Mol. Biol. 2016, 19, 13–29. [Google Scholar] [CrossRef]
- Zeiss, D.R.; Mhlongo, M.I.; Tugizimana, F.; Steenkamp, P.A.; Dubery, I.A. Metabolomic Profiling of the Host Response of Tomato (Solanum lycopersicum) Following Infection by Ralstonia solanacearum. Int. J. Mol. Sci. 2019, 20, 3945. [Google Scholar] [CrossRef] [Green Version]
- Jia, H.; Li, T.; Haider, M.S.; Sadeghnezhad, E.; Pang, Q.; Han, J.; Zhang, P.; Su, L.; Jia, H.; Fang, J. Comparative Transcriptomic and Metabolomic Profiling of Grapevine Leaves (cv. Kyoho) upon Infestation of Grasshopper and Botrytis cinerea. Plant Mol. Biol. Rep. 2022, 40, 539–555. [Google Scholar] [CrossRef]
- Manresa-Grao, M.; Pastor-Fernandez, J.; Sanchez-Bel, P.; Jaques, J.A.; Pastor, V.; Flors, V. Mycorrhizal Symbiosis Triggers Local Resistance in Citrus Plants Against Spider Mites. Front. Plant Sci. 2022, 13, 867778. [Google Scholar] [CrossRef]
- Nehela, Y.; Killiny, N. Melatonin Is Involved in Citrus Response to the Pathogen Huanglongbing via Modulation of Phytohormonal Biosynthesis. Plant Physiol. 2020, 184, 2216–2239. [Google Scholar] [CrossRef]
- Yang, X.; Huang, J.; Yang, H.; Liao, H.; Xu, F.; Zhu, B.; Xu, X.; Zhang, L.; Huang, H.; Du, Q.; et al. Transcriptome and metabolome profiling in different stages of infestation of Eucalyptus urophylla clones by Ralstonia solanacearum. Mol. Genet. Genomic 2022, 297, 1081–1100. [Google Scholar] [CrossRef]
- Yadeta, K.; Thomma, B. The xylem as battleground for plant hosts and vascular wilt pathogens. Front. Plant Sci. 2013, 4, 97. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bae, C.; Han, S.W.; Song, Y.R.; Kim, B.Y.; Lee, H.J.; Lee, J.M.; Yeam, I.; Heu, S.; Oh, C.S. Infection processes of xylem-colonizing pathogenic bacteria: Possible explanations for the scarcity of qualitative disease resistance genes against them in crops. Theor. Appl. Genet. 2015, 128, 1219–1229. [Google Scholar] [CrossRef] [PubMed]
- Martin Mantz, G.; Ruben Rossi, F.; Esteban Viretto, P.; Cristina Noelting, M.; Javier Maiale, S. Stem canker caused by Phomopsis spp. Induces changes in polyamine levels and chlorophyll fluorescence parameters in pecan leaves. Plant Physiol. Bioch. 2021, 166, 761–769. [Google Scholar] [CrossRef]
- Nagy, N.E.; Franceschi, V.R.; Solheim, H.; Krekling, T.; Christiansen, E. Wound-induced traumatic resin duct development in stems of Norway spruce (Pinaceae): Anatomy and cytochemical traits. Am. J. Bot. 2000, 87, 302–313. [Google Scholar] [CrossRef]
- Pickard, W.F. Laticifers and secretory ducts: Two other tube systems in plants. New Phytol. 2008, 177, 877–887. [Google Scholar] [CrossRef]
- Bowers, W.H.; Williams, D.G. Isotopic Heterogeneity of Stem Water in Conifers Is Correlated to Xylem Hydraulic Traits and Supports Multiple Residence Times. Front. Water 2022, 4, 46. [Google Scholar] [CrossRef]
- Holtta, T.; Cochard, H.; Nikinmaa, E.; Mencuccini, M. Capacitive effect of cavitation in xylem conduits: Results from a dynamic model. Plant Cell Environ. 2009, 32, 10–21. [Google Scholar] [CrossRef] [PubMed]
