Discovering the Biological Significance and Therapeutic Potential of miR-29b-3p in Triple-Negative Breast Cancer
Abstract
1. Introduction
2. Results
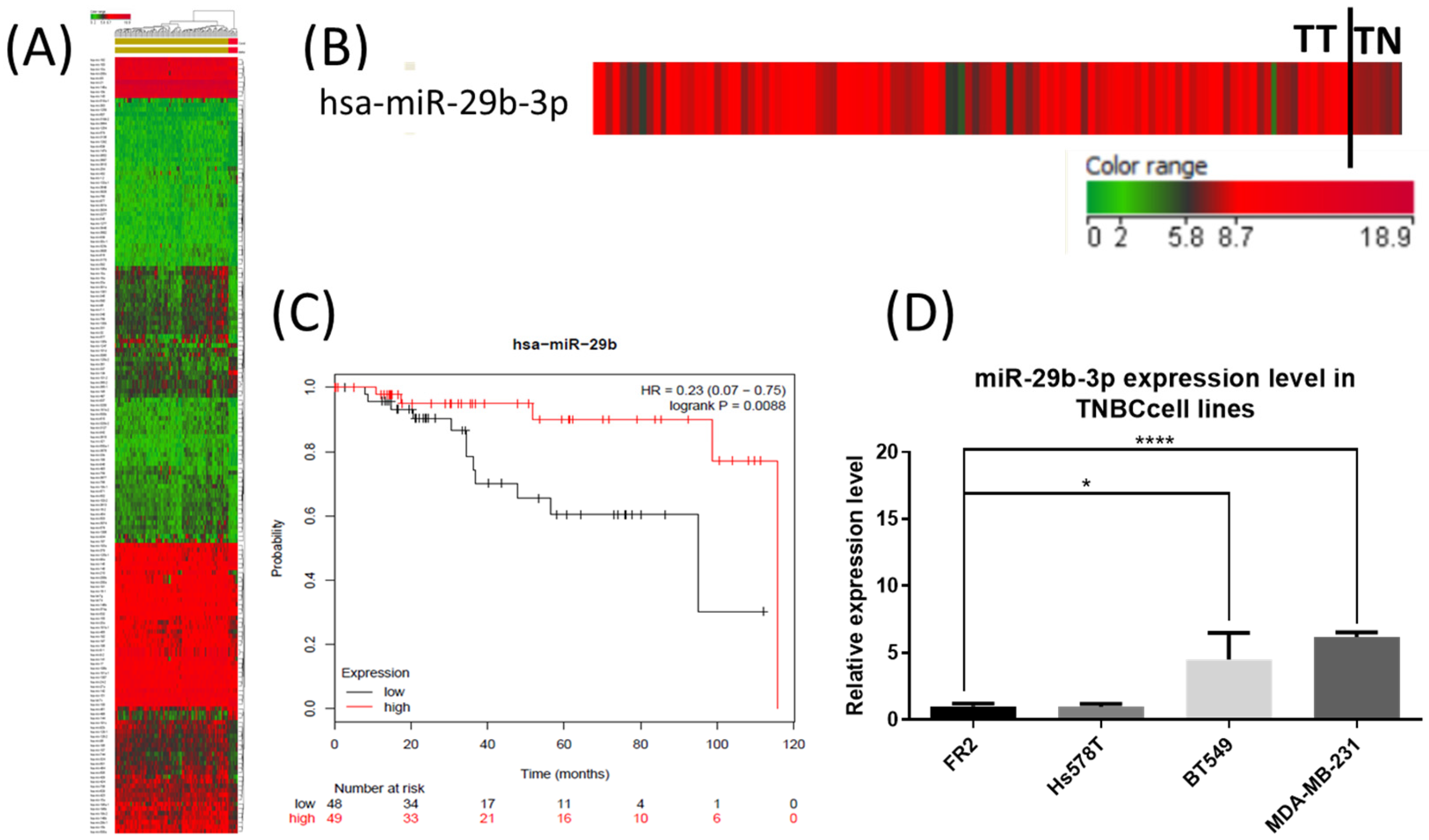
2.1. The Expression Level of miR-29b-3p in TNBC Tumor Tissue and Cell Lines
2.2. miR-29b-3p Inhibits Mitochondrial Activity and Activates Autophagy and Apoptosis
2.3. Identification of Differentially Expressed miRNAs as an Effect of miR-29b-3p Inhibition on TNBC Cell Lines Using Microarray Technology
2.4. The miR-29b-3p Expression Level Validated in Both TNBC Cell Lines Using qRT-PCR
2.5. Evaluation by qRT-PCR of Key Genes Targeted by miR-29b-3p
2.6. Quantifying IL6 in Cell Culture Medium for Both TNBC Cell Lines Using ELISA
3. Discussion
4. Materials and Methods
4.1. miR-29b-3p Expression Levels in TNBC
4.2. Cell Culture
4.3. miRNA Transfection
4.4. Cell Proliferation Assay
4.5. Colony Formation Assay
4.6. The Assessment of Mitochondrial Activity in Transfected TNBC Cell Lines
4.7. Apoptosis and Autophagic Vacuoles Assessment in Transfected TNBC Cell Lines
4.8. miRNA Altered Pattern as Effect of miR-29b Transfection in TNBC Cells
4.9. Gene and miRNA Expression Evaluation Using qRT-PCR
4.10. IL6 Quantification from the Cell Culture Medium
4.11. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Kumar, P.; Aggarwal, R. An overview of triple-negative breast cancer. Arch. Gynecol. Obstet. 2016, 293, 247–269. [Google Scholar] [CrossRef] [PubMed]
- Polyak, K. Heterogeneity in breast cancer. J. Clin. Investig. 2011, 121, 3786–3788. [Google Scholar] [CrossRef] [PubMed]
- Gurzu, S.; Banias, L.; Bara, T.; Feher, I.; Bara, T.; Jung, I. The Epithelial-Mesenchymal Transition Pathway in Two Cases with Gastric Metastasis Originating from Breast Carcinoma, One with a Metachronous Primary Gastric Cancer. Recent Patient Anticancer Drug Discov. 2018, 13, 118–124. [Google Scholar] [CrossRef]
- Bray, F.; Ferlay, J.; Soerjomataram, I.; Siegel, R.L.; Torre, L.A.; Jemal, A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA A Cancer J. Clin. 2018, 68, 394–424. [Google Scholar] [CrossRef] [PubMed]
- Chiorean, R.; Braicu, C.; Berindan-Neagoe, I. Another review on triple negative breast cancer. Are we on the right way towards the exit from the labyrinth? Breast 2013, 22, 1026–1033. [Google Scholar] [CrossRef]
- Choi, Y.L.; Oh, E.; Park, S.; Kim, Y.; Park, Y.H.; Song, K.; Cho, E.Y.; Hong, Y.C.; Choi, J.S.; Lee, J.E.; et al. Triple-negative, basal-like, and quintuple-negative breast cancers: Better prediction model for survival. BMC Cancer 2010, 10, 507. [Google Scholar] [CrossRef] [PubMed]
