Full-Length Transcriptome of the Great Himalayan Leaf-Nosed Bats (Hipposideros armiger) Optimized Genome Annotation and Revealed the Expression of Novel Genes
Abstract
:1. Introduction
2. Results
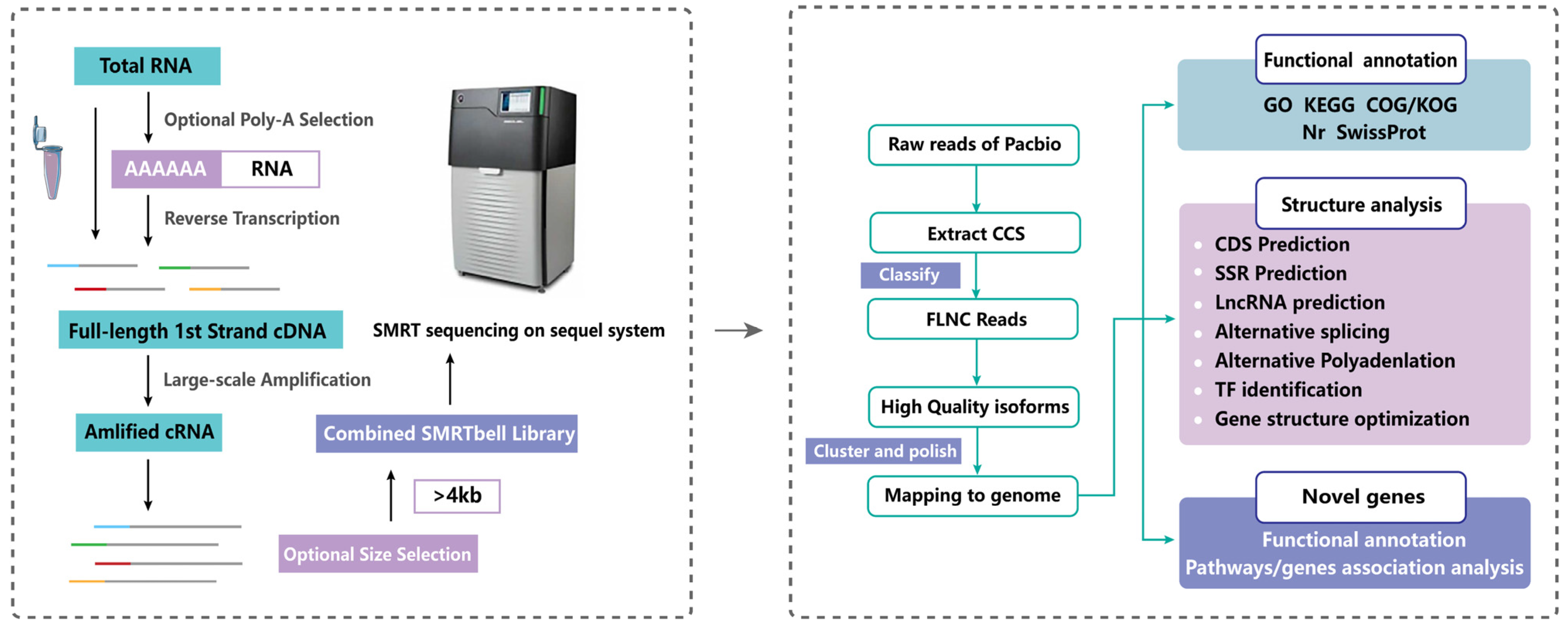
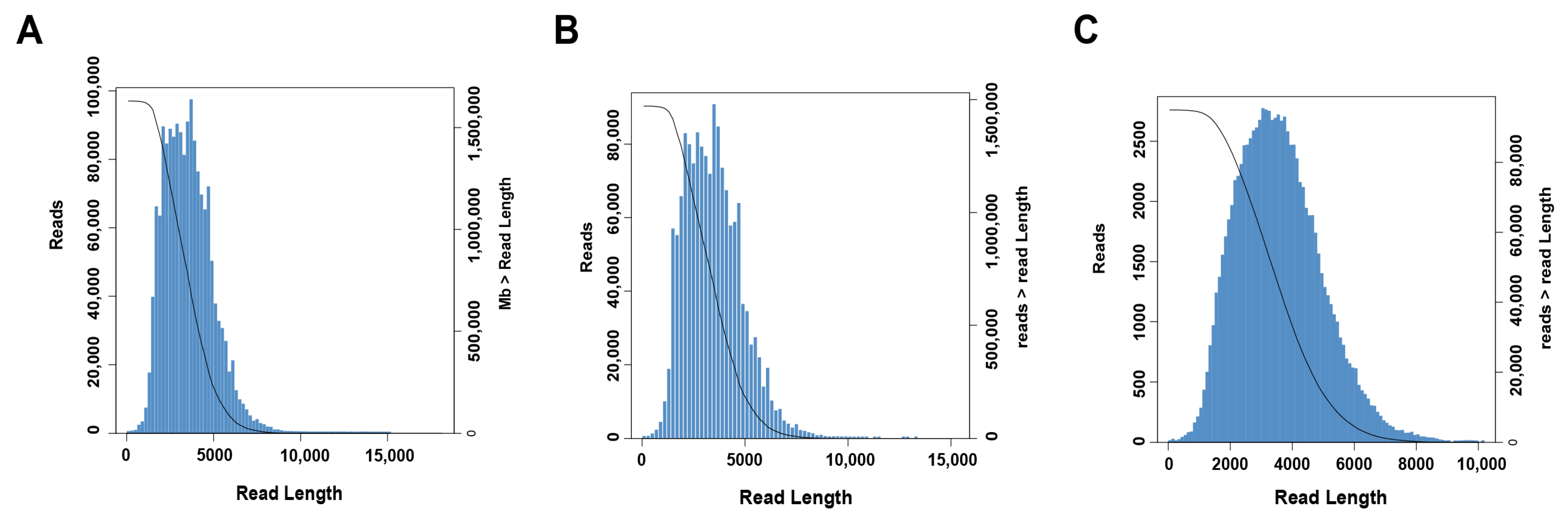
2.1. PacBio Iso-Seq Sequencing Data Analysis
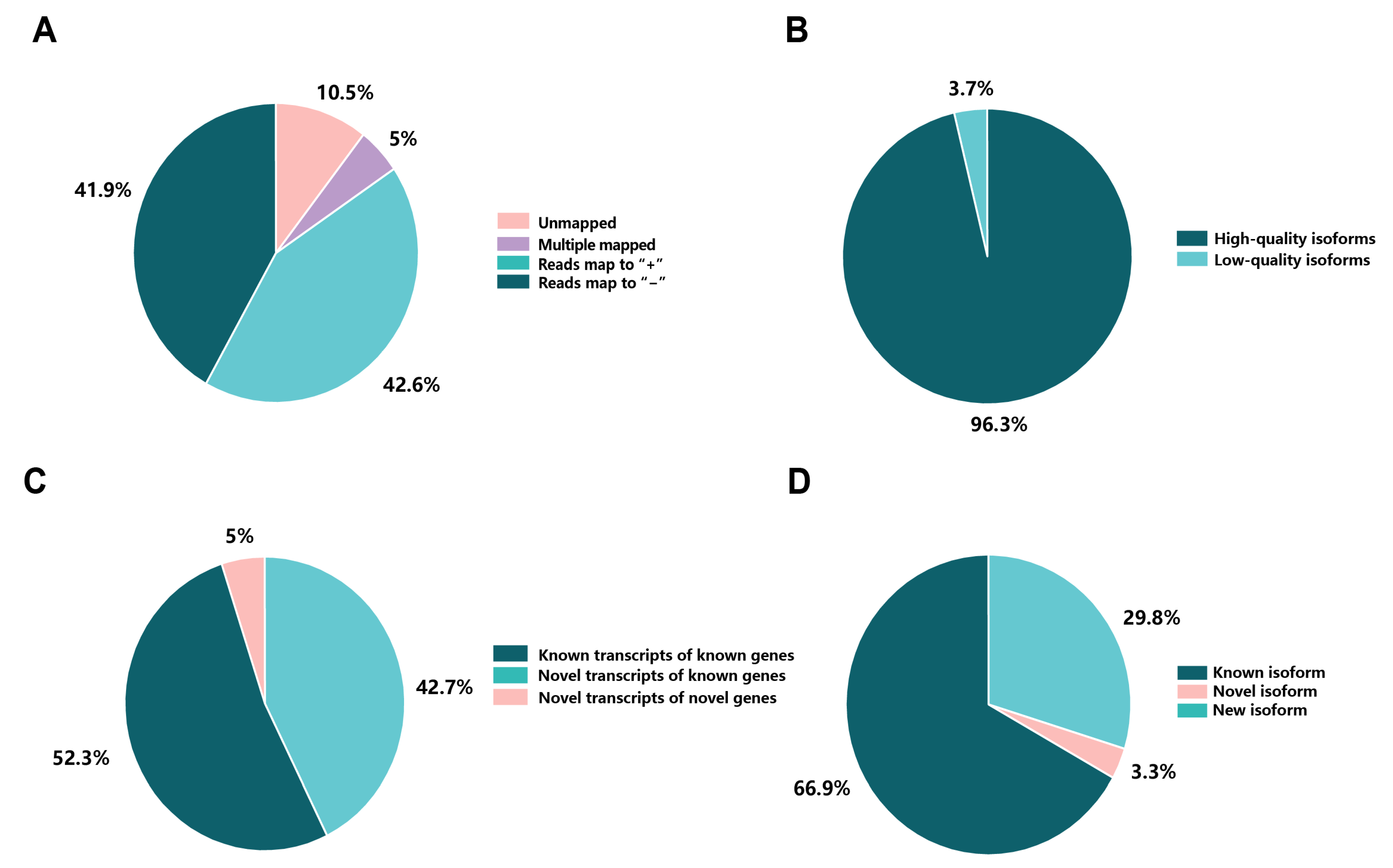
2.2. Mapping to the Reference Genome and Identification of Novel Isoforms
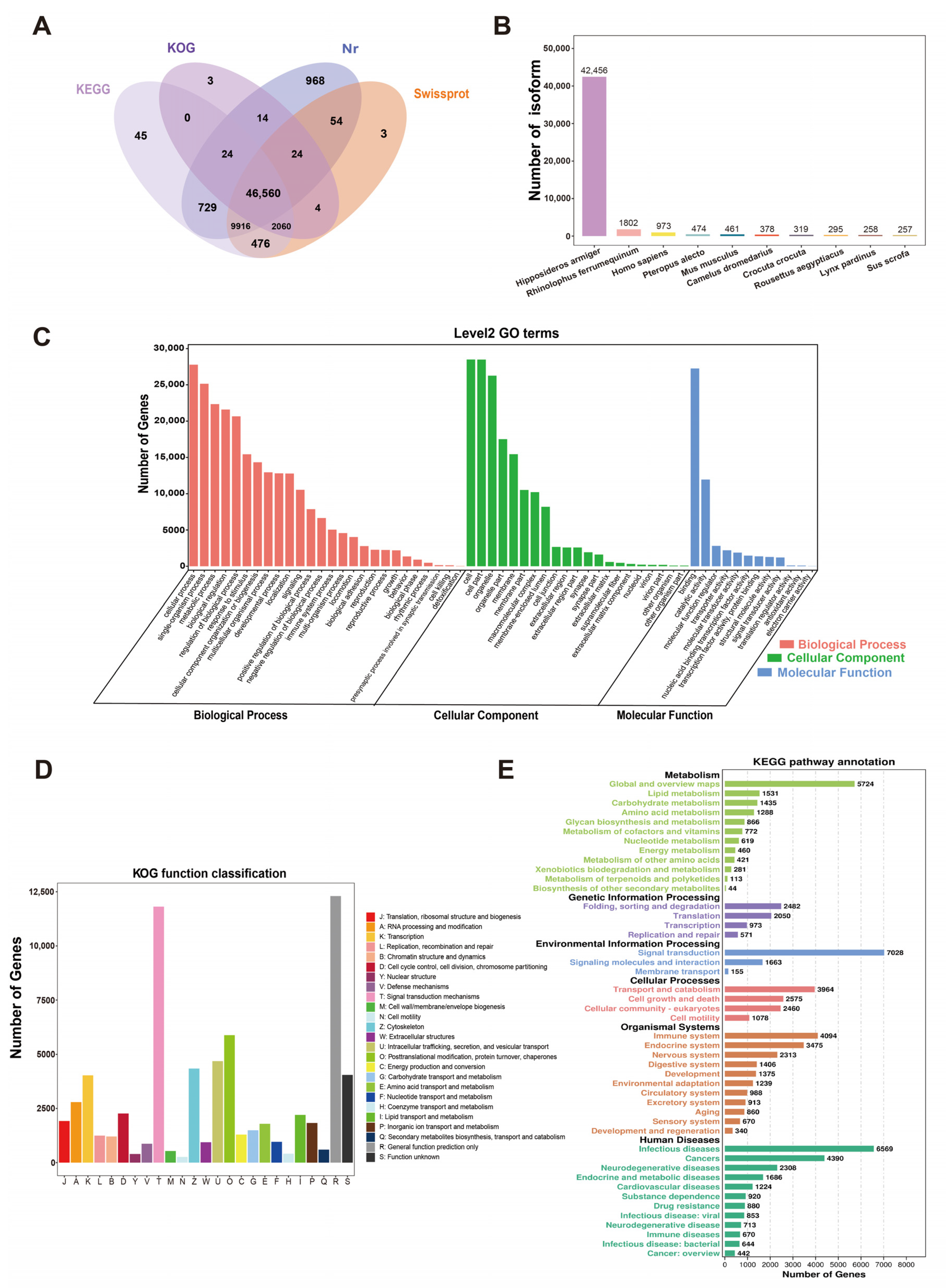
2.3. Functional Annotation of Full-Length Transcriptome of H. armiger
2.4. Prediction and Functional Annotation of Novel Genes
2.5. Structure Analysis
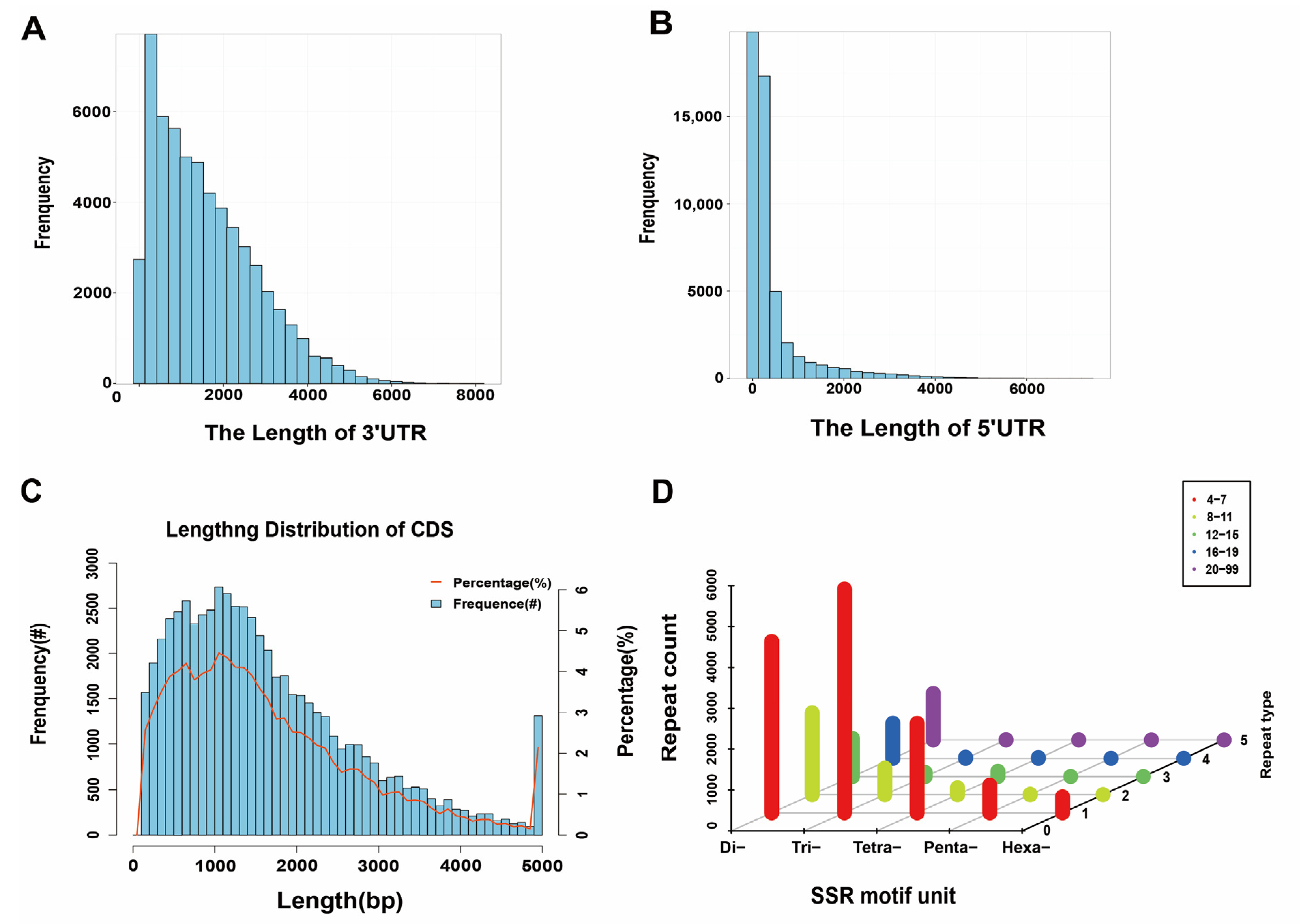
2.5.1. Prediction of Coding Sequences
2.5.2. SSR Analysis
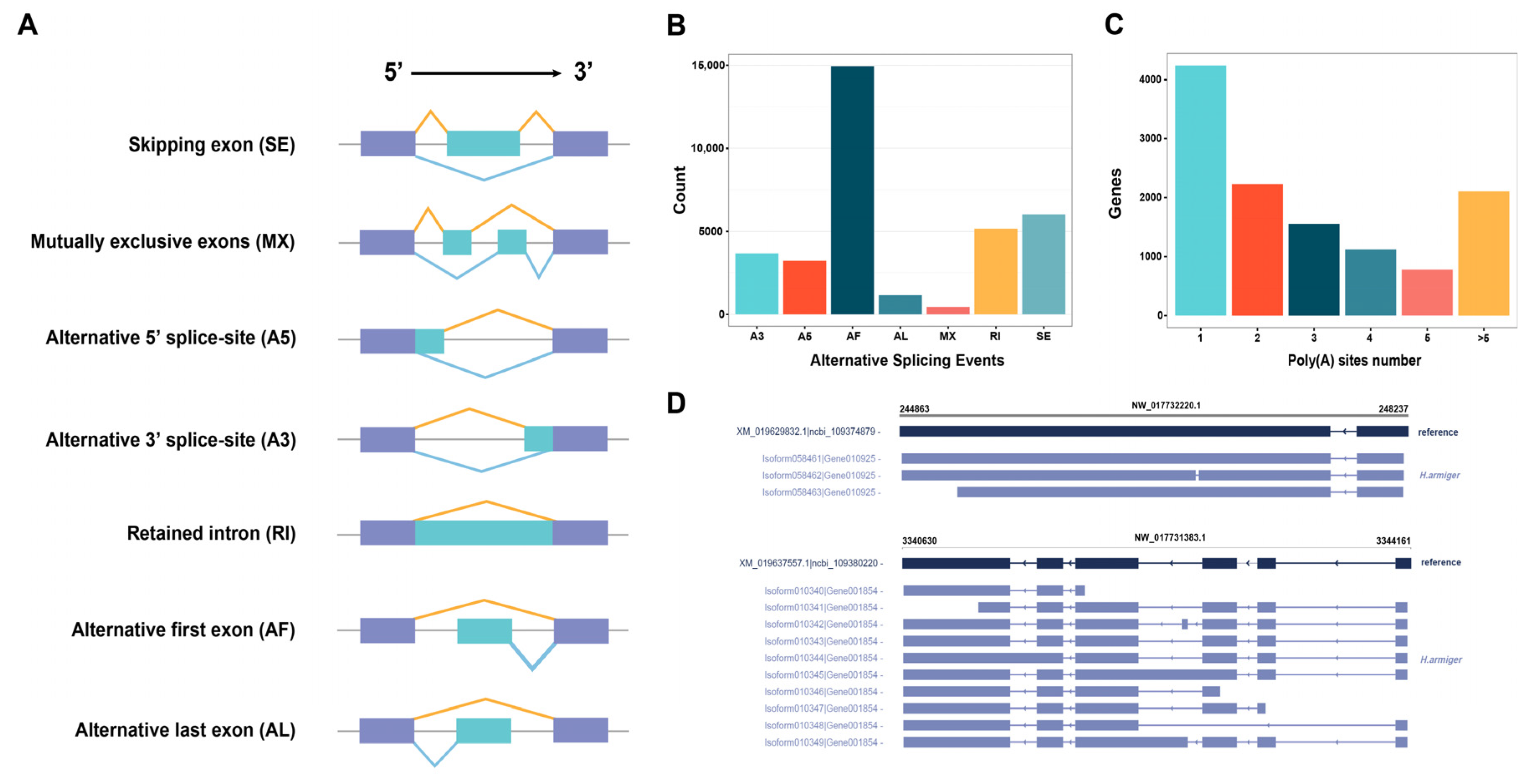
2.5.3. Characterization and Analysis of AS and APA Events
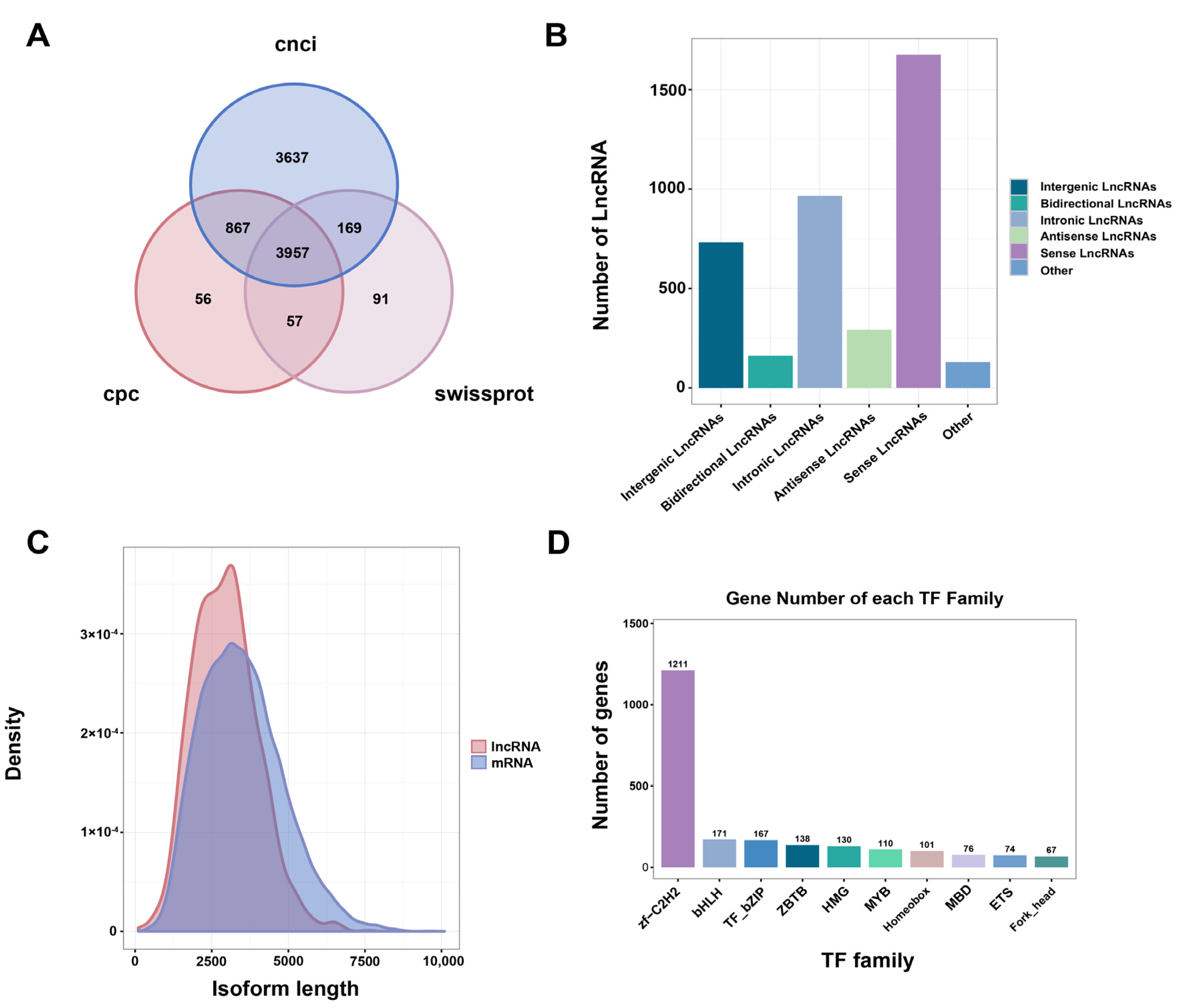
2.5.4. LncRNA and TF Identification
2.6. Gene Structure Optimization
3. Discussion
3.1. Prediction and Functional Annotation of Optimized Isoforms
3.2. Identification of Novel Genes Potential Related with Immune, Nervous and Signal Transduction
3.3. Gene Structures Increased the Complexity and Diversity of Transcripts
3.4. PacBio Sequencing Optimized Genome Annotation
4. Materials and Methods
4.1. Sample Collection and RNA Preparation
4.2. Library Construction and Single-Molecule Real-Time (SMRT) Sequencing
4.3. PacBio Sequencing Processing
4.4. Mapping to the Reference Genome
4.5. Functional Annotation of Isoforms and Novel Genes
4.6. Analysis of AS and APA
4.7. Simple Sequence Repeats (SSR) Prediction
4.8. Prediction and Analysis of ORF, LncRNA, and TFs
4.9. Gene Structure Optimization
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Schoeppler, D.; Schnitzler, H.U.; Denzinger, A. Precise Doppler shift compensation in the hipposiderid bat, Hipposideros armiger. Sci. Rep. 2018, 8, 4598. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Feng, J.; Metzner, W. Different auditory feedback control for echolocation and communication in horseshoe bats. PLoS ONE 2013, 8, e62710. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Warnecke, M.; Falk, B.; Moss, C.F. Echolocation and flight behavior of the bat Hipposideros armiger terasensis in a structured corridor. J. Acoust. Soc. Am. 2018, 144, 806. [Google Scholar] [CrossRef]