- Lintunen, A.; Lindfors, L.; Nikinmaa, E.; Holtta, T. Xylem diameter changes during osmotic stress, desiccation and freezing in Pinus sylvestris and Populus tremula. Tree Physiol. 2017, 37, 491–500. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cornelis, S.; Hazak, O. Understanding the root xylem plasticity for designing resilient crops. Plant Cell Environ. 2022, 45, 664–676. [Google Scholar] [CrossRef] [PubMed]
- Petit, G.; Bleve, G.; Gallo, A.; Mita, G.; Montanaro, G.; Nuzzo, V.; Zambonini, D.; Pitacco, A. Susceptibility to Xylella fastidiosa and functional xylem anatomy in Olea europaea: Revisiting a tale of plant-pathogen interaction. AoB Plants 2021, 13, plab027. [Google Scholar] [CrossRef] [PubMed]
- Kashyap, A.; Planas-Marques, M.; Capellades, M.; Valls, M.; Coll, N.S. Blocking intruders: Inducible physico-chemical barriers against plant vascular wilt pathogens. J. Exp. Bot. 2021, 72, 184–198. [Google Scholar] [CrossRef]
- Campos, F.G.; Ribeiro Vieira, M.A.; Esteves Amaro, A.C.; DelaCruz-Chacon, I.; Mayo Marques, M.O.; Ferreira, G.; Fernandes Boaro, C.S. Nitrogen in the defense system of Annona emarginata (Schltdl.) H. Rainer. PLoS ONE 2019, 14, e0217930. [Google Scholar] [CrossRef]
- de Oliveira, C.S.; Liao, L.M.; Alcantara, G.B. Metabolic response of soybean plants to Sclerotinia sclerotiorum infection. Phytochemistry 2019, 167, 112099. [Google Scholar] [CrossRef]
- Li, P.; Liu, W.; Zhang, Y.; Xing, J.; Li, J.; Feng, J.; Su, X.; Zhao, J. Fungal canker pathogens trigger carbon starvation by inhibiting carbon metabolism in poplar stems. Sci. Rep. 2019, 9, 10111. [Google Scholar] [CrossRef] [Green Version]
- Ramu, V.S.; Dawane, A.; Lee, S.; Oh, S.; Lee, H.K.; Sun, L.; Senthil-Kumar, M.; Mysore, K.S. Ribosomal protein QM/RPL10 positively regulates defence and protein translation mechanisms during nonhost disease resistance. Mol. Plant Pathol. 2020, 21, 1481–1494. [Google Scholar] [CrossRef]
- Chouhan, R.; Ahmed, S.; Gandhi, S.G. Over-expression of PR proteins with chitinase activity in transgenic plants for alleviation of fungal pathogenesis. J. Plant Pathol. 2022, 1–13. [Google Scholar] [CrossRef]
- Lin, D.R.; Xiao, M.S.; Zhao, J.J.; Li, Z.H.; Xing, B.S.; Li, X.D.; Kong, M.Z.; Li, L.Y.; Zhang, Q.; Liu, Y.W.; et al. An Overview of Plant Phenolic Compounds and Their Importance in Human Nutrition and Management of Type 2 Diabetes. Molecules 2016, 21, 1374. [Google Scholar] [CrossRef]
- Wilson, R.A.; Gibson, R.P.; Quispe, C.F.; Littlechild, J.A.; Talbot, N.J. An NADPH-dependent genetic switch regulates plant infection by the rice blast fungus. Proc. Natl. Acad. Sci. USA 2010, 107, 21902–21907. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, S.; Ren, J.; Zhao, H.; Wang, X.; Wang, T.; Jin, S.; Wang, Z.; Li, C.; Liu, A.; Lin, X.; et al. Trichoderma harzianum Improves Defense Against Fusarium oxysporum by Regulating ROS and RNS Metabolism, Redox Balance, and Energy Flow in Cucumber Roots. Phytopathology 2019, 109, 972–982. [Google Scholar] [CrossRef] [PubMed]