- Masuda, H.; Baggerly, K.A.; Wang, Y.; Zhang, Y.; Gonzalez-Angulo, A.M.; Meric-Bernstam, F.; Valero, V.; Lehmann, B.D.; Pietenpol, J.A.; Hortobagyi, G.N.; et al. Differential response to neoadjuvant chemotherapy among 7 triple-negative breast cancer molecular subtypes. Clin. Cancer Res. Off. J. Am. Assoc. Cancer Res. 2013, 19, 5533–5540. [Google Scholar] [CrossRef] [PubMed]
- Denkert, C.; Liedtke, C.; Tutt, A.; von Minckwitz, G. Molecular alterations in triple-negative breast cancer-the road to new treatment strategies. Lancet 2017, 389, 2430–2442. [Google Scholar] [CrossRef]
- Turashvili, G.; Brogi, E. Tumor Heterogeneity in Breast Cancer. Front. Med. 2017, 4, 227. [Google Scholar] [CrossRef]
- Irimie, A.I.; Braicu, C.; Sonea, L.; Zimta, A.A.; Cojocneanu-Petric, R.; Tonchev, K.; Mehterov, N.; Diudea, D.; Buduru, S.; Berindan-Neagoe, I. A Looking-Glass of Non-coding RNAs in oral cancer. Int. J. Mol. Sci. 2017, 18, 2620. [Google Scholar] [CrossRef]
- Zhang, L.L.; Wu, J.; Liu, Q.; Zhang, Y.; Sun, Z.L.; Jing, H. MiR-886-5p inhibition inhibits growth and induces apoptosis of MCF7 cells. Asian Pac. J. Cancer Prev. 2014, 15, 1511–1515. [Google Scholar] [CrossRef]
- Berindan-Neagoe, I.; Calin, G.A. Molecular pathways: MicroRNAs, cancer cells, and microenvironment. Clin. Cancer Res. 2014, 20, 6247–6253. [Google Scholar] [CrossRef]
- Cho, W.C. MicroRNAs: Potential biomarkers for cancer diagnosis, prognosis and targets for therapy. Int. J. Biochem. Cell Biol. 2010, 42, 1273–1281. [Google Scholar] [CrossRef]
- Yan, B.; Guo, Q.; Fu, F.J.; Wang, Z.; Yin, Z.; Wei, Y.B.; Yang, J.R. The role of miR-29b in cancer: Regulation, function, and signaling. Onco Targets Ther. 2015, 8, 539–548. [Google Scholar] [CrossRef] [PubMed]
- Kole, A.J.; Swahari, V.; Hammond, S.M.; Deshmukh, M. miR-29b is activated during neuronal maturation and targets BH3-only genes to restrict apoptosis. Genes Dev. 2011, 25, 125–130. [Google Scholar] [CrossRef] [PubMed]
- Zhang, B.; Shetti, D.; Fan, C.; Wei, K. miR-29b-3p promotes progression of MDA-MB-231 triple-negative breast cancer cells through downregulating TRAF3. Biol. Res. 2019, 52, 38. [Google Scholar] [CrossRef] [PubMed]
- Stenvang, J.; Petri, A.; Lindow, M.; Obad, S.; Kauppinen, S. Inhibition of microRNA function by antimiR oligonucleotides. Silence 2012, 3, 1–7. [Google Scholar] [CrossRef]
