Characterization of the Seed Biopriming, Plant Growth-Promoting and Salinity-Ameliorating Potential of Halophilic Fungi Isolated from Hypersaline Habitats
Abstract
1. Introduction
2. Results
2.1. Microbial Isolation from Saline Soils
2.2. Assessment of IAA in Fungal Culture Filtrates
2.3. Effect of Isolated Microbes on Wheat Seed Germination and Seedling Length
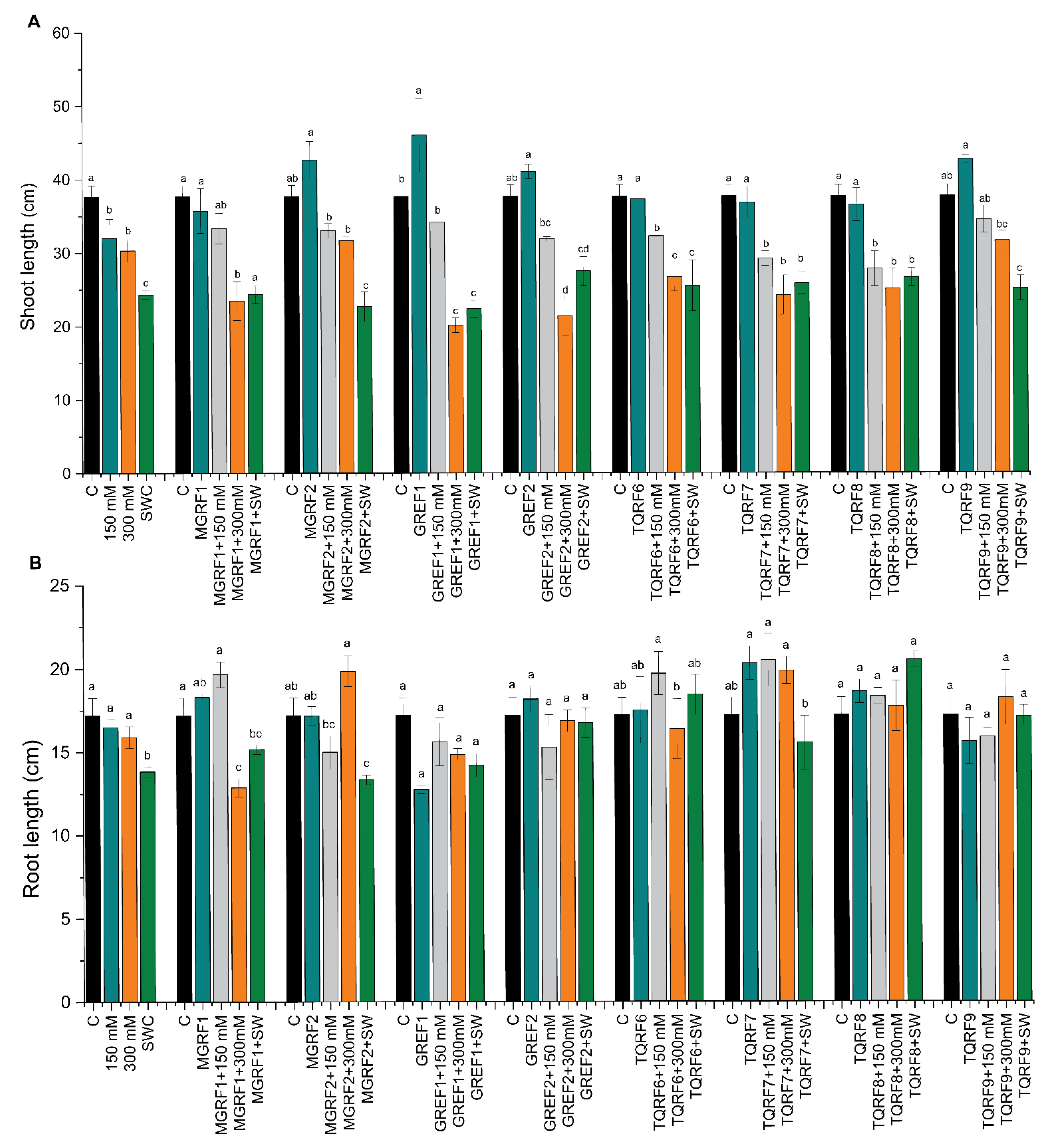
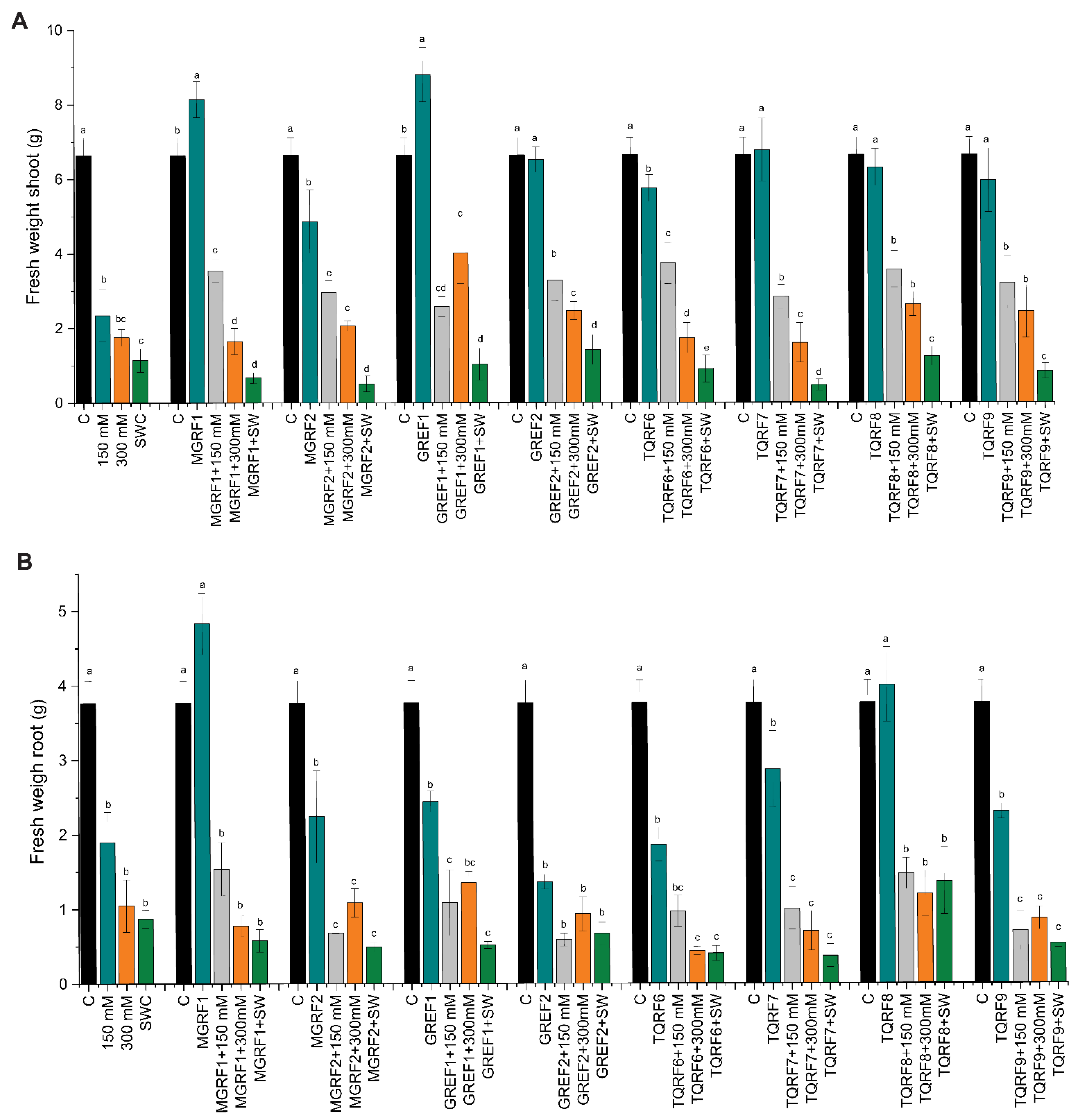
2.4. Effect of Halophilic Fungi on Wheat Seedling Growth and Biomass
2.5. Effect of Fungal Cultural Filtrates on Wheat Plants Growth under Salinity Stress
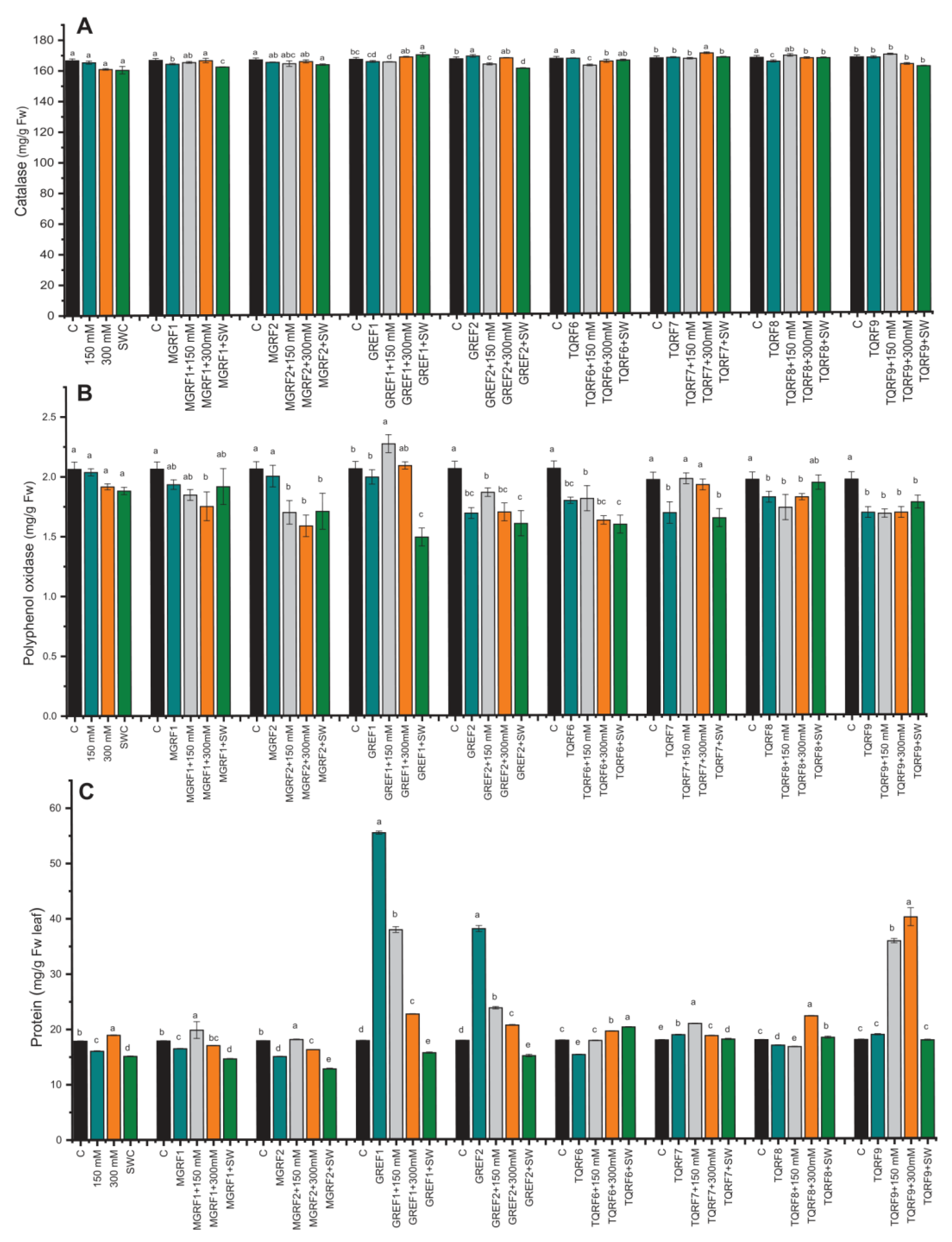
2.6. Regulation of Enzymatic and Non-Enzymatic Antioxidants under Salinity Stress
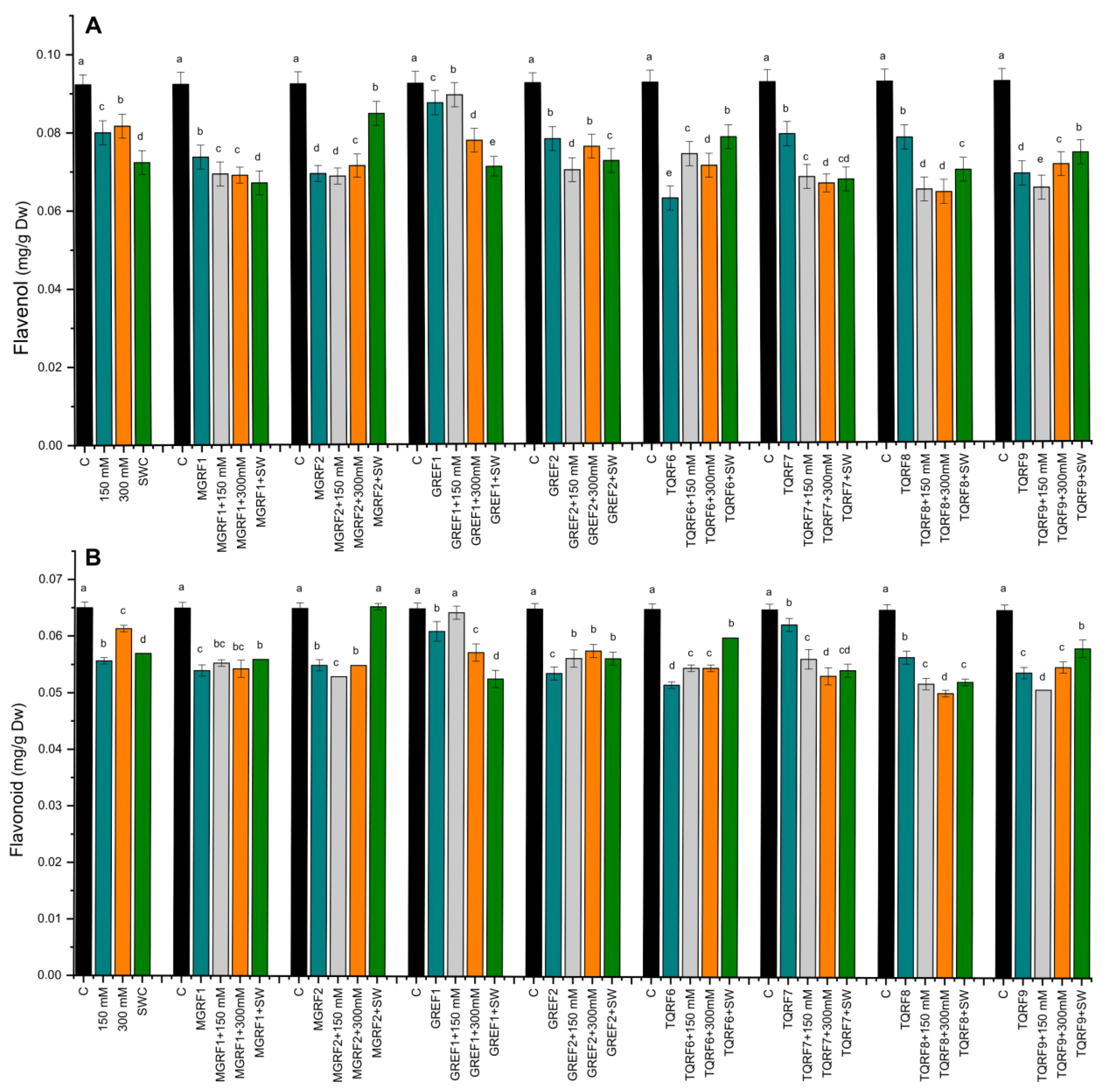
2.7. Determination of Flavonoids and Flavonols Contents
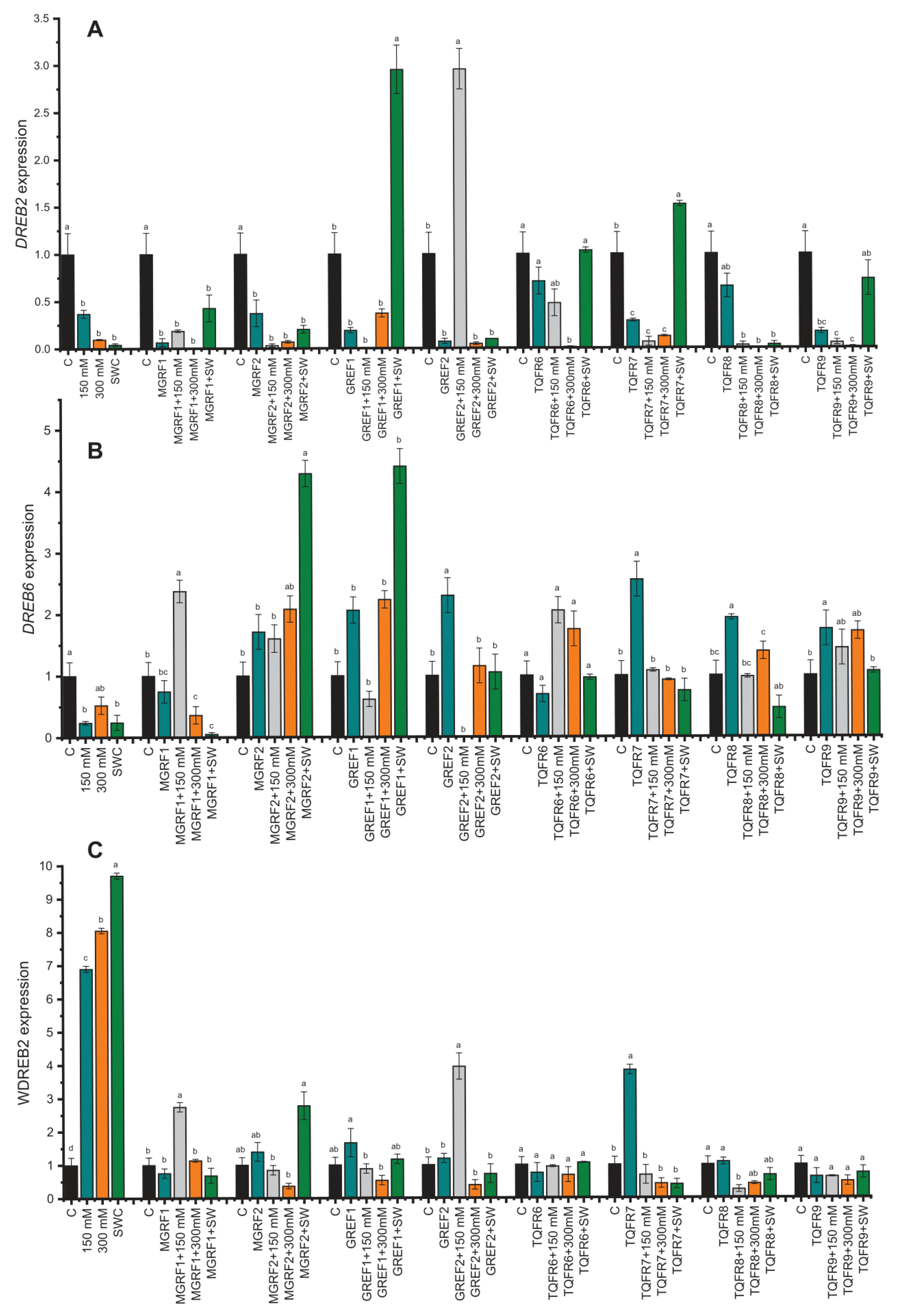
2.8. Gene Expression under Salinity Stress and Fungal Inoculation
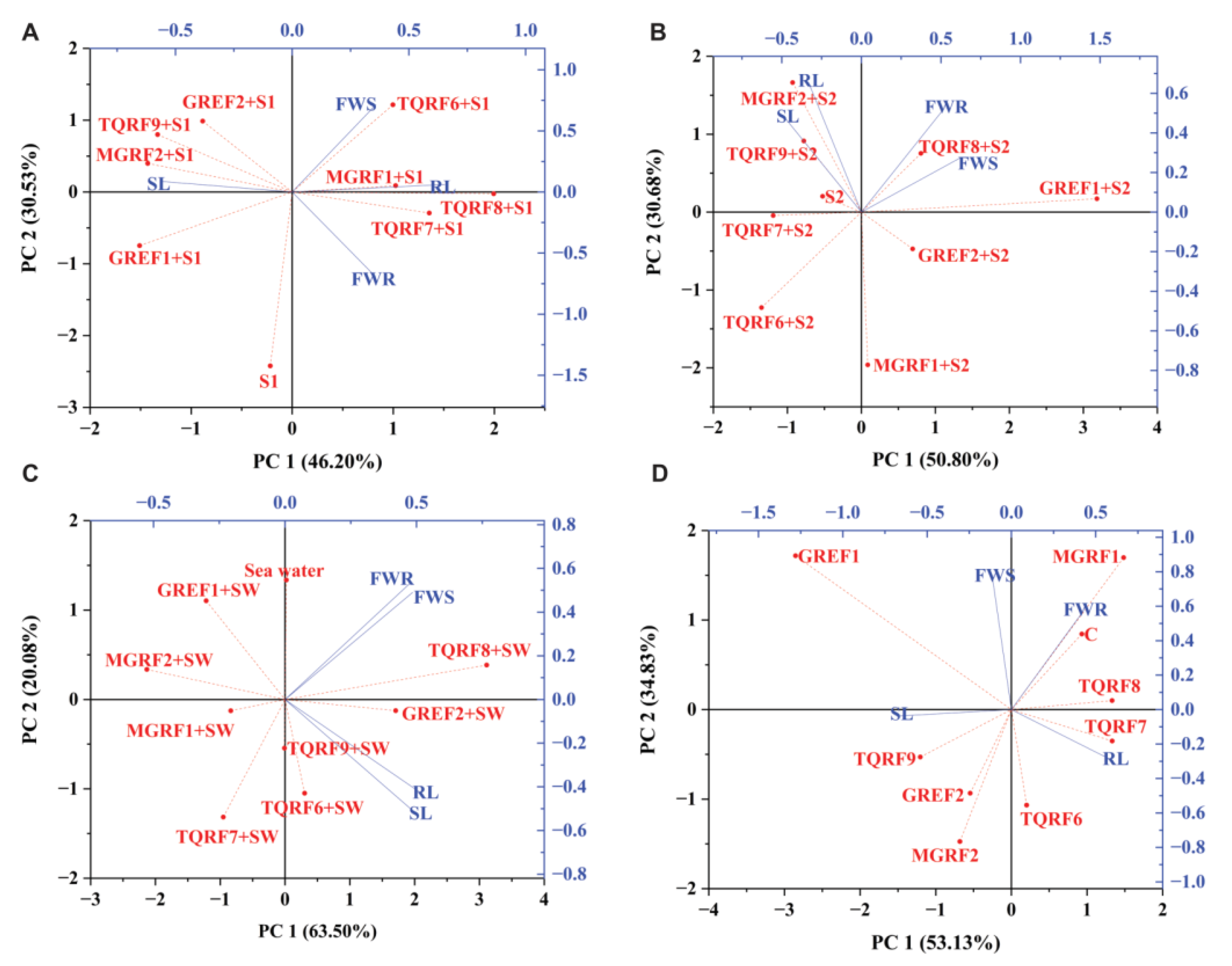
2.9. PCA Biplot Analysis
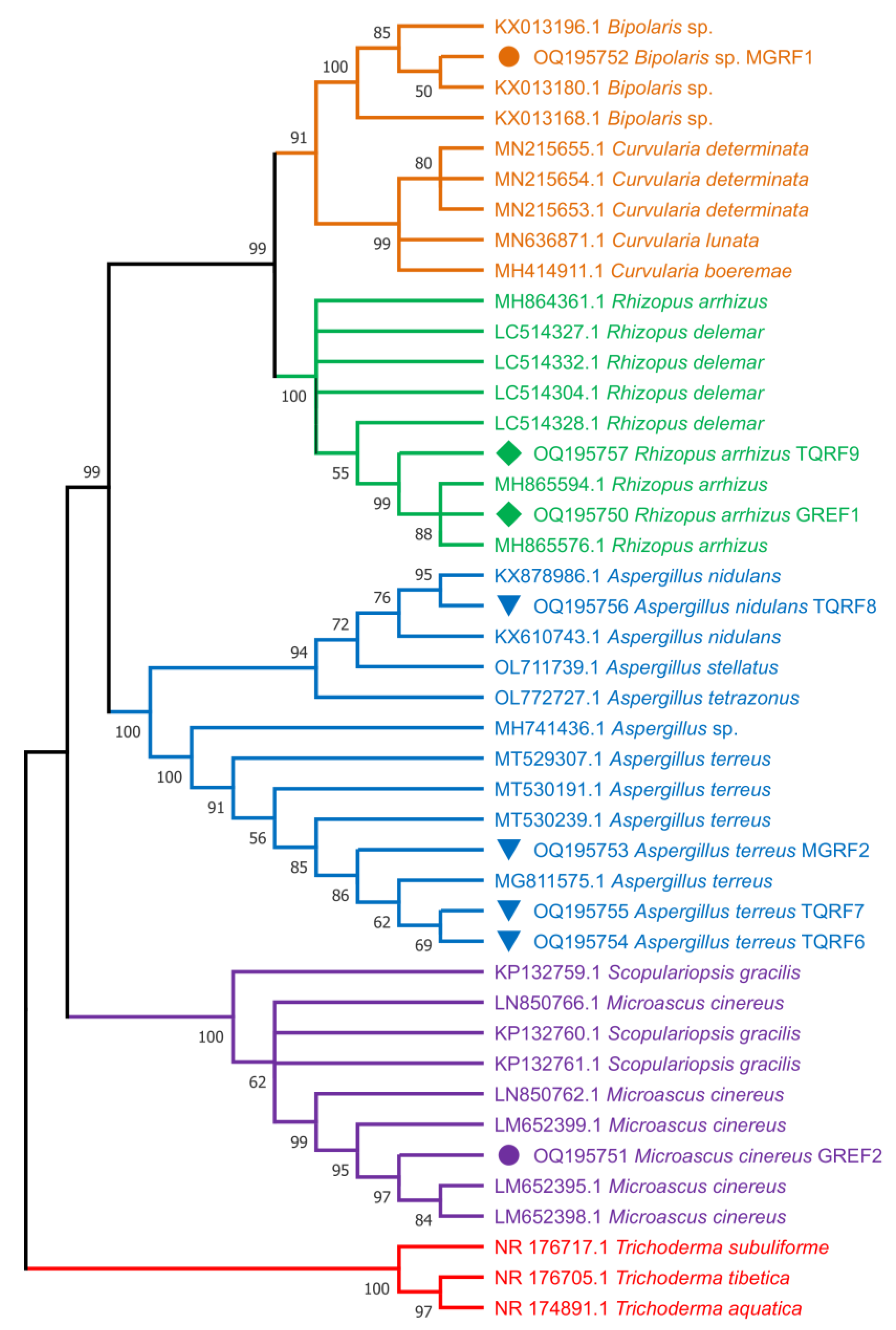
2.10. Fungal Strain Identification and Phylogenetic Analysis
3. Discussion
4. Materials and Methods
4.1. Isolation of Rhizospheric, Endophytic and Soil Fungi
4.2. Screening Fungal Strains for IAA Production
4.3. Fungi Identification and Phylogenetic Analysis
4.4. Seed Biopriming and Germination with Fungal Strain
4.5. Fungal Strain’s Interaction with Wheat Plants under NaCl Stress
4.6. Determination of Protein and Catalase Activity
4.7. Determination of Total Polyphenol, Polyphenol Oxidase, Flavonoid, and Flavonol Activity
4.8. RNA Isolation and RT–qPCR
4.9. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| GR | Grass (Paspalum vaginatum), |
| TQ | Tetraena qatarensis, |
| SA | Sueda Aegyptiaca, |
| MG | Mangrove (Avicennia marina) |
| WS | Water sand |
| S1 | 150 mM |
| S2 | 300 mM |
| SW | Sea water |
References
- Arti, D.; Choudhary, M.; Sourirajan, A. Salt tolerant bacteria for crop improvement in saline agriculture fields: Development, challenges and opportunities. Plant Arch. 2020, 20, 7139–7155. [Google Scholar]
- Khan, N.; Bano, A.; Rahman, M.A.; Guo, J.; Kang, Z.; Babar, M. Comparative physiological and metabolic analysis reveals a complex mechanism involved in drought tolerance in chickpea (Cicer arietinum L.) induced by PGPR and PGRs. Sci. Rep. 2019, 9, 1–19. [Google Scholar] [CrossRef] [PubMed]
- de Oliveira, A.B.; Alencar, N.L.M.; Gomes-Filho, E. Comparison between the water and salt stress effects on plant growth and development. Responses Org. Water Stress 2013, 4, 67–94. [Google Scholar]
- Munns, R.; Tester, M. Mechanisms of salinity tolerance. Annu. Rev. Plant Biol. 2008, 59, 651. [Google Scholar] [CrossRef] [PubMed]
- Prasad, S.R.; Bagali, P.G.; Hittalmani, S.; Shashidhar, H. Molecular mapping of quantitative trait loci associated with seedling tolerance to salt stress in rice (Oryza sativa L.). Curr. Sci. 2000, 78, 162–164. [Google Scholar]
- Neumann, P.M. Coping mechanisms for crop plants in drought-prone environments. Ann. Bot. 2008, 101, 901–907. [Google Scholar] [CrossRef] [PubMed]