- Fahrendorf, T.; Ni, W.T.; Shorrosh, B.S.; Dixon, R.A. Stress Responses in alfalfa (Medicago-sativa L.). 19. Transcriptional activation of oxidative pentose-phosphate pathway genes at the onset of the isoflavonoid phytoalexin response. Plant Mol. Biol. 1995, 28, 885–900. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lee, O.; Kwon, Y.; Apostolidis, E.; Shetty, K.; Kim, Y. Rhodiola-induced Inhibition of Adipogenesis Involves Antioxidant Enzyme Response Associated with Pentose Phosphate Pathway. Phytother. Res. 2011, 25, 106–115. [Google Scholar] [CrossRef]
- Xie, Y.; Hu, L.; Du, Z.; Sun, X.; Amombo, E.; Fan, J.; Fu, J. Effects of Cadmium Exposure on Growth and Metabolic Profile of Bermudagrass [Cynodon dactylon (L.) Pers.]. PLoS ONE 2014, 9, e115279. [Google Scholar] [CrossRef] [PubMed]
- Corredor-Moreno, P.; Minter, F.; Davey, P.E.; Wegel, E.; Kular, B.; Brett, P.; Lewis, C.M.; Morgan, Y.M.L.; Perez, L.A.M.; Korolev, A.V.; et al. The branched-chain amino acid aminotransferase TaBCAT1 modulates amino acid metabolism and positively regulates wheat rust susceptibility. Plant Cell 2021, 33, 1728–1747. [Google Scholar] [CrossRef]
- Li, Y.; Li, S.; Du, R.; Wang, J.; Li, H.; Xie, D.; Yan, J. Isoleucine Enhances Plant Resistance Against Botrytis cinerea via Jasmonate Signaling Pathway. Front. Plant Sci. 2021, 12, 1738. [Google Scholar] [CrossRef]
- Zhao, P.; Gu, S.; Han, C.; Lu, Y.; Ma, C.; Tian, J.; Bi, J.; Deng, Z.; Wang, Q.; Xu, Q. Targeted and Untargeted Metabolomics Profiling of Wheat Reveals Amino Acids Increase Resistance to Fusarium Head Blight. Front. Plant Sci. 2021, 12, 762605. [Google Scholar] [CrossRef]
- Samsatly, J.; Bayen, S.; Jabaji, S.H. Vitamin B6 Is Under a Tight Balance During Disease Development by Rhizoctonia solanion Different Cultivars of Potato and on Arabidopsis thaliana Mutants. Front. Plant Sci. 2020, 11, 875. [Google Scholar] [CrossRef]
- Liu, H.; Lu, C.; Li, Y.; Wu, T.; Zhang, B.; Liu, B.; Feng, W.; Xu, Q.; Dong, H.; He, S.; et al. The bacterial effector AvrRxo1 inhibits vitamin B6 biosynthesis to promote infection in rice. Plant Commun. 2022, 3, 100324. [Google Scholar] [CrossRef]
- Gutensohn, M.; Hartzell, E.; Dudareva, N. Another level of complex-ity: The role of metabolic channeling and metabolons in plant terpenoid metabolism. Front. Plant Sci. 2022, 13, 954083. [Google Scholar] [CrossRef]
- Zhao, Y.; Cui, J.; Liu, M.; Zhao, L. Progress on Terpenoids with Biological Activities Produced by Plant Endophytic Fungi in China between 2017 and 2019. Nat. Prod. Commun. 2020, 15, 1934578X20937204. [Google Scholar] [CrossRef]
- Ruan, X.; Ma, L.; Zhang, Y.; Wang, Q.; Gao, X. Dissection of the Complex Transcription and Metabolism Regulation Networks Associated with Maize Resistance to Ustilago maydis. Genes 2021, 12, 1789. [Google Scholar] [CrossRef]
- Rani, M.; Jha, G. Host Gamma-Aminobutyric Acid Metabolic Pathway Is Involved in Resistance Against Rhizoctonia solani. Phytopathology 2021, 111, 1207–1218. [Google Scholar] [CrossRef] [PubMed]
- Wang, C.; Tang, D.; Gao, Y.; Zhang, L. Succinic Semialdehyde Promotes Prosurvival Capability of Agrobacterium tumefaciens. J. Bacteriol. 2016, 198, 930–940. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gurtler, J.B.; Keller, S.E.; Fan, X.; Olanya, O.M.; Jin, T.; Camp, M.J. Survival of Salmonella during Apple Dehydration as Affected by Apple Cultivar and Antimicrobial Pretreatment. J. Food Protect. 2020, 83, 902–909. [Google Scholar] [CrossRef] [PubMed]
- Khanna, K.; Jamwal, V.L.; Sharma, A.; Gandhi, S.G.; Ohri, P.; Bhardwaj, R.; Al-Huqail, A.A.; Siddiqui, M.H.; Marraiki, N.; Ahmad, P. Evaluation of the role of rhizobacteria in controlling root-knot nematode infection in Lycopersicon esculentum plants by modulation in the secondary metabolite profiles. AoB Plants 2019, 11, plz069. [Google Scholar] [CrossRef]
- Khanna, K.; Sharma, A.; Ohri, P.; Bhardwaj, R.; Abd Allah, E.F.; Hashem, A.; Ahmad, P. Impact of Plant Growth Promoting Rhizobacteria in the Orchestration of Lycopersicon esculentum Mill. Resistance to Plant Parasitic Nematodes: A Metabolomic Approach to Evaluate Defense Responses Under Field Conditions. Biomolecules 2019, 9, 676. [Google Scholar] [CrossRef] [Green Version]
- Zaveri, A.; Edwards, J.; Rochfort, S. Production of Primary Metabolites by Rhizopus stolonifer, Causal Agent of Almond Hull Rot Disease. Molecules 2022, 27, 7199. [Google Scholar] [CrossRef]
- Ma, L.; Wang, W.; Shi, R.; Zhang, X.; Li, X.; Yang, Y.; Mo, M.H. Effects of organic acids on the chemotaxis profiles and biocontrol traits of antagonistic bacterial endophytes against root-rot disease in Panax notoginseng. Anton. Leeuw. Int. J. G. 2021, 114, 1771–1789. [Google Scholar] [CrossRef] [PubMed]
- Kojima, M.; Kimura, N.; Miura, R. Regulation of Primary Metabolic Pathways in Oyster Mushroom Mycelia Induced by Blue Light Stimulation: Accumulation of Shikimic Acid. Sci. Rep. 2015, 5, 8630. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, Y.; Huang, L.; Du, F.; Wang, J.; Zhao, X.; Li, Z.; Wang, W.; Xu, J.; Fu, B. Comparative transcriptome and metabolome profiling reveal molecular mechanisms underlying OsDRAP1-mediated salt tolerance in rice. Sci. Rep. 2021, 11, 5166. [Google Scholar] [CrossRef] [PubMed]
- Zhao, X.; Wang, W.; Xie, Z.; Gao, Y.; Wang, C.; Rashid, M.M.; Islam, M.R.; Fu, B.; Li, Z. Comparative analysis of metabolite changes in two contrasting rice genotypes in response to low-nitrogen stress. Crop J. 2018, 6, 464–474. [Google Scholar] [CrossRef]


| Sample | Raw Reads | Clean Reads | Error Rate | Q20 (%) | Q30 (%) | GC pct (%) | Total Map |
|---|---|---|---|---|---|---|---|
| Infected1 | 45,456,664 | 44,189,378 | 0.02 | 98.16 | 94.49 | 46.33 | 38,911,491 (88.06%) |
| Infected2 | 46,940,018 | 45,063,852 | 0.03 | 97.78 | 93.85 | 46.03 | 42,362,147 (94.0%) |
| Infected3 | 45,204,774 | 43,300,026 | 0.02 | 98.07 | 94.30 | 46.45 | 38,563,666 (89.06%) |
| CK1 | 45,829,520 | 43,916,056 | 0.02 | 98.11 | 94.36 | 45.55 | 41,962,692 (95.55%) |
| CK2 | 46,463,026 | 44,739,028 | 0.02 | 98.12 | 94.31 | 45.22 | 42,973,409 (96.05%) |
| CK3 | 42,767,614 | 41,123,598 | 0.03 | 97.83 | 93.93 | 45.55 | 39,110,575 (95.1%) |
| Category | Pathways | DEM Down | DEM Up |
|---|---|---|---|
| Carbohydrate metabolism | Pyruvate metabolism | Fumaric acid; Phosphoenolpyruvic acid | |
| C5-branched dibasic acid metabolism | Itaconic acid; cis-Aconitic acid | ||
| Ascorbate and aldarate metabolism | Ascorbic acid; D-Xylulose 5-phosphate; D-Glucarate | ||
| Fructose and mannose metabolism | α-D-Mannose 1-phosphate | D-Sorbitol | |
| Pentose phosphate pathway | D-Gluconic acid; D-Xylulose 5-phosphate; D-Sedoheptulose 7-phosphate; Gluconolactone | ||
| Butanoate metabolism | Fumaric acid; Succinic semialdehyde | ||
| Inositol phosphate metabolism | D-myo-Inositol 1,4-bisphosphate | ||
| Citrate cycle (TCA cycle) | Fumaric acid; cis-Aconitic acid; Phosphoenolpyruvic acid | ||
| Pentose and glucuronate interconversions | D-Galacturonic acid; D-Xylulose; D-Xylulose 5-phosphate; | Xylitol | |
| Amino sugar and nucleotide sugar metabolism | D-Galacturonic acid | ||
| Amino acid metabolism | Tyrosine metabolism | Fumaric acid; Succinic semialdehyde | |
| Cysteine and methionine metabolism | Glutathione | ||
| Alanine, aspartate, and glutamate metabolism | Fumaric acid; Succinic semialdehyde | ||
| Phenylalanine metabolism | Fumaric acid; N-Acetyl-L-phenylalanine; | Phenylacetaldehyde | |
| Tryptophan metabolism | 3-Hydroxyanthranilic acid | ||
| Lysine degradation | L-Saccharopine; 2-Oxoadipic acid; Glutaric acid | N6-Acetyl-L-lysine; Pipecolic acid; | |
| Biosynthesis of other secondary metabolites | Phenylpropanoid biosynthesis | Scopoletin; Scopolin; Chlorogenic acid | Sinapyl Alcohol; Cinnamaldehyde; |
| Flavonoid biosynthesis | Chlorogenic acid; | Neohesperidin; (−)-Epigallocatechin; Pinocembrin; Prunin; Taxifolin; Naringin; Dihydromyricetin; 5,7,3′,4′,5′-Pentahydroxyflavone; Kaempferol; Luteolin | |
| Stilbenoid, diarylheptanoid, and gingerol biosynthesis | Bisdemethoxycurcumin; Chlorogenic acid; | Resveratrol | |
| Isoquinoline alkaloid biosynthesis | Tyramine; p-Coumaric acid | Corydaline; 3,4-Dihydroxybenzaldehyde; Lycorine | |
| Metabolism of cofactors and vitamins | Nicotinate and nicotinamide metabolism | Fumaric acid; Succinic semialdehyde | |
| Vitamin B6 metabolism | Succinic semialdehyde | ||
| Ubiquinone and other terpenoid–quinone biosynthesis | p-Coumaric acid; Shikonin | Phylloquinone | |
| One carbon pool by folate | Folinic acid |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhou, L.; Zhu, T.; Han, S.; Li, S.; Liu, Y.; Lin, T.; Qiao, T. Changes in the Histology of Walnut (Juglans regia L.) Infected with Phomopsis capsici and Transcriptome and Metabolome Analysis. Int. J. Mol. Sci. 2023, 24, 4879. https://doi.org/10.3390/ijms24054879
Zhou L, Zhu T, Han S, Li S, Liu Y, Lin T, Qiao T. Changes in the Histology of Walnut (Juglans regia L.) Infected with Phomopsis capsici and Transcriptome and Metabolome Analysis. International Journal of Molecular Sciences. 2023; 24(5):4879. https://doi.org/10.3390/ijms24054879
Chicago/Turabian StyleZhou, Leming, Tianhui Zhu, Shan Han, Shujiang Li, Yinggao Liu, Tiantian Lin, and Tianmin Qiao. 2023. "Changes in the Histology of Walnut (Juglans regia L.) Infected with Phomopsis capsici and Transcriptome and Metabolome Analysis" International Journal of Molecular Sciences 24, no. 5: 4879. https://doi.org/10.3390/ijms24054879
APA StyleZhou, L., Zhu, T., Han, S., Li, S., Liu, Y., Lin, T., & Qiao, T. (2023). Changes in the Histology of Walnut (Juglans regia L.) Infected with Phomopsis capsici and Transcriptome and Metabolome Analysis. International Journal of Molecular Sciences, 24(5), 4879. https://doi.org/10.3390/ijms24054879