- Braicu, C.; Catana, C.; Calin, G.A.; Berindan-Neagoe, I. NCRNA combined therapy as future treatment option for cancer. Curr. Pharm. Des. 2014, 20, 6565–6574. [Google Scholar] [CrossRef]
- Hozaka, Y.; Seki, N.; Tanaka, T.; Asai, S.; Moriya, S.; Idichi, T.; Wada, M.; Tanoue, K.; Kawasaki, Y.; Mataki, Y.; et al. Molecular Pathogenesis and Regulation of the miR-29-3p-Family: Involvement of ITGA6 and ITGB1 in Intra-Hepatic Cholangiocarcinoma. Cancers 2021, 13, 2804. [Google Scholar] [CrossRef]
- Ciocan-Cartita, C.A.; Jurj, A.; Zanoaga, O.; Cojocneanu, R.; Pop, L.A.; Moldovan, A.; Moldovan, C.; Zimta, A.A.; Raduly, L.; Pop-Bica, C.; et al. New insights in gene expression alteration as effect of doxorubicin drug resistance in triple negative breast cancer cells. J. Exp. Clin. Cancer Res. 2020, 39, 241. [Google Scholar] [CrossRef]
- Ciocan-Cȃrtiţă, C.A.; Jurj, A.; Raduly, L.; Cojocneanu, R.; Moldovan, A.; Pileczki, V.; Pop, L.A.; Budişan, L.; Braicu, C.; Korban, S.S.; et al. New perspectives in triple-negative breast cancer therapy based on treatments with TGFβ1 siRNA and doxorubicin. Mol. Cell Biochem. 2020, 475, 285–299. [Google Scholar] [CrossRef] [PubMed]
- Jurj, A.; Pop, L.A.; Zanoaga, O.; Ciocan-Cârtiţă, C.A.; Cojocneanu, R.; Moldovan, C.; Raduly, L.; Pop-Bica, C.; Trif, M.; Irimie, A.; et al. New Insights in Gene Expression Alteration as Effect of Paclitaxel Drug Resistance in Triple Negative Breast Cancer Cells. Cell Physiol. Biochem. 2020, 54, 648–664. [Google Scholar] [CrossRef] [PubMed]
- Muluhngwi, P.; Alizadeh-Rad, N.; Vittitow, S.L.; Kalbfleisch, T.S.; Klinge, C.M. The miR-29 transcriptome in endocrine-sensitive and resistant breast cancer cells. Sci. Rep. 2017, 7, 5205. [Google Scholar] [CrossRef] [PubMed]
- Pan, D.; Du, Y.; Li, R.; Shen, A.; Liu, X.; Li, C.; Hu, B. miR-29b-3p Increases Radiosensitivity in Stemness Cancer Cells via Modulating Oncogenes Axis. Front. Cell Dev. Biol. 2021, 9, 741074. [Google Scholar] [CrossRef]
- Grassilli, S.; Vezzali, F.; Cairo, S.; Brugnoli, F.; Volinia, S.; De Mattei, M.; Judde, J.G.; Bertagnolo, V. Targeting the Vav1/miR-29b axis as a potential approach for treating selected molecular subtypes of triple-negative breast cancer. Oncol. Rep. 2021, 45, 1–7. [Google Scholar] [CrossRef]
- De Blasio, A.; Di Fiore, R.; Pratelli, G.; Drago-Ferrante, R.; Saliba, C.; Baldacchino, S.; Grech, G.; Scerri, C.; Vento, R.; Tesoriere, G. A loop involving NRF2, miR-29b-1-5p and AKT, regulates cell fate of MDA-MB-231 triple-negative breast cancer cells. J. Cell. Physiol. 2020, 235, 629–637. [Google Scholar] [CrossRef]