- Sharma, A.; Dev, K.; Sourirajan, A.; Choudhary, M. Isolation and characterization of salt-tolerant bacteria with plant growth-promoting activities from saline agricultural fields of Haryana, India. J. Genet. Eng. Biotechnol. 2021, 19, 1–10. [Google Scholar] [CrossRef]
- Gangwar, P.; Singh, R.; Trivedi, M.; Tiwari, R.K. Sodic soil: Management and reclamation strategies. In Environmental Concerns and Sustainable Development; Springer: Berlin/Heidelberg, Germany, 2020; pp. 175–190. [Google Scholar]
- Hasanuzzaman, M.; Nahar, K.; Rahman, A.; Anee, T.I.; Alam, M.U.; Bhuiyan, T.F.; Oku, H.; Fujita, M. Approaches to enhance salt stress tolerance in wheat. In Wheat Improvement, Management and Utilization; IntechOpen: London, UK, 2017; pp. 151–187. [Google Scholar]
- World Health Organization. Preventing Disease through Healthy Environments: Exposure to Highly Hazardous Pesticides: A Major Public Health Concern; World Health Organization: Geneva, Switzerland, 2019. [Google Scholar]
- Seleiman, M.F.; Aslam, M.T.; Alhammad, B.A.; Hassan, M.U.; Maqbool, R.; Chattha, M.U.; Khan, I.; Gitari, H.I.; Uslu, O.S.; Rana, R. Salinity stress in wheat: Effects, mechanisms and management strategies. Phyton 2022, 91, 667. [Google Scholar]
- Mishra, P.; Singh, P.P.; Singh, S.K.; Verma, H. Sustainable agriculture and benefits of organic farming to special emphasis on PGPR. In Role of Plant Growth Promoting Microorganisms in Sustainable Agriculture and Nanotechnology; Elsevier: Amsterdam, The Netherlands, 2019; pp. 75–87. [Google Scholar]
- Flowers, T.J.; Colmer, T.D. Plant salt tolerance: Adaptations in halophytes. Ann. Bot. 2015, 115, 327–331. [Google Scholar] [CrossRef]
- Gul, B.; Ansari, R.; Khan, M.A. Salt tolerance of Salicornia utahensis from the great basin desert. Pak. J. Bot. 2009, 41, 2925–2932. [Google Scholar]
- Leach, J.E.; Triplett, L.R.; Argueso, C.T.; Trivedi, P. Communication in the phytobiome. Cell 2017, 169, 587–596. [Google Scholar] [CrossRef] [PubMed]
- Chauhan, H.; Bagyaraj, D.; Selvakumar, G.; Sundaram, S. Novel plant growth promoting rhizobacteria—Prospects and potential. Appl. Soil Ecol. 2015, 95, 38–53. [Google Scholar] [CrossRef]
- Numan, M.; Bashir, S.; Khan, Y.; Mumtaz, R.; Shinwari, Z.K.; Khan, A.L.; Khan, A.; Ahmed, A.-H. Plant growth promoting bacteria as an alternative strategy for salt tolerance in plants: A review. Microbiol. Res. 2018, 209, 21–32. [Google Scholar] [CrossRef]
- Pudake, R.N.; Mehta, C.M.; Mohanta, T.K.; Sharma, S.; Varma, A.; Sharma, A.K. Expression of four phosphate transporter genes from Finger millet (Eleusine coracana L.) in response to mycorrhizal colonization and Pi stress. 3 Biotech 2017, 7, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Vandenkoornhuyse, P.; Quaiser, A.; Duhamel, M.; Le Van, A.; Dufresne, A. The importance of the microbiome of the plant holobiont. New Phytol. 2015, 206, 1196–1206. [Google Scholar] [CrossRef] [PubMed]
- Radhakrishnan, R.; Shim, K.-B.; Lee, B.-W.; Hwang, C.-D.; Pae, S.-B.; Park, C.-H.; Kim, S.-U.; Lee, C.-K.; Baek, I.-Y. IAA-producing Penicillium sp. NICS01 triggers plant growth and suppresses Fusarium sp.-induced oxidative stress in sesame (Sesamum indicum L.). J. Microbiol. Biotechnol. 2013, 23, 856–863. [Google Scholar] [CrossRef]