- Xu, L.; He, C.; Shen, C.; Jiang, T.; Shi, L.; Sun, K.; Berquist, S.W.; Feng, J. Phylogeography and population genetic structure of the great leaf-nosed bat (Hipposideros armiger) in China. J. Hered. 2010, 101, 562–572. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, Y.; Liu, Q.; Su, Q.; Sun, Y.; Peng, X.; He, X.; Zhang, L. ‘Compromise’ in Echolocation Calls between Different Colonies of the Intermediate Leaf-Nosed Bat (Hipposideros larvatus). PLoS ONE 2016, 11, e0151382. [Google Scholar] [CrossRef] [Green Version]
- Lin, A.; Jiang, T.; Feng, J.; Kanwal, J.S. Acoustically diverse vocalization repertoire in the Himalayan leaf-nosed bat, a widely distributed Hipposideros species. J. Acoust. Soc. Am. 2016, 140, 3765. [Google Scholar] [CrossRef]
- Lin, A.Q.; Jin, L.R.; Shi, L.M.; Sun, K.P.; Berquist, S.W.; Liu, Y.; Feng, J. Postnatal development in Andersen’s leaf-nosed bat Hipposideros pomona: Flight, wing shape, and wing bone lengths. Zoology 2011, 114, 69–77. [Google Scholar] [CrossRef]
- Sun, C.; Zhang, C.; Lucas, J.R.; Gu, H.; Feng, J.; Jiang, T. Vocal performance reflects individual quality in male Great Himalayan leaf-nosed bats (Hipposideros armiger). Integr. Zool. 2022, 17, 731–740. [Google Scholar] [CrossRef]
- Baroja, U.; Garin, I.; Aihartza, J.; Arrizabalaga-Escudero, A.; Vallejo, N.; Aldasoro, M.; Goiti, U. Pest consumption in a vineyard system by the lesser horseshoe bat (Rhinolophus hipposideros). PLoS ONE 2019, 14, e0219265. [Google Scholar] [CrossRef] [Green Version]
- Gerbáčová, K.; Maliničová, L.; Kisková, J.; Maslišová, V.; Uhrin, M.; Pristaš, P. The Faecal Microbiome of Building-Dwelling Insectivorous Bats (Myotis myotis and Rhinolophus hipposideros) also Contains Antibiotic-Resistant Bacterial Representatives. Curr. Microbiol. 2020, 77, 2333–2344. [Google Scholar] [CrossRef]
- Lin, C.L.; Hsiao, C.J.; Hsu, C.H.; Wang, S.E.; Jen, P.H.; Wu, C.H. Hypothermic neuroprotections in the brain of an echolocation bat, Hipposideros terasensis. Neuroreport 2017, 28, 956–962. [Google Scholar] [CrossRef]
- Liu, S.; Sun, K.; Jiang, T.; Ho, J.P.; Liu, B.; Feng, J. Natural epigenetic variation in the female great roundleaf bat (Hipposideros armiger) populations. Mol. Genet. Genom. MGG 2012, 287, 643–650. [Google Scholar] [CrossRef]