- Song, Y.; Zeng, S.; Zheng, G.; Chen, D.; Li, P.; Yang, M.; Luo, K.; Yin, J.; Gu, Y.; Zhang, Z.; et al. FOXO3a-driven miRNA signatures suppresses VEGF-A/NRP1 signaling and breast cancer metastasis. Oncogene 2021, 40, 777–790. [Google Scholar] [CrossRef]
- Wang, H.; An, X.; Yu, H.; Zhang, S.; Tang, B.; Zhang, X.; Li, Z. MiR-29b/TET1/ZEB2 signaling axis regulates metastatic properties and epithelial-mesenchymal transition in breast cancer cells. Oncotarget 2017, 8, 102119–102133. [Google Scholar] [CrossRef]
- Grassilli, S.; Bertagnolo, V.; Brugnoli, F. Mir-29b in Breast Cancer: A Promising Target for Therapeutic Approaches. Diagnostics 2022, 12, 2139. [Google Scholar] [CrossRef]
- Muluhngwi, P.; Klinge, C.M. Identification and Roles of miR-29b-1-3p and miR29a-3p-Regulated and Non-Regulated lncRNAs in Endocrine-Sensitive and Resistant Breast Cancer Cells. Cancers 2021, 13, 3530. [Google Scholar] [CrossRef]
- Chung, A.W.; Kozielski, A.J.; Qian, W.; Zhou, J.; Anselme, A.C.; Chan, A.A.; Pan, P.-Y.; Lee, D.J.; Chang, J.C. Tocilizumab overcomes chemotherapy resistance in mesenchymal stem-like breast cancer by negating autocrine IL-1A induction of IL-6. NPJ Breast Cancer 2022, 8, 30. [Google Scholar] [CrossRef] [PubMed]
- Hartman, Z.C.; Poage, G.M.; den Hollander, P.; Tsimelzon, A.; Hill, J.; Panupinthu, N.; Zhang, Y.; Mazumdar, A.; Hilsenbeck, S.G.; Mills, G.B.; et al. Growth of triple-negative breast cancer cells relies upon coordinate autocrine expression of the proinflammatory cytokines IL-6 and IL-8. Cancer Res. 2013, 73, 3470–3480. [Google Scholar] [CrossRef]
- Campbell, K.J.; Mason, S.M.; Winder, M.L.; Willemsen, R.B.E.; Cloix, C.; Lawson, H.; Rooney, N.; Dhayade, S.; Sims, A.H.; Blyth, K.; et al. Breast cancer dependence on MCL-1 is due to its canonical anti-apoptotic function. Cell Death Differ. 2021, 28, 2589–2600. [Google Scholar] [CrossRef]
- Wagner, K.-U. Know thy cells: Commonly used triple-negative human breast cancer cell lines carry mutations in RAS and effectors. Breast Cancer Res. 2022, 24, 44. [Google Scholar] [CrossRef] [PubMed]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Vandesompele, J.; De Preter, K.; Pattyn, F.; Poppe, B.; Van Roy, N.; De Paepe, A.; Speleman, F. Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol. 2002, 3, 00341–003411. [Google Scholar] [CrossRef] [PubMed]