- Bilal, L.; Asaf, S.; Hamayun, M.; Gul, H.; Iqbal, A.; Ullah, I.; Lee, I.-J.; Hussain, A. Plant growth promoting endophytic fungi Asprgillus fumigatus TS1 and Fusarium proliferatum BRL1 produce gibberellins and regulates plant endogenous hormones. Symbiosis 2018, 76, 117–127. [Google Scholar] [CrossRef]
- Mbarki, S.; Cerdà, A.; Brestic, M.; Mahendra, R.; Abdelly, C.; Pascual, J.A. Vineyard compost supplemented with Trichoderma harzianum T78 improve saline soil quality. Land Degrad. Dev. 2017, 28, 1028–1037. [Google Scholar] [CrossRef]
- Radhakrishnan, R.; Kang, S.-M.; Baek, I.-Y.; Lee, I.-J. Characterization of plant growth-promoting traits of Penicillium species against the effects of high soil salinity and root disease. J. Plant Interact. 2014, 9, 754–762. [Google Scholar] [CrossRef]
- Lubna; Khan, M.A.; Asaf, S.; Jan, R.; Waqas, M.; Kim, K.-M.; Lee, I.-J. Endophytic fungus Bipolaris sp. CSL-1 induces salt tolerance in Glycine max. L via modulating its endogenous hormones, antioxidative system and gene expression. J. Plant Interact. 2022, 17, 319–332. [Google Scholar] [CrossRef]
- Asaf, S.; Jan, R.; Khan, M.A.; Khan, A.L.; Asif, S.; Bilal, S.; Ahmad, W.; Waqas, M.; Kim, K.-M.; Ahmed, A.-H. Unraveling the mutualistic interaction between endophytic Curvularia lunata CSL1 and tomato to mitigate cadmium (Cd) toxicity via transcriptomic insights. Sci. Total Environ. 2022, 861, 160542. [Google Scholar] [CrossRef] [PubMed]
- Asaf, S.; Jan, R.; Khan, A.L.; Bilal, S.; Asif, S.; Al-Harrasi, A.; Kim, K.-M. Unraveling the Genome Sequence of Plant Growth Promoting Aspergillus niger (CSR3) Provides Insight into the Synthesis of Secondary Metabolites and Its Comparative Genomics. J. Fungi 2022, 8, 107. [Google Scholar]
- Bilal, S.; Shahzad, R.; Asaf, S.; Imran, M.; Al-Harrasi, A.; Lee, I.-J. Efficacy of endophytic SB10 and glycine betaine duo in alleviating phytotoxic impact of combined heat and salinity in Glycine max L. via regulation of redox homeostasis and physiological and molecular responses. Environ. Pollut. 2023, 316, 120658. [Google Scholar] [CrossRef]
- Dodd, I.; Zinovkina, N.; Safronova, V.; Belimov, A. Rhizobacterial mediation of plant hormone status. Ann. Appl. Biol. 2010, 157, 361–379. [Google Scholar] [CrossRef]
- Spaepen, S.; Bossuyt, S.; Engelen, K.; Marchal, K.; Vanderleyden, J. Phenotypical and molecular responses of A rabidopsis thaliana roots as a result of inoculation with the auxin-producing bacterium A zospirillum brasilense. New Phytol. 2014, 201, 850–861. [Google Scholar] [CrossRef] [PubMed]
- Verbon, E.H.; Liberman, L.M. Beneficial microbes affect endogenous mechanisms controlling root development. Trends Plant Sci. 2016, 21, 218–229. [Google Scholar] [CrossRef]
- Demeulenaere, M.J.; Beeckman, T. The interplay between auxin and the cell cycle during plant development. In Auxin and Its Role in Plant Development; Springer: Berlin/Heidelberg, Germany, 2014; pp. 119–141. [Google Scholar]
- Spaepen, S.; Vanderleyden, J.; Remans, R. Indole-3-acetic acid in microbial and microorganism-plant signaling. FEMS Microbiol. Rev. 2007, 31, 425–448. [Google Scholar] [CrossRef]
