Combinatorial Network of Transcriptional and miRNA Regulation in Colorectal Cancer
Abstract
1. Introduction
2. Material and Methods
2.1. Network Statistical Analysis of Colorectal Cancer
2.1.1. Degree (k) and Probability of Degree Distribution (P(k))
2.1.2. Clustering Coefficient C(k)
2.1.3. Neighborhood Connectivity
2.1.4. Betweenness Centrality
2.1.5. Closeness Centrality
2.1.6. Eigenvector Centrality
2.2. Tracing of Bottleneck-Hubs
2.3. Detection of Subnetwork and Key Mediator Bottleneck-Hub
2.4. Functional Analysis of Subnetworks
2.5. Pathway Analysis of Bn-Hs
2.6. Construction of a Bn-H Regulatory Network
Coherent and Incoherent Feed-Forward Loops
3. Results
3.1. Hierarchal Scale-Free CRC-PPIN Topology
3.2. Central Bottleneck-Hubs
3.3. Subnetworks and Their Cross-Talk with Bottleneck-Hub
3.4. Gene Ontology (GO) Analysis of Subnetworks
3.5. Highly Enriched Pathway Associated with Bn-Hs
3.6. Combinatorial Regulatory Network of Bottleneck-Hubs
3.7. Coherent and Incoherent Type Feed-forward Loops in the CRC Bn-H Regulatory Network
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
References
- Jemal, A.; Siegel, R.; Xu, J.Q.; Ward, E. Cancer Statistics, 2010. CA Cancer J. Clin. 2010, 60, 277–300. [Google Scholar] [CrossRef] [PubMed]
- Yamagishi, H.; Kuroda, H.; Imai, Y.; Hiraishi, H. Molecular pathogenesis of sporadic colorectal cancers. Chin. J. Cancer 2016, 35, 4. [Google Scholar] [CrossRef] [PubMed]
- Alzahrani, S.M.; Al Doghaither, H.A.; Al-Ghafari, A.B. General insight into cancer: An overview of colorectal cancer (Review). Mol. Clin. Oncol. 2021, 15, 271. [Google Scholar] [CrossRef] [PubMed]
- Pino, M.S.; Chung, D.C. The chromosomal instability pathway in colon cancer. Gastroenterology 2010, 138, 2059–2072. [Google Scholar] [CrossRef] [PubMed]
- Allison, D.B.; Cui, X.; Page, G.; Sabripour, M. Microarray data analysis: From disarray to consolidation and consensus. Nat. Rev. Genet. 2006, 7, 55–65. [Google Scholar] [CrossRef]
- Chen, Y.; Gao, Z.; Wang, B.; Xu, R. Towards precision medicine-based therapies for glioblastoma: Interrogating human disease genomics and mouse phenotypes. BMC Genom. 2016, 17 (Suppl. S7), 516. [Google Scholar] [CrossRef]
- Chen, Y.; Xu, R. Context-sensitive network-based disease genetics prediction and its implications in drug discovery. Bioinformatics 2017, 33, 1031–1039. [Google Scholar] [CrossRef]
- Moreau, Y.; Tranchevent, L.C. Computational tools for prioritizing candidate genes: Boosting disease gene discovery. Nat. Rev. Genet. 2012, 13, 523–536. [Google Scholar] [CrossRef]
- Sonachalam, M.; Shen, J.; Huang, H.; Huang, H.; Wu, X.; Wu, X. Systems biology approach to identify gene network signatures for colorectal cancer. Front. Genet. 2012, 3, 80. [Google Scholar] [CrossRef]
- Khan, M.M.; Serajuddin, M.; Malik, M.Z. Identification of microRNA and gene interactions through bioinformatic integrative analysis for revealing candidate signatures in prostate cancer. Gene Rep. 2022, 27, 101607. [Google Scholar] [CrossRef]
- Barabasi, A.L.; Oltvai, Z.N. Network biology: Understanding the cell’s functional organization. Nat. