Identification and Empiric Evaluation of New Inhibitors of the Multidrug Transporter P-Glycoprotein (ABCB1)
Abstract
1. Introduction
2. Results
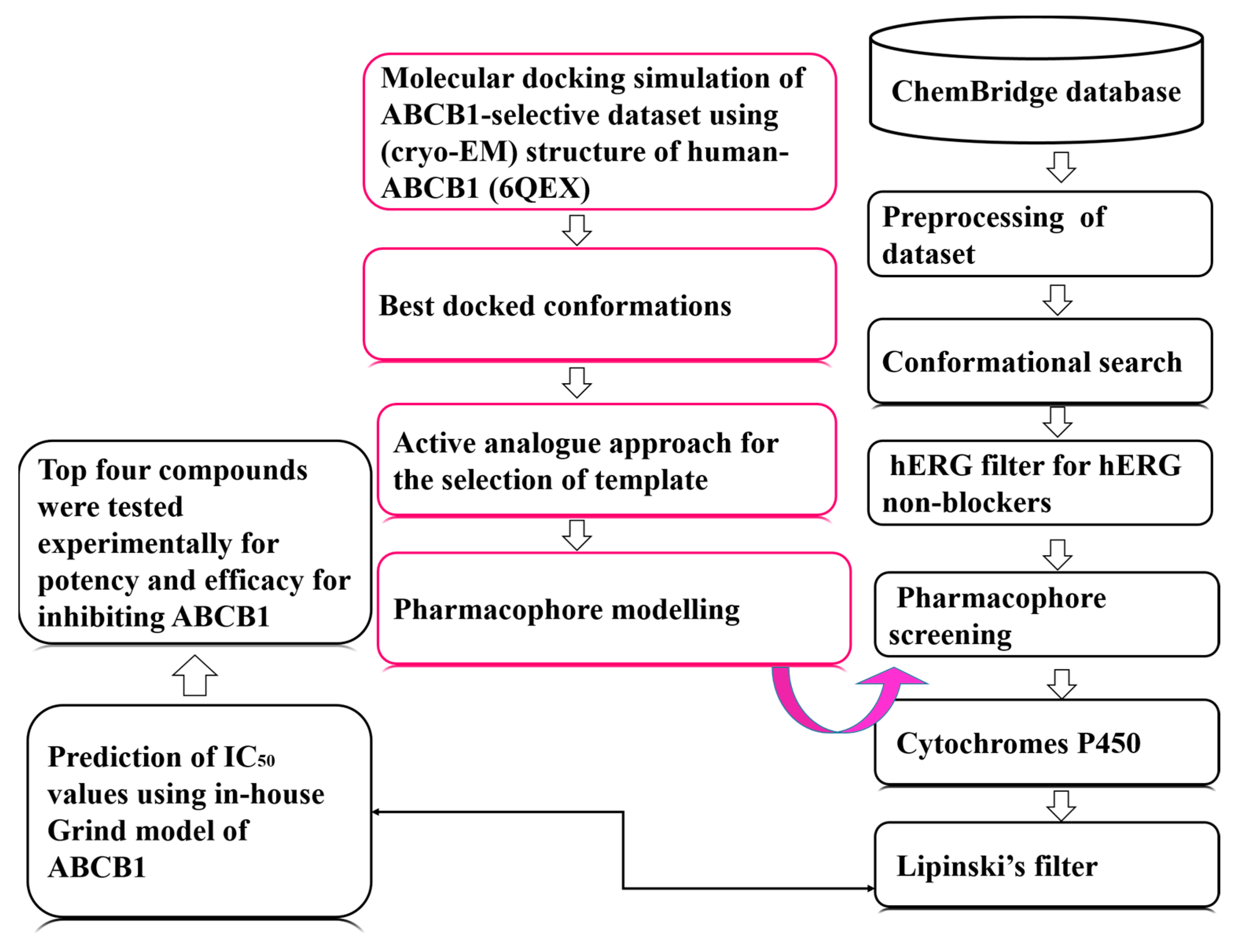
2.1. Data Collection and Template Selection
2.2. Pharmacophore Model Generation and Validation
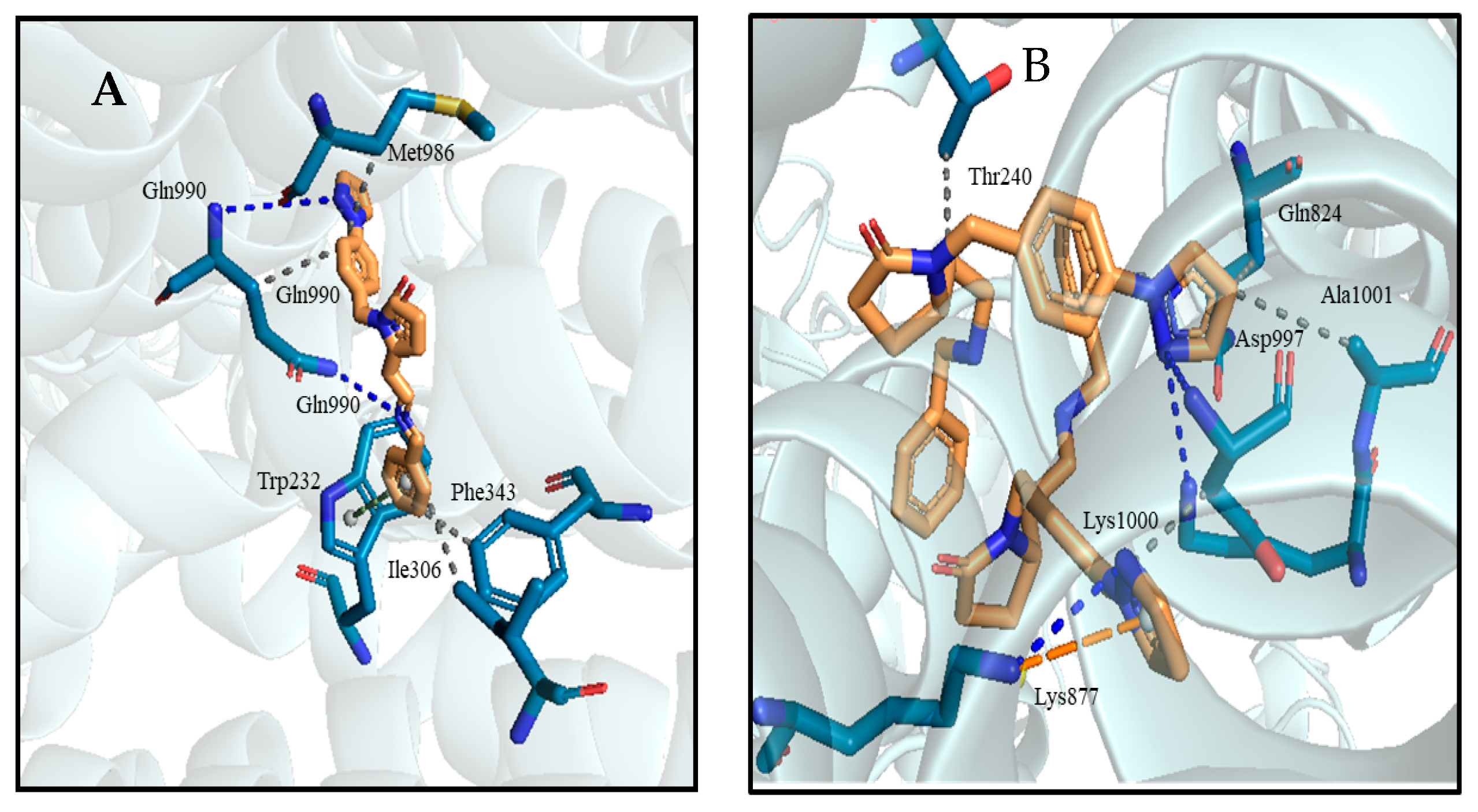
2.3. Virtual Screening and Hit Identification
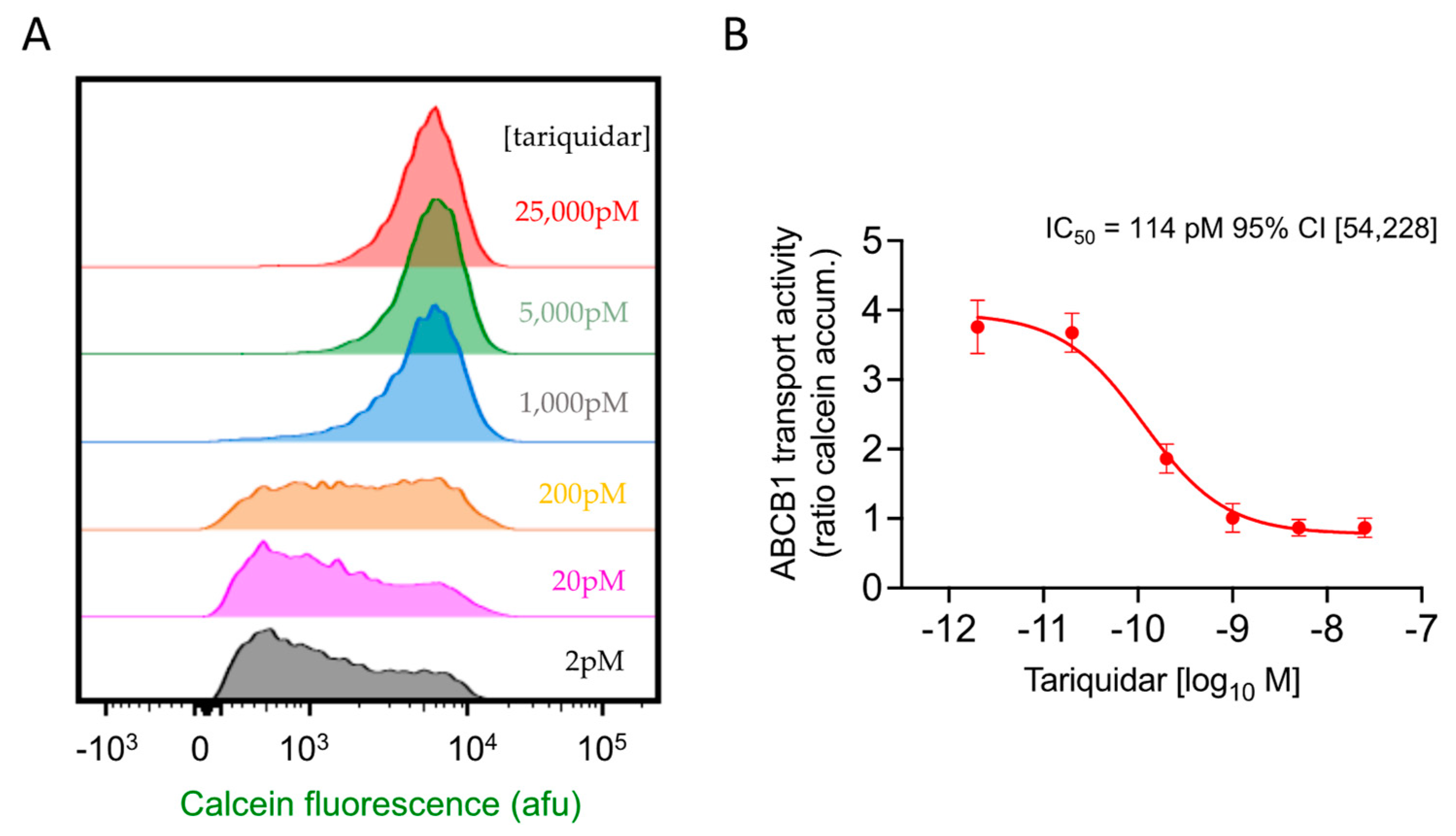
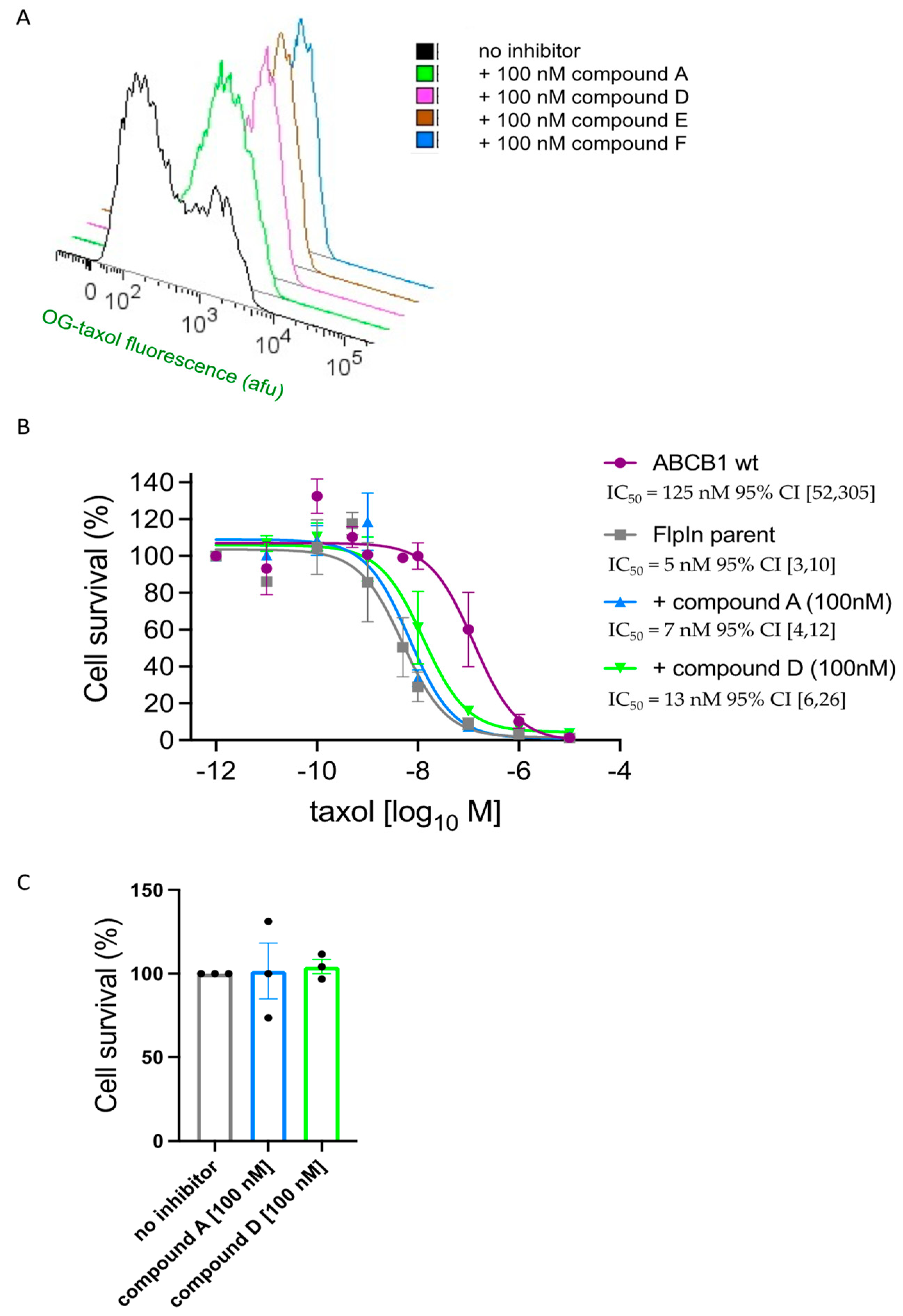
2.4. Flow Cytometric Assay of Drug Efficacy and Potency: ABCB1 Inhibition Validated Using Tariquidar
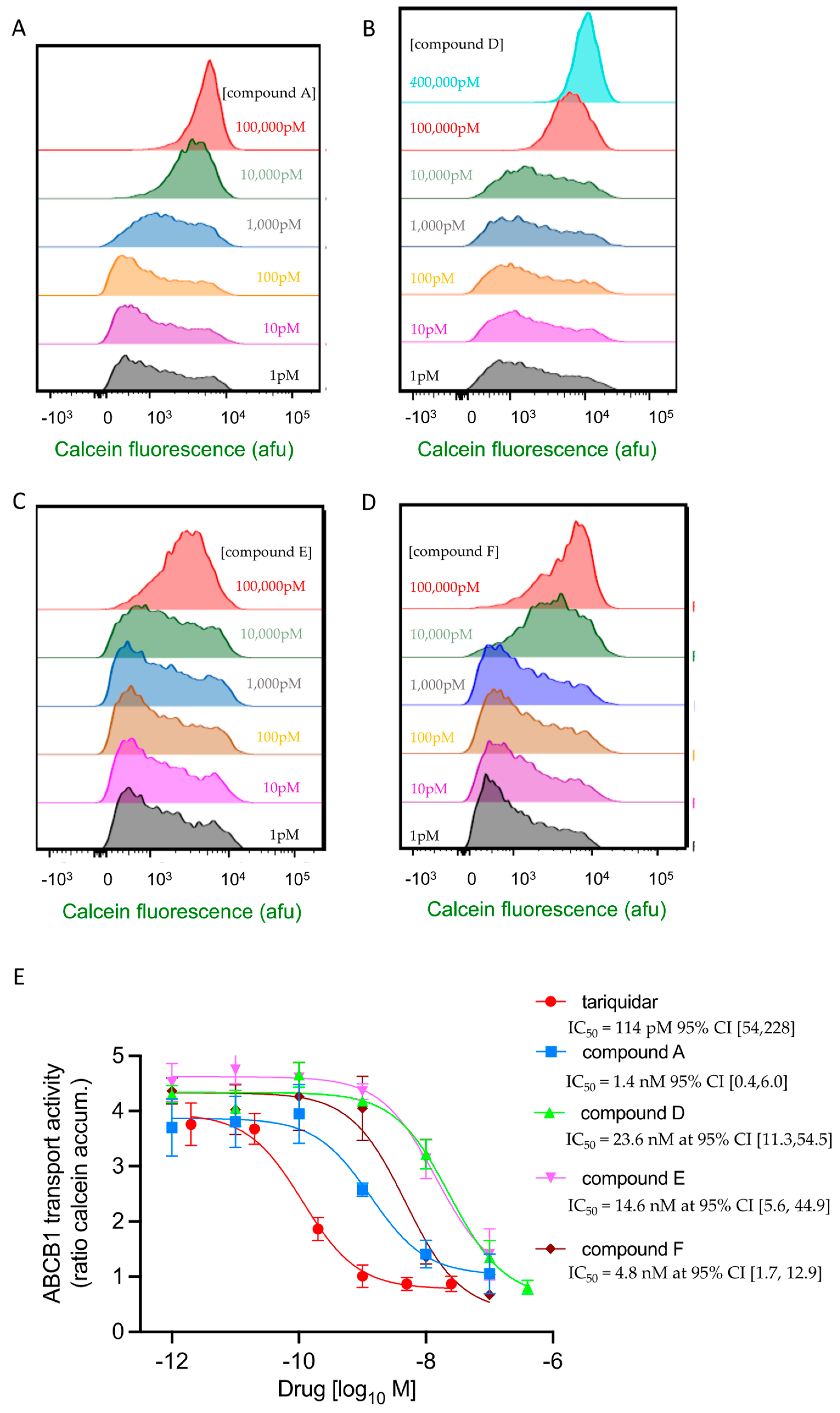
2.5. Four of the Six Test Compounds Fully Inhibit ABCB1 for the Transport of Calcein-AM but with Different Potencies
2.6. Compounds A and D Potentiate the Cytotoxicity of Taxol in ABCB1-Expressing Cells
3. Discussion
4. Materials and Methods
4.1. Computational
4.1.1. Dataset Collection
4.1.2. Molecular Docking Simulation
4.2. Template Selection and Pharmacophore Modelling
4.3. Pharmacophore Model Validation
4.4. Pharmacophore-Based Virtual Screening
4.5. In Vitro Transport Inhibition Studies
4.6. Taxol Challenge Assay
4.7. Lipophilic Efficiency of Potential Hits
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Alessia, C.; Iacopetta, D.; Ceramella, J.; Domenica, S.; Federica, G.; Carmela, S.; Aquaro, S.; Camillo, R.; Sinicropi, M.S. Multidrug Resistance (MDR): A Widespread Phenomenon in Pharmacological Therapies. Molecules 2022, 27, 616. [Google Scholar] [CrossRef]
- Zhang, Y.; Li, C.; Xia, C.; Wah To, K.K.; Guo, Z.; Ren, C.; Wen, L.; Wang, F.; Fu, L.; Liao, N. Adagrasib, a KRAS G12C inhibitor, reverses the multidrug resistance mediated by ABCB1 in vitro and in vivo. Cell Commun. Signal. 2022, 20, 142. [Google Scholar] [CrossRef]
- Engle, K.; Kumar, G. Cancer multidrug-resistance reversal by ABCB1 inhibition: A recent update. Eur. J. Med. Chem. 2022, 239, 114542. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Yang, D.-H.; Yang, Y.; Wang, J.-Q.; Cai, C.-Y.; Lei, Z.-N.; Teng, Q.-X.; Wu, Z.-X.; Zhao, L.; Chen, Z.-S. Overexpression of ABCB1 transporter confers resistance to mTOR inhibitor WYE-354 in cancer cells. Int. J. Mol. Sci. 2020, 21, 1387. [Google Scholar] [CrossRef] [PubMed]