- Rakotoarivelo, A.R.; Goodman, S.M.; Schoeman, M.C.; Willows-Munro, S. Phylogeography and population genetics of the endemic Malagasy bat, Macronycteris commersoni s.s. (Chiroptera: Hipposideridae). PeerJ 2019, 7, e5866. [Google Scholar] [CrossRef] [Green Version]
- Xu, H.; Yuan, Y.; He, Q.; Wu, Q.; Yan, Q.; Wang, Q. Complete mitochondrial genome sequences of two Chiroptera species (Rhinolophus luctus and Hipposideros armiger). Mitochondrial DNA 2012, 23, 327–328. [Google Scholar] [CrossRef]
- Guo, T.T.; Hua, P.Y.; Lin, L.K.; Zhang, S.Y. Characterization of novel microsatellite loci in the great leaf-nosed bat, Hipposideros armiger and cross-amplification in other related species. Conserv. Genet. 2008, 9, 1063–1065. [Google Scholar] [CrossRef]
- Lin, A.-Q.; Csorba, G.; Li, L.-F.; Jiang, T.-L.; Lu, G.-J.; Thong, V.D.; Soisook, P.; Sun, K.-P.; Feng, J. Phylogeography of Hipposideros armiger (Chiroptera: Hipposideridae) in the Oriental Region: The contribution of multiple Pleistocene glacial refugia and intrinsic factors to contemporary population genetic structure. J. Biogeogr. 2014, 41, 317–327. [Google Scholar] [CrossRef]
- Dong, D.; Lei, M.; Hua, P.; Pan, Y.H.; Mu, S.; Zheng, G.; Pang, E.; Lin, K.; Zhang, S. The Genomes of Two Bat Species with Long Constant Frequency Echolocation Calls. Mol. Biol. Evol. 2017, 34, 20–34. [Google Scholar] [CrossRef]
- Niedringhaus, T.P.; Milanova, D.; Kerby, M.B.; Snyder, M.P.; Barron, A.E. Landscape of next-generation sequencing technologies. Anal. Chem. 2011, 83, 4327–4341. [Google Scholar] [CrossRef] [Green Version]
- Van Dijk, E.L.; Auger, H.; Jaszczyszyn, Y.; Thermes, C. Ten years of next-generation sequencing technology. Trends Genet. TIG 2014, 30, 418–426. [Google Scholar] [CrossRef]
- Zhang, J.; Chiodini, R.; Badr, A.; Zhang, G. The impact of next-generation sequencing on genomics. J. Genet. Genom. = Yi Chuan Xue Bao 2011, 38, 95–109. [Google Scholar] [CrossRef] [Green Version]
- Zhou, X.; Ren, L.; Meng, Q.; Li, Y.; Yu, Y.; Yu, J. The next-generation sequencing technology and application. Protein Cell 2010, 1, 520–536. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gordon, S.P.; Tseng, E.; Salamov, A.; Zhang, J.; Meng, X.; Zhao, Z.; Kang, D.; Underwood, J.; Grigoriev, I.V.; Figueroa, M.; et al. Widespread Polycistronic Transcripts in Fungi Revealed by Single-Molecule mRNA Sequencing. PLoS ONE 2015, 10, e0132628. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tilgner, H.; Grubert, F.; Sharon, D.; Snyder, M.P. Defining a personal, allele-specific, and single-molecule long-read transcriptome. Proc. Natl. Acad. Sci. USA 2014, 111, 9869–9874. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Treutlein, B.; Gokce, O.; Quake, S.R.; Südhof, T.C. Cartography of neurexin alternative splicing mapped by single-molecule long-read mRNA sequencing. Proc. Natl. Acad. Sci. USA 2014, 111, E1291–E1299. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Steijger, T.; Abril, J.F.; Engström, P.G.; Kokocinski, F.; Hubbard, T.J.; Guigó, R.; Harrow, J.; Bertone, P. Assessment of transcript reconstruction methods for RNA-seq. Nat. Methods 2013, 10, 1177–1184. [Google Scholar] [CrossRef] [Green Version]
- Sharon, D.; Tilgner, H.; Grubert, F.; Snyder, M. A single-molecule long-read survey of the human transcriptome. Nat. Biotechnol. 2013, 31, 1009–1014. [Google Scholar] [CrossRef] [Green Version]
- Wang, B.; Tseng, E.; Regulski, M.; Clark, T.A.; Hon, T.; Jiao, Y.; Lu, Z.; Olson, A.; Stein, J.C.; Ware, D. Unveiling the complexity of the maize transcriptome by single-molecule long-read sequencing. Nat. Commun. 2016, 7, 11708. [Google Scholar] [CrossRef] [Green Version]
- Kawamoto, M.; Jouraku, A.; Toyoda, A.; Yokoi, K.; Minakuchi, Y.; Katsuma, S.; Fujiyama, A.; Kiuchi, T.; Yamamoto, K.; Shimada, T. High-quality genome assembly of the silkworm, Bombyx mori. Insect Biochem. Mol. Biol. 2019, 107, 53–62. [Google Scholar] [CrossRef]
- Suetsugu, Y.; Futahashi, R.; Kanamori, H.; Kadono-Okuda, K.; Sasanuma, S.; Narukawa, J.; Ajimura, M.; Jouraku, A.; Namiki, N.; Shimomura, M.; et al. Large scale full-length cDNA sequencing reveals a unique genomic landscape in a lepidopteran model insect, Bombyx mori. G3 2013, 3, 1481–1492. [Google Scholar] [CrossRef] [Green Version]
- Gagan, J.; Van Allen, E.M. Next-generation sequencing to guide cancer therapy. Genome Med. 2015, 7, 80. [Google Scholar] [CrossRef] [Green Version]
- McDaniel, A.S.; Stall, J.N.; Hovelson, D.H.; Cani, A.K.; Liu, C.J.; Tomlins, S.A.; Cho, K.R. Next-Generation Sequencing of Tubal Intraepithelial Carcinomas. JAMA Oncol. 2015, 1, 1128–1132. [Google Scholar] [CrossRef]