- Vlachos, I.S.; Zagganas, K.; Paraskevopoulou, M.D.; Georgakilas, G.; Karagkouni, D.; Vergoulis, T.; Dalamagas, T.; Hatzigeorgiou, A.G. DIANA-miRPath v3.0: Deciphering microRNA function with experimental support. Nucleic Acids Res. 2015, 43, W460–W466. [Google Scholar] [CrossRef]







| BT549 | MDA-MB-231 | ||||
|---|---|---|---|---|---|
| Systematic_Name | FC | Corr p Value | Systematic_Name | FC | Corr p Value |
| hsa-miR-29c-3p | −96.24 | 0.0001 | hsa-miR-6889-3p | −15.41 | 0.011 |
| hsa-miR-185-5p | −16.09 | 0.0002 | hsa-miR-6508-5p | −6.93 | 0.025 |
| hsa-miR-6512-5p | −15.81 | 0.0003 | hsa-miR-1228-3p | −6.12 | 0.036 |
| hsa-miR-101-3p | −14.33 | 0.0004 | hsa-miR-4433a-5p | −5.19 | 0.013 |
| hsa-miR-3156-5p | −13.59 | 0.0008 | hsa-miR-29a-3p | −5.01 | 0.045 |
| hsa-miR-660-5p | −11.97 | 0.0000 | hsa-miR-29b-3p | −3.62 | 0.043 |
| hsa-miR-455-3p | −11.63 | 0.0003 | hsa-miR-6737-3p | −3.37 | 0.006 |
| hsa-miR-1914-3p | −11.62 | 0.0020 | hsa-miR-940 | −3.10 | 0.047 |
| hsa-miR-29a-3p | −11.24 | 0.0016 | hsa-miR-6797-3p | −2.85 | 0.002 |
| hsa-miR-29b-3p | −7.17 | 0.0004 | hsa-miR-6766-3p | −2.69 | 0.009 |
| hsa-miR-1271-5p | −5.83 | 0.0284 | hsa-miR-6800-5p | 19.82 | 0.000 |
| hsa-miR-4800-5p | 19.97 | 0.0044 | hsa-miR-3125 | 19.51 | 0.000 |
| hsa-miR-3648 | 16.12 | 0.0091 | hsa-miR-4466 | 17.33 | 0.000 |
| hsa-miR-765 | 14.67 | 0.0004 | hsa-miR-1914-3p | 15.88 | 0.000 |
| hsa-miR-7110-5p | 3.74 | 0.0433 | hsa-miR-3141 | 15.33 | 0.000 |
| hsa-miR-1229-5p | 2.96 | 0.0060 | hsa-miR-5787 | 14.21 | 0.004 |
| hsa-miR-6087 | 2.79 | 0.0478 | hsa-miR-6858-5p | 14.05 | 0.024 |
| hsa-miR-4433a-5p | 2.12 | 0.0433 | hsa-miR-3665 | 13.60 | 0.001 |
| hsa-miR-6740-5p | 1.36 | 0.0284 | hsa-miR-423-5p | 13.54 | 0.001 |
| hsa-miR-3156-5p | 13.54 | 0.000 | |||
| hsa-miR-4298 | 12.98 | 0.000 | |||
| hsa-miR-7107-5p | 12.87 | 0.016 | |||
| hsa-miR-1268a | 12.83 | 0.001 | |||
| hsa-miR-6769b-5p | 10.73 | 0.006 | |||
| hsa-miR-1229-5p | 10.08 | 0.018 | |||
| hsa-miR-1288-3p | 9.33 | 0.001 | |||
| hsa-miR-6891-5p | 8.77 | 0.004 | |||
| hsa-miR-4653-3p | 8.51 | 0.042 | |||
| hsa-miR-2861 | 8.15 | 0.035 | |||
| hsa-miR-7847-3p | 8.01 | 0.002 | |||
| hsa-miR-483-5p | 7.97 | 0.014 | |||
| hsa-miR-6780a-5p | 7.70 | 0.006 | |||
| hsa-miR-3656 | 7.55 | 0.004 | |||
| hsa-miR-1915-3p | 7.09 | 0.028 | |||
| hsa-miR-138-5p | 6.71 | 0.042 | |||
| hsa-miR-4669 | 6.43 | 0.001 | |||
| hsa-miR-7150 | 5.09 | 0.018 | |||