- Han, K.; Kahng, J.; Kim, M.; Lim, J.; Kim, Y.; Cho, B.; Kim, H.K.; Min, W.S.; Kim, C.C.; Lee, K.Y. Expression of functional markers in acute nonlymphoblastic leukemia. Acta Haematol. 2000, 104, 174–180. [Google Scholar] [CrossRef] [PubMed]
- Leith, C.P.; Kopecky, K.J.; Chen, I.-M.; Eijdems, L.; Slovak, M.L.; McConnell, T.S.; Head, D.R.; Weick, J.; Grever, M.R.; Appelbaum, F.R. Frequency and clinical significance of the expression of the multidrug resistance proteins MDR1/P-glycoprotein, MRP1, and LRP in acute myeloid leukemia. A Southwest Oncology Group Study. Blood J. Am. Soc. Hematol. 1999, 94, 1086–1099. [Google Scholar]
- Trock, B.J.; Leonessa, F.; Clarke, R. Multidrug resistance in breast cancer: A meta-analysis of MDR1/gp170 expression and its possible functional significance. J. Natl. Cancer Inst. 1997, 89, 917–931. [Google Scholar] [CrossRef]
- Triller, N.; Korošec, P.; Kern, I.; Košnik, M.; Debeljak, A. Multidrug resistance in small cell lung cancer: Expression of P-glycoprotein, multidrug resistance protein 1 and lung resistance protein in chemo-naive patients and in relapsed disease. Lung Cancer 2006, 54, 235–240. [Google Scholar] [CrossRef]
- Fletcher, J.I.; Williams, R.T.; Henderson, M.J.; Norris, M.D.; Haber, M. ABC transporters as mediators of drug resistance and contributors to cancer cell biology. Drug Resist. Updat. 2016, 26, 1–9. [Google Scholar] [CrossRef]
- Robey, R.W.; Pluchino, K.M.; Hall, M.D.; Fojo, A.T.; Bates, S.E.; Gottesman, M.M. Revisiting the role of ABC transporters in multidrug-resistant cancer. Nat. Rev. Cancer 2018, 18, 452–464. [Google Scholar] [CrossRef]
- Juliano, R.L.; Ling, V. A surface glycoprotein modulating drug permeability in Chinese hamster ovary cell mutants. Biochim. Et Biophys. Acta (BBA)-Biomembr. 1976, 455, 152–162. [Google Scholar] [CrossRef]
- Chen, C.-J.; Chin, J.E.; Ueda, K.; Clark, D.P.; Pastan, I.; Gottesman, M.M.; Roninson, I.B. Internal duplication and homology with bacterial transport proteins in the mdr1 (P-glycoprotein) gene from multidrug-resistant human cells. Cell 1986, 47, 381–389. [Google Scholar] [CrossRef]