- Mutz, K.O.; Heilkenbrinker, A.; Lönne, M.; Walter, J.G.; Stahl, F. Transcriptome analysis using next-generation sequencing. Curr. Opin. Biotechnol. 2013, 24, 22–30. [Google Scholar] [CrossRef]
- Anvar, S.Y.; Allard, G.; Tseng, E.; Sheynkman, G.M.; de Klerk, E.; Vermaat, M.; Yin, R.H.; Johansson, H.E.; Ariyurek, Y.; den Dunnen, J.T.; et al. Full-length mRNA sequencing uncovers a widespread coupling between transcription initiation and mRNA processing. Genome Biol. 2018, 19, 46. [Google Scholar] [CrossRef] [Green Version]
- Byrne, A.; Cole, C.; Volden, R.; Vollmers, C. Realizing the potential of full-length transcriptome sequencing. Philos. Trans. R. Soc. Lond. Ser. B Biol. Sci. 2019, 374, 20190097. [Google Scholar] [CrossRef] [Green Version]
- Wang, H.; Zhao, H.; Huang, X.; Sun, K.; Feng, J. Comparative cochlear transcriptomics of echolocating bats provides new insights into different nervous activities of CF bat species. Sci. Rep. 2018, 8, 15934. [Google Scholar] [CrossRef] [Green Version]
- Moreno-Santillán, D.D.; Machain-Williams, C.; Hernández-Montes, G.; Ortega, J. De Novo Transcriptome Assembly and Functional Annotation in Five Species of Bats. Sci. Rep. 2019, 9, 6222. [Google Scholar] [CrossRef] [Green Version]
- Zhao, H.; Wang, H.; Liu, T.; Liu, S.; Jin, L.; Huang, X.; Dai, W.; Sun, K.; Feng, J. Gene expression vs. sequence divergence: Comparative transcriptome sequencing among natural Rhinolophus ferrumequinum populations with different acoustic phenotypes. Front. Zool. 2019, 16, 37. [Google Scholar] [CrossRef]
- Nakano, K.; Shiroma, A.; Shimoji, M.; Tamotsu, H.; Ashimine, N.; Ohki, S.; Shinzato, M.; Minami, M.; Nakanishi, T.; Teruya, K.; et al. Advantages of genome sequencing by long-read sequencer using SMRT technology in medical area. Hum. Cell 2017, 30, 149–161. [Google Scholar] [CrossRef] [Green Version]
- Ma, L.; Sun, H.; Mao, X. Transcriptome sequencing of cochleae from constant-frequency and frequency-modulated echolocating bats. Sci. Data 2020, 7, 341. [Google Scholar] [CrossRef]
- Ashburner, M.; Ball, C.A.; Blake, J.A.; Botstein, D.; Butler, H.; Cherry, J.M.; Davis, A.P.; Dolinski, K.; Dwight, S.S.; Eppig, J.T.; et al. Gene ontology: Tool for the unification of biology. The Gene Ontology Consortium. Nat. Genet. 2000, 25, 25–29. [Google Scholar] [CrossRef] [Green Version]