- Poupin, M.J.; Greve, M.; Carmona, V.; Pinedo, I. A complex molecular interplay of auxin and ethylene signaling pathways is involved in Arabidopsis growth promotion by Burkholderia phytofirmans PsJN. Front. Plant Sci. 2016, 7, 492. [Google Scholar] [CrossRef]
- Iqbal, A.; Hasnain, S. Auxin producing Pseudomonas strains: Biological candidates to modulate the growth of Triticum aestivum beneficially. Am. J. Plant Sci. 2013, 4, 1693. [Google Scholar] [CrossRef]
- Johnson, J.M.; Alex, T.; Oelmüller, R. Piriformospora indica: The versatile and multifunctional root endophytic fungus for enhanced yield and tolerance to biotic and abiotic stress in crop plants. J. Trop. Agric. 2014, 52, 103–122. [Google Scholar]
- Khan, A.L.; Hussain, J.; Al-Harrasi, A.; Al-Rawahi, A.; Lee, I.-J. Endophytic fungi: Resource for gibberellins and crop abiotic stress resistance. Crit. Rev. Biotechnol. 2015, 35, 62–74. [Google Scholar] [CrossRef] [PubMed]
- Abeer, H.; Abd_Allah, E.; Alqarawi, A.; El-Didamony, G.; Alwhibi, M.; Egamberdieva, D.; Ahmad, P. Alleviation of adverse impact of salinity on faba bean (Vicia faba L.) by arbuscular mycorrhizal fungi. Pak. J. Bot 2014, 46, 2003–2013. [Google Scholar]
- Contreras-Cornejo, H.A.; Macías-Rodríguez, L.; Cortés-Penagos, C.; López-Bucio, J. Trichoderma virens, a plant beneficial fungus, enhances biomass production and promotes lateral root growth through an auxin-dependent mechanism in Arabidopsis. Plant Physiol. 2009, 149, 1579–1592. [Google Scholar] [CrossRef] [PubMed]
- Knief, C. Analysis of plant microbe interactions in the era of next generation sequencing technologies. Front. Plant Sci. 2014, 5, 216. [Google Scholar] [CrossRef] [PubMed]
- Sharma, M.; Sudheer, S.; Usmani, Z.; Rani, R.; Gupta, P. Deciphering the omics of plant-microbe interaction: Perspectives and new insights. Curr. Genom. 2020, 21, 343–362. [Google Scholar] [CrossRef]
- Hamayun, M.; Khan, S.A.; Khan, A.L.; Shin, J.-H.; Ahmad, B.; Shin, D.-H.; Lee, I.-J. Exogenous gibberellic acid reprograms soybean to higher growth and salt stress tolerance. J. Agric. Food Chem. 2010, 58, 7226–7232. [Google Scholar] [CrossRef]
- Waqas, M.; Khan, A.L.; Kamran, M.; Hamayun, M.; Kang, S.-M.; Kim, Y.-H.; Lee, I.-J. Endophytic fungi produce gibberellins and indoleacetic acid and promotes host-plant growth during stress. Molecules 2012, 17, 10754–10773. [Google Scholar] [CrossRef]
- Souza, R.d.; Ambrosini, A.; Passaglia, L.M. Plant growth-promoting bacteria as inoculants in agricultural soils. Genet. Mol. Biol. 2015, 38, 401–419. [Google Scholar] [CrossRef]
- Hmaeid, N.; Wali, M.; Mahmoud, O.M.-B.; Pueyo, J.J.; Ghnaya, T.; Abdelly, C. Efficient rhizobacteria promote growth and alleviate NaCl-induced stress in the plant species Sulla carnosa. Appl. Soil Ecol. 2019, 133, 104–113. [Google Scholar] [CrossRef]
- Sadak, M.S.; El-Hameid, A.; Asmaa, R.; Zaki, F.S.; Dawood, M.G.; El-Awadi, M.E. Physiological and biochemical responses of soybean (Glycine max L.) to cysteine application under sea salt stress. Bull. Natl. Res. Cent. 2020, 44, 1–10. [Google Scholar] [CrossRef]
- Porcel, R.; Aroca, R.; Ruiz-Lozano, J.M. Salinity stress alleviation using arbuscular mycorrhizal fungi. A review. Agron. Sustain. Dev. 2012, 32, 181–200. [Google Scholar] [CrossRef]