- Kaczor, A.; Szemerédi, N.; Kucwaj-Brysz, K.; Dąbrowska, M.; Starek, M.; Latacz, G.; Spengler, G.; Handzlik, J. Computer-aided search for 5-arylideneimidazolone anticancer agents able to overcome ABCB1-based multidrug resistance. ChemMedChem 2021, 16, 2386–2401. [Google Scholar] [CrossRef] [PubMed]
- Pan, M.; Cui, J.; Jiao, L.; Ghaleb, H.; Liao, C.; Zhou, J.; Kairuki, M.; Lin, H.; Huang, W.; Qian, H. Synthesis and biological evaluation of JL-A7 derivatives as potent ABCB1 inhibitors. Bioorg. Med. Chem. 2017, 25, 4194–4202. [Google Scholar] [CrossRef]
- Shawky, A.M.; Abdalla, A.N.; Ibrahim, N.A.; Abourehab, M.A.; Gouda, A.M. Discovery of new pyrimidopyrrolizine/indolizine-based derivatives as P-glycoprotein inhibitors: Design, synthesis, cytotoxicity, and MDR reversal activities. Eur. J. Med. Chem. 2021, 218, 113403. [Google Scholar] [CrossRef] [PubMed]
- Jeevitha Priya, M.; Vidyalakshmi, S.; Rajeswari, M. Study on reversal of ABCB1 mediated multidrug resistance in Colon cancer by acetogenins: An in-silico approach. J. Biomol. Struct. Dyn. 2022, 40, 4273–4284. [Google Scholar] [CrossRef]
- Alam, A.; Kowal, J.; Broude, E.; Roninson, I.; Locher, K.P. Structural insight into substrate and inhibitor discrimination by human P-glycoprotein. Science 2019, 363, 753–756. [Google Scholar] [CrossRef] [PubMed]
- Kim, Y.; Chen, J. Molecular structure of human P-glycoprotein in the ATP-bound, outward-facing conformation. Science 2018, 359, 915–919. [Google Scholar] [CrossRef] [PubMed]
- Nosol, K.; Romane, K.; Irobalieva, R.N.; Alam, A.; Kowal, J.; Fujita, N.; Locher, K.P. Cryo-EM structures reveal distinct mechanisms of inhibition of the human multidrug transporter ABCB1. Proc. Natl. Acad. Sci. USA 2020, 117, 26245–26253. [Google Scholar] [CrossRef]
- Żesławska, E.; Tejchman, W.; Kincses, A.; Spengler, G.; Nitek, W.; Żuchowski, G.; Szymańska, E. 5-Arylidenerhodanines as P-gp Modulators: An Interesting Effect of the Carboxyl Group on ABCB1 Function in Multidrug-Resistant Cancer Cells. Int. J. Mol. Sci. 2022, 23, 10812. [Google Scholar] [CrossRef]