| hsa-miR-484 | 4.49 | 0.017 | |||
| hsa-miR-186-5p | 3.45 | 0.000 | |||
| hsa-miR-6727-5p | 3.14 | 0.006 | |||
| hsa-miR-584-5p | 3.09 | 0.004 | |||
| hsa-miR-937-5p | 2.71 | 0.048 | |||
| hsa-miR-23a-5p | 2.11 | 0.023 | |||
| Demographics | TNBC |
| No. of cases | |
| Females | 112 |
| Age | |
| Median, Range | 54 (29–90) |
| TNM | |
| T1 | 27 |
| T2 | 70 |
| T3 | 11 |
| T4 | 4 |
| Tx | - |
| N0 | 72 |
| N1 | 25 |
| N2 | 11 |
| N3 | 4 |
| Nx | - |
| M0 | 95 |
| Mx | 17 |
| Tumor grade (I–IV) | |
| I: | 20 |
| II: | 70 |
| III: | 18 |
| IV: | 1 |
| Unknown: | 3 |
| Primer | Sequence |
|---|---|
| MCL1 | FW-TGTCCAGTTCCGAAGCAT/ RV-AAGCGAATGGGCAGGTCGT |
| BCL2 | FW-GCGCTACAGTTCCACAAAGG/ RV-AGTACCTGAACCGGCACCT |
| TP53 | FW: CCC TTT TTG GAC TTC AGG TG/ RV: AGG CCT TGG AAC TCA AGG AT |
| Caspase 3 | FW-GCTTGTCGGCATACTGTTTCAG/ RV-AGAACTGGACTGTGGCATTGAG |
| TGFβ1 | FW-ACTACTACGCCAAGGAGGTCAC/ RV-TGCTTGAACTTGTCATAGATTTCG |
| TGFΒR2 | FW-CACCGCACGTTCAGAAGTC/ RV-TGGATGGGCAGTCCTATTACA |
| B2M | FW-CACCCCCACTGAAAAAGATGAG/ RV-CCTCCATGATGCTGCTTACATG |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Jurj, A.; Zanoaga, O.; Raduly, L.; Morhan, V.; Papi, Z.; Ciocan, C.; Pop, L.-A.; Berindan-Neagoe, I.; Braicu, C. Discovering the Biological Significance and Therapeutic Potential of miR-29b-3p in Triple-Negative Breast Cancer. Int. J. Mol. Sci. 2023, 24, 5048. https://doi.org/10.3390/ijms24055048
Jurj A, Zanoaga O, Raduly L, Morhan V, Papi Z, Ciocan C, Pop L-A, Berindan-Neagoe I, Braicu C. Discovering the Biological Significance and Therapeutic Potential of miR-29b-3p in Triple-Negative Breast Cancer. International Journal of Molecular Sciences. 2023; 24(5):5048. https://doi.org/10.3390/ijms24055048
Chicago/Turabian StyleJurj, Ancuta, Oana Zanoaga, Lajos Raduly, Vlad Morhan, Zsofia Papi, Cristina Ciocan, Laura-Ancuta Pop, Ioana Berindan-Neagoe, and Cornelia Braicu. 2023. "Discovering the Biological Significance and Therapeutic Potential of miR-29b-3p in Triple-Negative Breast Cancer" International Journal of Molecular Sciences 24, no. 5: 5048. https://doi.org/10.3390/ijms24055048
APA StyleJurj, A., Zanoaga, O., Raduly, L., Morhan, V., Papi, Z., Ciocan, C., Pop, L.-A., Berindan-Neagoe, I., & Braicu, C. (2023). Discovering the Biological Significance and Therapeutic Potential of miR-29b-3p in Triple-Negative Breast Cancer. International Journal of Molecular Sciences, 24(5), 5048. https://doi.org/10.3390/ijms24055048