- Mo, W.; Tang, W.; Du, Y.; Jing, Y.; Bu, Q.; Lin, R. PHYTOCHROME-INTERACTING FACTOR-LIKE14 and SLENDER RICE1 interaction controls seedling growth under salt stress. Plant Physiol. 2020, 184, 506–517. [Google Scholar] [CrossRef] [PubMed]
- Metwali, E.M.; Abdelmoneim, T.S.; Bakheit, M.A.; Kadasa, N.M. Alleviation of salinity stress in faba bean (Vicia faba L.) plants by inoculation with plant growth promoting rhizobacteria (PGPR). Plant Omics 2015, 8, 449–460. [Google Scholar]
- Hajiboland, R.; Aliasgharzadeh, N.; Laiegh, S.F.; Poschenrieder, C. Colonization with arbuscular mycorrhizal fungi improves salinity tolerance of tomato (Solanum lycopersicum L.) plants. Plant Soil 2010, 331, 313–327. [Google Scholar] [CrossRef]
- Borde, M.; Dudhane, M.; Jite, P. Growth photosynthetic activity and antioxidant responses of mycorrhizal and non-mycorrhizal bajra (Pennisetum glaucum) crop under salinity stress condition. Crop Prot. 2011, 30, 265–271. [Google Scholar] [CrossRef]
- Khan, A.L.; Hamayun, M.; Kim, Y.-H.; Kang, S.-M.; Lee, I.-J. Ameliorative symbiosis of endophyte (Penicillium funiculosum LHL06) under salt stress elevated plant growth of Glycine max L. Plant Physiol. Biochem. 2011, 49, 852–861. [Google Scholar] [CrossRef]
- Kohler, J.; Hernández, J.A.; Caravaca, F.; Roldán, A. Induction of antioxidant enzymes is involved in the greater effectiveness of a PGPR versus AM fungi with respect to increasing the tolerance of lettuce to severe salt stress. Environ. Exp. Bot. 2009, 65, 245–252. [Google Scholar] [CrossRef]
- Wu, Q.; Zou, Y.; Liu, W.; Ye, X.; Zai, H.; Zhao, L. Alleviation of salt stress in citrus seedlings inoculated with mycorrhiza: Changes in leaf antioxidant defense systems. Plant Soil Environ. 2010, 56, 470–475. [Google Scholar] [CrossRef]
- Khan, A.L.; Waqas, M.; Hussain, J.; Al-Harrasi, A.; Hamayun, M.; Lee, I.-J. Phytohormones enabled endophytic fungal symbiosis improve aluminum phytoextraction in tolerant Solanum lycopersicum: An examples of Penicillium janthinellum LK5 and comparison with exogenous GA3. J. Hazard. Mater. 2015, 295, 70–78. [Google Scholar] [CrossRef]
- Radi, A.A.; Farghaly, F.A.; Hamada, A.M. Physiological and biochemical responses of salt-tolerant and salt-sensitive wheat and bean cultivars to salinity. J. Biol. Earth Sci. 2013, 3, 72–88. [Google Scholar]
- Sarkar, A.; Ghosh, P.K.; Pramanik, K.; Mitra, S.; Soren, T.; Pandey, S.; Mondal, M.H.; Maiti, T.K. A halotolerant Enterobacter sp. displaying ACC deaminase activity promotes rice seedling growth under salt stress. Res. Microbiol. 2018, 169, 20–32. [Google Scholar] [CrossRef] [PubMed]
- Cheplick, G.P. Recovery from drought stress in Lolium perenne (Poaceae): Are fungal endophytes detrimental? Am. J. Bot. 2004, 91, 1960–1968. [Google Scholar] [CrossRef] [PubMed]
- Yeon-Sik, C.; In-Jung, L.; Jong-Myeong, K.; Seon-Kap, H.; Seok-Jong, S.; Ho-Youn, K.; Hyeokjun, Y.; Muhammad, H.; Sumera, K.; Ung-Han, Y. Plant growth promotion and Penicillium citrinum. BMC Microbiol 2008, 8, 231. [Google Scholar]
- Waller, F.; Achatz, B.; Baltruschat, H.; Fodor, J.; Becker, K.; Fischer, M.; Heier, T.; Hückelhoven, R.; Neumann, C.; von Wettstein, D. The endophytic fungus Piriformospora indica reprograms barley to salt-stress tolerance, disease resistance, and higher yield. Proc. Natl. Acad. Sci. USA 2005, 102, 13386–13391. [Google Scholar] [CrossRef]