- Ji, N.; Yang, Y.; Cai, C.-Y.; Wang, J.-Q.; Lei, Z.-N.; Wu, Z.-X.; Cui, Q.; Yang, D.-H.; Chen, Z.-S.; Kong, D. Midostaurin reverses ABCB1-mediated multidrug resistance, an in vitro study. Front. Oncol. 2019, 9, 514. [Google Scholar] [CrossRef]
- Kamrani, S.; Mosaffa, F.; Behravan, J.; Hadizadeh, F. P-gp and BCRP inhibition induced by some new 1, 4-Dihydropyridine and Dihydropyrimidines. Int. J. Bioinform. Res. Appl. 2017, 13, 62–74. [Google Scholar] [CrossRef]
- Allen, F.H.; Motherwell, W.S. Applications of the Cambridge Structural Database in organic chemistry and crystal chemistry. Acta Crystallogr. Sect. B Struct. Sci. 2002, 58, 407–422. [Google Scholar] [CrossRef] [PubMed]
- Teodori, E.; Dei, S.; Bartolucci, G.; Perrone, M.G.; Manetti, D.; Romanelli, M.N.; Contino, M.; Colabufo, N.A. Structure–Activity Relationship Studies on 6, 7-Dimethoxy-2-phenethyl-1, 2, 3, 4-tetrahydroisoquinoline Derivatives as Multidrug Resistance Reversers. ChemMedChem 2017, 12, 1369–1379. [Google Scholar] [CrossRef] [PubMed]
- Pellicani, R.Z.; Stefanachi, A.; Niso, M.; Carotti, A.; Leonetti, F.; Nicolotti, O.; Perrone, R.; Berardi, F.; Cellamare, S.; Colabufo, N.A. Potent galloyl-based selective modulators targeting multidrug resistance associated protein 1 and P-glycoprotein. J. Med. Chem. 2012, 55, 424–436. [Google Scholar] [CrossRef]
- Pajeva, I.K.; Globisch, C.; Wiese, M. Combined pharmacophore modeling, docking, and 3D QSAR studies of ABCB1 and ABCC1 transporter inhibitors. ChemMedChem Chem. Enabling Drug Discov. 2009, 4, 1883–1896. [Google Scholar] [CrossRef]
- Cramer, J.; Kopp, S.; Bates, S.E.; Chiba, P.; Ecker, G.F. Multispecificity of drug transporters: Probing inhibitor selectivity for the human drug efflux transporters ABCB1 and ABCG2. ChemMedChem Chem. Enabling Drug Discov. 2007, 2, 1783–1788. [Google Scholar] [CrossRef]
- Kühnle, M.; Egger, M.; Müller, C.; Mahringer, A.; Bernhardt, G.n.; Fricker, G.; König, B.; Buschauer, A. Potent and selective inhibitors of breast cancer resistance protein (ABCG2) derived from the p-glycoprotein (ABCB1) modulator tariquidar. J. Med. Chem. 2009, 52, 1190–1197. [Google Scholar] [CrossRef]