- Marquez, Y.; Brown, J.W.; Simpson, C.; Barta, A.; Kalyna, M. Transcriptome survey reveals increased complexity of the alternative splicing landscape in Arabidopsis. Genome Res. 2012, 22, 1184–1195. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pan, Q.; Shai, O.; Lee, L.J.; Frey, B.J.; Blencowe, B.J. Deep surveying of alternative splicing complexity in the human transcriptome by high-throughput sequencing. Nat. Genet. 2008, 40, 1413–1415. [Google Scholar] [CrossRef] [PubMed]
- Grau-Bové, X.; Ruiz-Trillo, I.; Irimia, M. Origin of exon skipping-rich transcriptomes in animals driven by evolution of gene architecture. Genome Biol. 2018, 19, 135. [Google Scholar] [CrossRef] [PubMed]
- Patthy, L. Exon skipping-rich transcriptomes of animals reflect the significance of exon-shuffling in metazoan proteome evolution. Biol. Direct 2019, 14, 2. [Google Scholar] [CrossRef]
- Bentley, D.L. Coupling mRNA processing with transcription in time and space. Nat. Rev. Genet. 2014, 15, 163–175. [Google Scholar] [CrossRef] [Green Version]
- Moehle, E.A.; Braberg, H.; Krogan, N.J.; Guthrie, C. Adventures in time and space: Splicing efficiency and RNA polymerase II elongation rate. RNA Biol. 2014, 11, 313–319. [Google Scholar] [CrossRef] [Green Version]
- Tranbarger, T.J.; Kluabmongkol, W.; Sangsrakru, D.; Morcillo, F.; Tregear, J.W.; Tragoonrung, S.; Billotte, N. SSR markers in transcripts of genes linked to post-transcriptional and transcriptional regulatory functions during vegetative and reproductive development of Elaeis guineensis. BMC Plant Biol. 2012, 12, 1. [Google Scholar] [CrossRef]
- Jia, D.; Wang, Y.; Liu, Y.; Hu, J.; Guo, Y.; Gao, L.; Ma, R. SMRT sequencing of full-length transcriptome of flea beetle Agasicles hygrophila (Selman and Vogt). Sci. Rep. 2018, 8, 2197. [Google Scholar] [CrossRef] [Green Version]
- Kuo, R.I.; Tseng, E.; Eory, L.; Paton, I.R.; Archibald, A.L.; Burt, D.W. Normalized long read RNA sequencing in chicken reveals transcriptome complexity similar to human. BMC Genom. 2017, 18, 323. [Google Scholar] [CrossRef] [Green Version]
- Wang, J.; He, Y.; Peng, X.; Bo, L.; Wang, Z.; Song, Q. Characterization of cadmium-responsive transcription factors in wolf spider Pardosa pseudoannulata. Chemosphere 2021, 268, 129239. [Google Scholar] [CrossRef]