- Alexander, A.; Singh, V.K.; Mishra, A. Halotolerant PGPR Stenotrophomonas maltophilia BJ01 induces salt tolerance by modulating physiology and biochemical activities of Arachis hypogaea. Front. Microbiol. 2020, 11, 568289. [Google Scholar] [CrossRef]
- Hernández, J.A.; Aguilar, A.B.; Portillo, B.; López-Gómez, E.; Beneyto, J.M.; García-Legaz, M.F. The effect of calcium on the antioxidant enzymes from salt-treated loquat and anger plants. Funct. Plant Biol. 2003, 30, 1127–1137. [Google Scholar] [CrossRef]
- Radhakrishnan, R.; Kumari, B.R. Protective role of pulsed magnetic field against salt stress effects in soybean organ culture. Plant Biosyst. —Int. J. Deal. All Asp. Plant Biol. 2013, 147, 135–140. [Google Scholar] [CrossRef]
- Abogadallah, G.M. Differential regulation of photorespiratory gene expression by moderate and severe salt and drought stress in relation to oxidative stress. Plant Sci. 2011, 180, 540–547. [Google Scholar] [CrossRef]
- Ahmad, P.; Hashem, A.; Abd-Allah, E.F.; Alqarawi, A.; John, R.; Egamberdieva, D.; Gucel, S. Role of Trichoderma harzianum in mitigating NaCl stress in Indian mustard (Brassica juncea L.) through antioxidative defense system. Front. Plant Sci. 2015, 6, 868. [Google Scholar] [CrossRef]
- Mondini, L.; Nachit, M.M.; Pagnotta, M.A. Allelic variants in durum wheat (Triticum turgidum L. var. durum) DREB genes conferring tolerance to abiotic stresses. Mol. Genet. Genom. 2015, 290, 531–544. [Google Scholar] [CrossRef]
- Kumar, R.; Masthigowda, M.H.; Kaur, A.; Bhusal, N.; Pandey, A.; Kumar, S.; Mishra, C.; Singh, G.; Singh, G.P. Identification and characterization of multiple abiotic stress tolerance genes in wheat. Mol. Biol. Rep. 2020, 47, 8629–8643. [Google Scholar] [CrossRef]
- Kobayashi, F.; Ishibashi, M.; Takumi, S. Transcriptional activation of Cor/Lea genes and increase in abiotic stress tolerance through expression of a wheat DREB2 homolog in transgenic tobacco. Transgenic Res. 2008, 17, 755–767. [Google Scholar] [CrossRef]
- Petrini, O.; Fisher, P. Fungal endophytes in Salicornia perennis. Trans. Br. Mycol. Soc. 1986, 87, 647–651. [Google Scholar] [CrossRef]
- Sandhu, S.S.; Kumar, S.; Aharwal, R.P. Isolation and identification of endophytic fungi from Ricinus communis Linn. and their antibacterial activity. Int. J. Res. Pharm. Chem. 2014, 4, 611–618. [Google Scholar]
- Gond, S.; Verma, V.; Kumar, A.; Kumar, V.; Kharwar, R. Study of endophytic fungal community from different parts of Aegle marmelos Correae (Rutaceae) from Varanasi (India). World J. Microbiol. Biotechnol. 2007, 23, 1371–1375. [Google Scholar] [CrossRef]
- Reyes, A.; Mitchell, J. Growth response of several isolates of Fusarium in rhizospheres of host and nonhost plants. Phytopathology 1962, 52, 1196. [Google Scholar]
- Waksman, S.A. Principles of Soil Microbiology; Williams & Wilkins: Philadelphia, PA, USA, 1927. [Google Scholar]
- Lubna, L.; Khan, M.A.; Asaf, S.; Jan, R.; Waqas, M.; Kim, K.; Lee, I.-J. Plant growth promoting Bipolaris sp. CSL-1 mitigate salinity stress in soybean via altering endogenous phytohormonal level, antioxidants and genes expression. Res. Sq 2020, 2, 1–23. [Google Scholar]