- Chicco, D.; Jurman, G. The advantages of the Matthews correlation coefficient (MCC) over F1 score and accuracy in binary classification evaluation. BMC Genom. 2020, 21, 6. [Google Scholar] [CrossRef]
- Munawar, S.; Windley, M.J.; Tse, E.G.; Todd, M.H.; Hill, A.P.; Vandenberg, J.I.; Jabeen, I. Experimentally validated pharmacoinformatics approach to predict hERG inhibition potential of new chemical entities. Front. Pharmacol. 2018, 9, 1035. [Google Scholar] [CrossRef]
- Sushko, I.; Novotarskyi, S.; Körner, R.; Pandey, A.K.; Rupp, M.; Teetz, W.; Brandmaier, S.; Abdelaziz, A.; Prokopenko, V.V.; Tanchuk, V.Y. Online chemical modeling environment (OCHEM): Web platform for data storage, model development and publishing of chemical information. J. Comput. Aided Mol. Des. 2011, 25, 533–554. [Google Scholar] [CrossRef] [PubMed]
- Shafi, T.; Jabeen, I. Grid-independent Descriptors (GRIND) analysis and SAR guided molecular docking studies to probe selectivity profiles of inhibitors of multidrug resistance transporters ABCB1 and ABCG2. Curr. Cancer Drug Targets 2017, 17, 177–190. [Google Scholar] [CrossRef] [PubMed]
- Akkaya, B.G.; Zolnerciks, J.K.; Ritchie, T.K.; Bauer, B.; Hartz, A.M.; Sullivan, J.A.; Linton, K.J. The multidrug resistance pump ABCB1 is a substrate for the ubiquitin ligase NEDD4-1. Mol. Membr. Biol. 2015, 32, 39–45. [Google Scholar] [CrossRef] [PubMed]
- van Roon, E.N.; Flikweert, S.; le Comte, M.; Langendijk, P.N.; Kwee-Zuiderwijk, W.J.; Smits, P.; Brouwers, J.R. Clinical relevance of drug-drug interactions. Drug Saf. 2005, 28, 1131–1139. [Google Scholar] [CrossRef] [PubMed]
- Holló, Z.; Homolya, L.; Davis, C.W.; Sarkadi, B. Calcein accumulation as a fluorometric functional assay of the multidrug transporter. Biochim. Et Biophys. Acta (BBA)-Biomembr. 1994, 1191, 384–388. [Google Scholar] [CrossRef]
- Homolya, L.; Holló, Z.; Germann, U.A.; Pastan, I.; Gottesman, M.M.; Sarkadi, B. Fluorescent cellular indicators are extruded by the multidrug resistance protein. J. Biol. Chem. 1993, 268, 21493–21496. [Google Scholar] [CrossRef]