- Yu, J.Q.; Gu, K.D.; Sun, C.H.; Zhang, Q.Y.; Wang, J.H.; Ma, F.F.; You, C.X.; Hu, D.G.; Hao, Y.J. The apple bHLH transcription factor MdbHLH3 functions in determining the fruit carbohydrates and malate. Plant Biotechnol. J. 2021, 19, 285–299. [Google Scholar] [CrossRef]
- Wu, T.D.; Watanabe, C.K. GMAP: A genomic mapping and alignment program for mRNA and EST sequences. Bioinformatics 2005, 21, 1859–1875. [Google Scholar] [CrossRef] [Green Version]
- Conesa, A.; Götz, S.; García-Gómez, J.M.; Terol, J.; Talón, M.; Robles, M. Blast2GO: A universal tool for annotation, visualization and analysis in functional genomics research. Bioinformatics 2005, 21, 3674–3676. [Google Scholar] [CrossRef] [Green Version]
- Ye, J.; Fang, L.; Zheng, H.; Zhang, Y.; Chen, J.; Zhang, Z.; Wang, J.; Li, S.; Li, R.; Bolund, L.; et al. WEGO: A web tool for plotting GO annotations. Nucleic Acids Res. 2006, 34, W293–W297. [Google Scholar] [CrossRef]
- Alamancos, G.P.; Pagès, A.; Trincado, J.L.; Bellora, N.; Eyras, E. Leveraging transcript quantification for fast computation of alternative splicing profiles. RNA 2015, 21, 1521–1531. [Google Scholar] [CrossRef] [Green Version]
- Shimizu, K.; Adachi, J.; Muraoka, Y. ANGLE: A sequencing errors resistant program for predicting protein coding regions in unfinished cDNA. J. Bioinform. Comput. Biol. 2006, 4, 649–664. [Google Scholar] [CrossRef]
- Sun, L.; Luo, H.; Bu, D.; Zhao, G.; Yu, K.; Zhang, C.; Liu, Y.; Chen, R.; Zhao, Y. Utilizing sequence intrinsic composition to classify protein-coding and long non-coding transcripts. Nucleic Acids Res. 2013, 41, e166. [Google Scholar] [CrossRef]
- Kong, L.; Zhang, Y.; Ye, Z.Q.; Liu, X.Q.; Zhao, S.Q.; Wei, L.; Gao, G. CPC: Assess the protein-coding potential of transcripts using sequence features and support vector machine. Nucleic Acids Res. 2007, 35, W345–W349. [Google Scholar] [CrossRef]
- Nawrocki, E.P.; Eddy, S.R. Infernal 1.1: 100-fold faster RNA homology searches. Bioinformatics 2013, 29, 2933–2935. [Google Scholar] [CrossRef] [Green Version]



| Categories | Datasets |
|---|---|
| Number of bases sequenced (bp) | 125,546,653,021 |
| Number of subreads | 40,724,724 |
| Number of subreads average length | 3082 |
| Number of N50 length | 3573 |
| Number of CCS bases | 5,765,831,264 |
| Number of CCS reads | 1,630,747 |
| Number of mean CCS reads length | 3535 |
| Number of mean passes | 22 |