- Tamura, K.; Stecher, G.; Peterson, D.; Filipski, A.; Kumar, S. MEGA6: Molecular evolutionary genetics analysis version 6.0. Mol. Biol. Evol. 2013, 30, 2725–2729. [Google Scholar] [CrossRef]
- Mitelut, A.C.; Popa, M.E. Seed germination bioassay for toxicity evaluation of different composting biodegradable materials. Rom. Biotechnol. Lett. 2011, 16, 121–129. [Google Scholar]
- Bradford, M.M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 1976, 72, 248–254. [Google Scholar] [CrossRef]
- Aebi, H. Catalase in vitro. In Methods in Enzymology; Elsevier: Amsterdam, The Netherlands, 1984; Volume 105, pp. 121–126. [Google Scholar]
- Haghighi, T.M.; Saharkhiz, M.J. Phytotoxic potential of Vitex pseudo-negundo leaf and flower extracts and analysis of phenolic compounds. Biocatal. Agric. Biotechnol. 2021, 34, 102018. [Google Scholar] [CrossRef]
- Kumar, K.B.; Khan, P.A. Peroxidase & Polyphenol Oxidase In Excised Ragi (Eleusine Coracana Cv Pr 202) Leaves During Senescence. Indian J. Exp. Biol. 1982, 20, 412–416. [Google Scholar]
- Taban, A.; Saharkhiz, M.J.; Kavoosi, G. Development of pre-emergence herbicide based on Arabic gum-gelatin, apple pectin and savory essential oil nano-particles: A potential green alternative to metribuzin. Int. J. Biol. Macromol. 2021, 167, 756–765. [Google Scholar] [CrossRef]
- Levon, V.; Klymenko, S. Content of anthocyanins and flavonols in the fruits of Cornus spp. Agrobiodiversity Improv. Nutr. Health Life Qual. 2021, 5. [Google Scholar]
- Liu, L.; Han, R.; Yu, N.; Zhang, W.; Xing, L.; Xie, D.; Peng, D. A method for extracting high-quality total RNA from plant rich in polysaccharides and polyphenols using Dendrobium huoshanense. PLoS ONE 2018, 13, e0196592. [Google Scholar] [CrossRef]

| Isolated Microbes | |||
|---|---|---|---|
| Endophytic Fungi | Rhizospheric Fungi | Soil Fungi Coastal Region | Sand Fungi Inside Water |
| TQEF1 | TQRF1 | SASF1 | WSF1 |
| TQEF2 | TQRF2 | TQRF4 | WSF2 |
| SAEF1 | TQRF3 | WSF3 | |
| SAEF2 | MGRF1 | ||
| GREF1 | MGRF2 | ||
| GREF2 | TQRF5 | ||
| GREF3 | TQRF6 | ||
| TQRF7 | |||
| TQRF8 | |||
| TQRF9 | |||
| SARF1 | |||
| SARF2 | |||
| WSF4 | |||
| WSF5 | |||
| S. No | Strain | IAA Production | S. No | Strain | IAA Production |
|---|---|---|---|---|---|
| 1 | TQEF1 | + | 14 | SARF1 | + |
| 2 | TQEF2 | − | 15 | SARF2 | + |
| 3 | TQRF1 | + | 16 | SASF1 | + |
| 4 | TQRF2 | − | 17 | MGRF1 | − |
| 5 | TQRF3 | − | 18 | MGRF2 | + |
| 6 | TQRF4 | + | 19 | GREF1 | + |
| 7 | TQRF5 | + | 20 | GREF2 | + |
| 8 | TQRF6 | + | 21 | GREF3 | + |
| 9 | TQRF7 | + | 22 | WSF1 | − |
| 10 | TQRF8 | + | 23 | WSF2 | − |
| 11 | TQRF9 | + | 24 | WSF3 | − |
| 12 | SAEF1 | − | 25 | WSF4 | + |
| 13 | SAEF2 | + | 26 | WSF5 | + |
| S.No | Strain | D1 | D2 | D3 | D4 | D5 | D6 |
|---|---|---|---|---|---|---|---|
| 1 | Control | 0 | 53.3 | 53.3 | 53.3 | 53.3 | 53.3 |
| 2 | TQEF1 | 6.6 | 66.6 | 73.3 | 73.3 | 0 | 73.3 |
| 3 | TQEF2 | 13.3 | 0 | 46.6 | 73.3 | 73.3 | 86.6 |
| 4 | TQRF1 | 0 | 0 | 0 | 0 | 0 | 0 |
| 5 | TQRF2 | 13.3 | 0 | 26.6 | 26.6 | 26.6 | 26.6 |
| 6 | TQRF3 | 6.6 | 0 | 6.6 | 13.3 | 26.6 | 33.3 |
| 7 | TQRF4 | 26.6 | 40 | 60 | 60 | 66.6 | 73.3 |