- Liao, D.; Zhang, W.; Gupta, P.; Lei, Z.-N.; Wang, J.-Q.; Cai, C.-Y.; Vera, A.A.D.; Zhang, L.; Chen, Z.-S.; Yang, D.-H. Tetrandrine interaction with ABCB1 reverses multidrug resistance in cancer cells through competition with anti-cancer drugs followed by downregulation of ABCB1 expression. Molecules 2019, 24, 4383. [Google Scholar] [CrossRef]
- Ferreira, R.J.; Baptista, R.; Moreno, A.; Madeira, P.G.; Khonkarn, R.; Baubichon-Cortay, H.; Dos Santos, D.J.; Falson, P.; Ferreira, M.-J.U. Optimizing the flavanone core toward new selective nitrogen-containing modulators of ABC transporters. Future Med. Chem. 2018, 10, 725–741. [Google Scholar] [CrossRef]
- Gao, Y.; Zhang, T.; Terai, H.; Ficarro, S.B.; Kwiatkowski, N.; Hao, M.-F.; Sharma, B.; Christensen, C.L.; Chipumuro, E.; Wong, K.-k. Overcoming resistance to the THZ series of covalent transcriptional CDK inhibitors. Cell Chem. Biol. 2018, 25, 135–142.e135. [Google Scholar] [CrossRef]
- Jabeen, I.; Pleban, K.; Rinner, U.; Chiba, P.; Ecker, G.F. Structure–activity relationships, ligand efficiency, and lipophilic efficiency profiles of benzophenone-type inhibitors of the multidrug transporter P-glycoprotein. J. Med. Chem. 2012, 55, 3261–3273. [Google Scholar] [CrossRef]
- Akhtar, N.; Jabeen, I.; Jalal, N.; Antilla, J. Structure-based pharmacophore models to probe anticancer activity of inhibitors of protein kinase B-beta (PKB β). Chem. Biol. Drug Des. 2019, 93, 325–336. [Google Scholar] [CrossRef]
- Zhang, S.L.; Wei, Y.X.; Li, Q.; Sun, H.P.; Peng, H.; You, Q.D. Pharmacophore-Based Drug Design and Biological Evaluation of Novel ABCB1 Inhibitors. Chem. Biol. Drug Des. 2013, 81, 349–358. [Google Scholar] [CrossRef] [PubMed]
- Ismatullah, H.; Jabeen, I. Combined Pharmacophore and Grid-Independent Molecular Descriptors (GRIND) Analysis to Probe 3D Features of Inositol 1, 4, 5-Trisphosphate Receptor (IP3R) Inhibitors in Cancer. Int. J. Mol. Sci. 2021, 22, 12993. [Google Scholar] [CrossRef] [PubMed]