| Number of full-length non-chimeric reads | 1,472,058 |
| Full-length non-chimeric percentage (FLNC%) | 90.27% |
| Number of mean Length of FLNC | 3440 |
| Number of consensus isoforms | 94,981 |
| Number of polished high-quality isoforms | 91,477 |
| Number of polished low-quality isoforms | 3504 |
| Terms | Number of Reads |
|---|---|
| All reference gene number | 18,949 |
| All reference transcript number | 47,179 |
| All mapped gene | 12,029 |
| All mapped Isoform | 63,419 |
| known isoform | 18,890 |
| novel isoform | 2112 |
| new isoform | 42,417 |
| known transcripts from known genes | 47,192 |
| novel transcripts from known genes | 57,908 |
| novel transcripts from novel genes | 5511 |
| Type | Transcript | Average Length | 3′-UTR | 5′-UTR |
|---|---|---|---|---|
| Before | 47,192 | 3317 | 43,557 | 44,526 |
| After | 110,611 | 3343 | 43,557 | 43,165 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bao, M.; Wang, X.; Sun, R.; Wang, Z.; Li, J.; Jiang, T.; Lin, A.; Wang, H.; Feng, J. Full-Length Transcriptome of the Great Himalayan Leaf-Nosed Bats (Hipposideros armiger) Optimized Genome Annotation and Revealed the Expression of Novel Genes. Int. J. Mol. Sci. 2023, 24, 4937. https://doi.org/10.3390/ijms24054937
Bao M, Wang X, Sun R, Wang Z, Li J, Jiang T, Lin A, Wang H, Feng J. Full-Length Transcriptome of the Great Himalayan Leaf-Nosed Bats (Hipposideros armiger) Optimized Genome Annotation and Revealed the Expression of Novel Genes. International Journal of Molecular Sciences. 2023; 24(5):4937. https://doi.org/10.3390/ijms24054937
Chicago/Turabian StyleBao, Mingyue, Xue Wang, Ruyi Sun, Zhiqiang Wang, Jiqian Li, Tinglei Jiang, Aiqing Lin, Hui Wang, and Jiang Feng. 2023. "Full-Length Transcriptome of the Great Himalayan Leaf-Nosed Bats (Hipposideros armiger) Optimized Genome Annotation and Revealed the Expression of Novel Genes" International Journal of Molecular Sciences 24, no. 5: 4937. https://doi.org/10.3390/ijms24054937
APA StyleBao, M., Wang, X., Sun, R., Wang, Z., Li, J., Jiang, T., Lin, A., Wang, H., & Feng, J. (2023). Full-Length Transcriptome of the Great Himalayan Leaf-Nosed Bats (Hipposideros armiger) Optimized Genome Annotation and Revealed the Expression of Novel Genes. International Journal of Molecular Sciences, 24(5), 4937. https://doi.org/10.3390/ijms24054937