| 8 | TQRF5 | 6.6 | 33.3 | 66.6 | 73.3 | 100 | 100 |
| 9 | TQRF6 | 80 | 80 | 100 | 100 | 100 | 100 |
| 10 | TQRF7 | 46.6 | 66.6 | 93.3 | 93.3 | 100 | 100 |
| 11 | TQRF8 | 80.3 | 80 | 100 | 100 | 100 | 100 |
| 12 | TQRF9 | 66.6 | 80 | 100 | 100 | 100 | 100 |
| 13 | SASF1 | 0 | 0 | 0 | 0 | 0 | 0 |
| 14 | SAEF1 | 0 | 6.6 | 13.3 | 13.3 | 13.3 | 13.3 |
| 15 | SAEF2 | 26.6 | 46.6 | 46.6 | 46.6 | 0 | 0 |
| 16 | SARF1 | 13.3 | 13.3 | 13.3 | 13.3 | 13.3 | 13.3 |
| 17 | SARF2 | 0 | 13.3 | 13.3 | 13.3 | 13.3 | 13.3 |
| 18 | GREF1 | 60 | 66.6 | 93.3 | 93.3 | 100 | 100 |
| 19 | GREF2 | 80 | 80 | 86.6 | 86.6 | 86.6 | 86.6 |
| 20 | GREF3 | 6.6 | 0 | 20 | 20 | 20 | 26.6 |
| 21 | WSF1 | 6.6 | 0 | 6.6 | 26.6 | 26.6 | 26.6 |
| 22 | WSF2 | 13.3 | 0 | 40 | 66.6 | 66.6 | 66.6 |
| 23 | WSF3 | 0 | 0 | 26.6 | 26.6 | 26.6 | 33.3 |
| 24 | WSF4 | 0 | 0 | 0 | 0 | 0 | 6.6 |
| 25 | WSF5 | 0 | 0 | 0 | 0 | 0 | 6.6 |
| 26 | MGRF1 | 46.6 | 100 | 100 | 100 | 100 | 100 |
| 27 | MGRF2 | 40 | 53.3 | 53.3 | 60 | 60 | 60 |
| Control | Eigenvalue | Percentage of Variance | Cumulative |
| 2.06504 | 51.63% | 51.63% | |
| 1.02057 | 25.51% | 77.14% | |
| 0.65773 | 16.44% | 93.58% | |
| 0.25666 | 6.42% | 100.00% | |
| 150 mm | 1.45911 | 36.48% | 36.48% |
| 1.09593 | 27.40% | 63.88% | |
| 1.01013 | 25.25% | 89.13% | |
| 0.43483 | 10.87% | 100.00% | |
| 300 mm | 1.99783 | 49.95% | 49.95% |
| 1.13771 | 28.44% | 78.39% | |
| 0.66465 | 16.62% | 95.00% | |
| 0.19982 | 5.00% | 100.00% | |
| 100% | 2.18834 | 54.71% | 54.71% |
| 0.99629 | 24.91% | 79.62% | |
| 0.58621 | 14.66% | 94.27% | |
| 0.22916 | 5.73% | 100.00% |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Aizaz, M.; Ahmad, W.; Asaf, S.; Khan, I.; Saad Jan, S.; Salim Alamri, S.; Bilal, S.; Jan, R.; Kim, K.-M.; Al-Harrasi, A. Characterization of the Seed Biopriming, Plant Growth-Promoting and Salinity-Ameliorating Potential of Halophilic Fungi Isolated from Hypersaline Habitats. Int. J. Mol. Sci. 2023, 24, 4904. https://doi.org/10.3390/ijms24054904
Aizaz M, Ahmad W, Asaf S, Khan I, Saad Jan S, Salim Alamri S, Bilal S, Jan R, Kim K-M, Al-Harrasi A. Characterization of the Seed Biopriming, Plant Growth-Promoting and Salinity-Ameliorating Potential of Halophilic Fungi Isolated from Hypersaline Habitats. International Journal of Molecular Sciences. 2023; 24(5):4904. https://doi.org/10.3390/ijms24054904
Chicago/Turabian StyleAizaz, Muhammad, Waqar Ahmad, Sajjad Asaf, Ibrahim Khan, Syed Saad Jan, Safiya Salim Alamri, Saqib Bilal, Rahmatullah Jan, Kyung-Min Kim, and Ahmed Al-Harrasi. 2023. "Characterization of the Seed Biopriming, Plant Growth-Promoting and Salinity-Ameliorating Potential of Halophilic Fungi Isolated from Hypersaline Habitats" International Journal of Molecular Sciences 24, no. 5: 4904. https://doi.org/10.3390/ijms24054904
APA StyleAizaz, M., Ahmad, W., Asaf, S., Khan, I., Saad Jan, S., Salim Alamri, S., Bilal, S., Jan, R., Kim, K.-M., & Al-Harrasi, A. (2023). Characterization of the Seed Biopriming, Plant Growth-Promoting and Salinity-Ameliorating Potential of Halophilic Fungi Isolated from Hypersaline Habitats. International Journal of Molecular Sciences, 24(5), 4904. https://doi.org/10.3390/ijms24054904