- Jones, G.; Willett, P.; Glen, R.C.; Leach, A.R.; Taylor, R. Development and validation of a genetic algorithm for flexible docking. J. Mol. Biol. 1997, 267, 727–748. [Google Scholar] [CrossRef] [PubMed]
- Güner, O.F. Pharmacophore Perception, Development, and Use in Drug Design; Internat’l University Line: La Jolla, CA, USA, 2000; Volume 2. [Google Scholar]
- Tebib, S.; Bourguignon, J.-J.; Wermuth, C.-G. The active analog approach applied to the pharmacophore identification of benzodiazepine receptor ligands. J. Comput. Aided Mol. Des. 1987, 1, 153–170. [Google Scholar] [CrossRef]
- Sasitharan, K.; Iqbal, H.A.; Bifsa, F.; Olszewska, A.; Linton, K.J. ABCB1 Does Not Require the Side-Chain Hydrogen-Bond Donors Gln347, Gln725, Gln990 to Confer Cellular Resistance to the Anticancer Drug Taxol. Int. J. Mol. Sci. 2021, 22, 8561. [Google Scholar] [CrossRef]
- Ivnitski-Steele, I.; Larson, R.S.; Lovato, D.M.; Khawaja, H.M.; Winter, S.S.; Oprea, T.I.; Sklar, L.A.; Edwards, B.S. High-throughput flow cytometry to detect selective inhibitors of ABCB1, ABCC1, and ABCG2 transporters. Assay Drug Dev. Technol. 2008, 6, 263–276. [Google Scholar] [CrossRef]
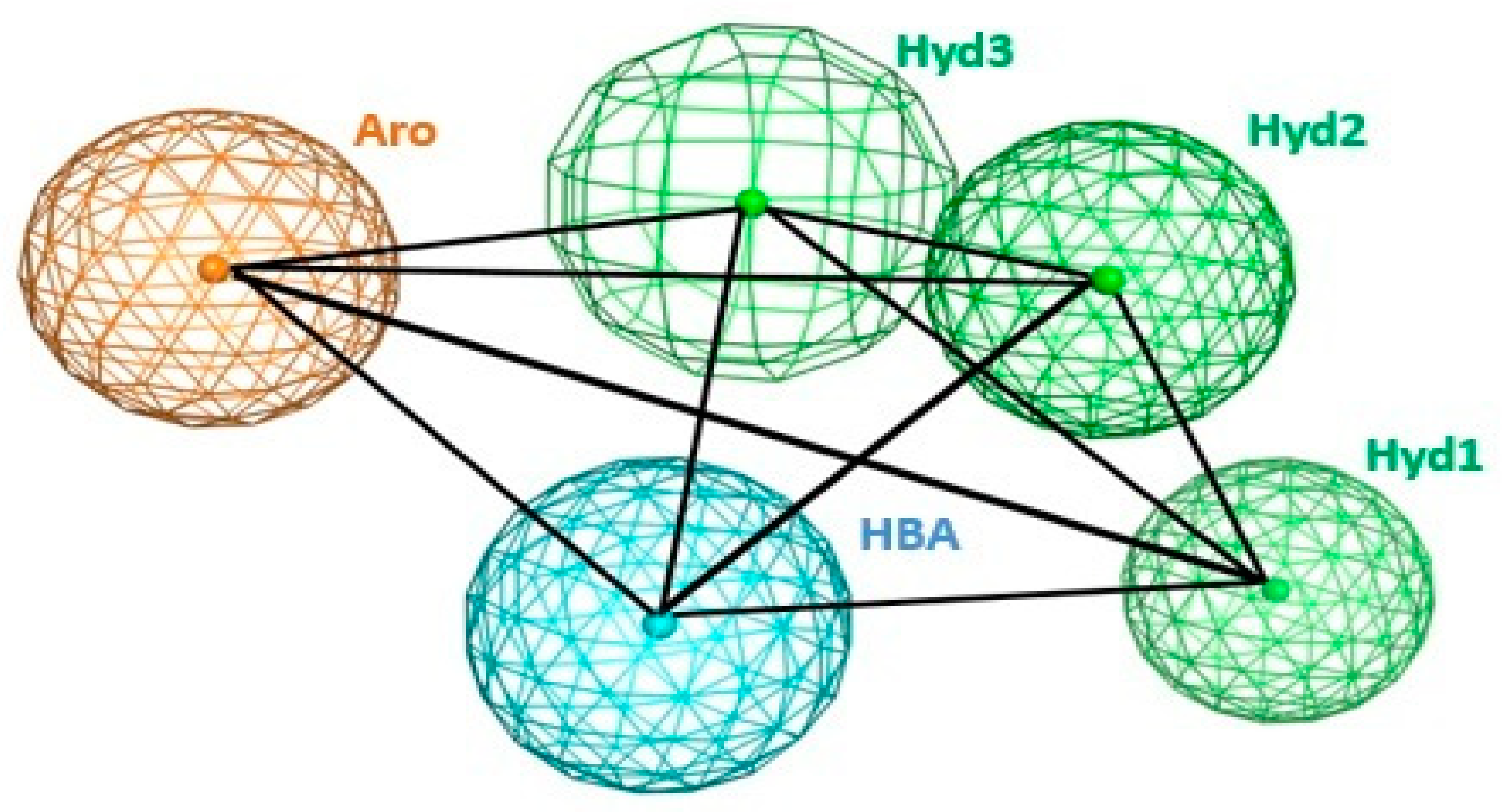

| Hyd1 | Hyd2 | Hyd3 | Aro | HBA | |
|---|---|---|---|---|---|
| Hyd1 | 0 | 6.05 | 5.56 | 4.32 | 7.74 |
| Hyd2 | 6.05 | 0 | 3.42 | 8.56 | 8.27 |
| Hyd3 | 5.56 | 3.42 | 0 | 5.39 | 4.64 |
| Aro | 4.32 | 8.56 | 5.39 | 0 | 6.00 |
| HBA | 7.74 | 8.27 | 4.64 | 6.00 | 0 |
| Molecular Formula | Structure | Characteristics | |
|---|---|---|---|
| A 14994098 | C23H26N4O 5-[2-(benzylamino)ethyl]-1-[4-(1H-pyrazol-1-yl)benzyl]-2-pyrrolidinone | 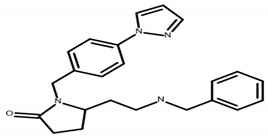 | MW 374.49 CLogP 3.13 LipE 5.69 |
| B 17027223 | C23H31N5O 1-[6-(3-aminocyclobutyl)-2-methylpyrimidin-4-yl]-N-(4-methylbenzyl)piperidine-4-carboxamide dihydrochloride | 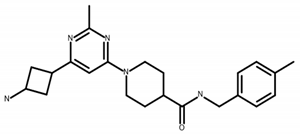 | MW 393.5 CLogP 1.13 |
| C 23677252 | C17H23N5O3S2 N-(5-methyl-1,3,4-thiadiazol-2-yl)-2-D-valyl-1,2,3,4-tetrahydroisoquinoline-7-sulfonamide |  | MW 409.5 CLogP 1.29 |
| D 57422280 | C19H28N6OS N-{5-[2-(dimethylamino)ethyl]-1,3,4-thiadiazol-2-yl}-2-[4-(3-methylphenyl)piperazin-1-yl]acetamide |  | MW 388.38 CLogP 2.35 LipE 5.37 |
| E 66597130 | C20H28N6 4-(3-aminocyclobutyl)-6-[4-(3,4-dimethylphenyl)piperazin-1-yl]pyrimidin-2-amine | 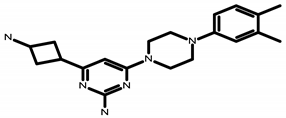 | MW 352.38 CLogP 1.99 LipE 6.01 |
| F 86609095 | C23H31N5 (3-{2-cyclopropyl-6-[4-(2,3-dimethylphenyl)-1-piperazinyl]-4-pyrimidinyl}cyclobutyl)amine dihydrochloride |  | MW 450.41 CLogP 2.53 LipE 5.89 |
| Tariquidar | C38H38N4O6 N-[2-[[4-[2-(6,7-dimethoxy-3,4-dihydro-1H-isoquinolin-2-yl)ethyl]phenyl]carbamoyl]-4,5-dimethoxyphenyl]quinoline-3-carboxamide | 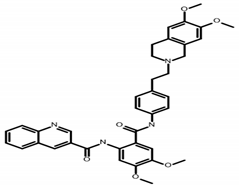 | MW 646.7 CLogP 5.55 LipE 4.39 |
| Tariquidar 0.114 nM CI [0.05, 0.23] | A 1.4 nM CI [0.4, 6.0] | D 23.6 nM CI [11.3, 54.5] | E 14.6 nM CI [5.6, 44.9] | F 4.8 nM CI [1.7, 12.8] | |
|---|---|---|---|---|---|
| Tar | n.s. | n.s. | n.s | n.s | |
| A | p = 0.0011 | n.s | n.s | n.s | |
| D | p =< 0.0001 | p = 0.0005 | n.s | n.s | |
| E | p =< 0.0001 | p = 0.0138 | n.s | n.s | |
| F | p =< 0.0001 | n.s | p = 0.0127 | n.s. |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Cheema, Y.; Kiani, Y.S.; Linton, K.J.; Jabeen, I. Identification and Empiric Evaluation of New Inhibitors of the Multidrug Transporter P-Glycoprotein (ABCB1). Int. J. Mol. Sci. 2023, 24, 5298. https://doi.org/10.3390/ijms24065298
Cheema Y, Kiani YS, Linton KJ, Jabeen I. Identification and Empiric Evaluation of New Inhibitors of the Multidrug Transporter P-Glycoprotein (ABCB1). International Journal of Molecular Sciences. 2023; 24(6):5298. https://doi.org/10.3390/ijms24065298
Chicago/Turabian StyleCheema, Yasmeen, Yusra Sajid Kiani, Kenneth J. Linton, and Ishrat Jabeen. 2023. "Identification and Empiric Evaluation of New Inhibitors of the Multidrug Transporter P-Glycoprotein (ABCB1)" International Journal of Molecular Sciences 24, no. 6: 5298. https://doi.org/10.3390/ijms24065298
APA StyleCheema, Y., Kiani, Y. S., Linton, K. J., & Jabeen, I. (2023). Identification and Empiric Evaluation of New Inhibitors of the Multidrug Transporter P-Glycoprotein (ABCB1). International Journal of Molecular Sciences, 24(6), 5298. https://doi.org/10.3390/ijms24065298









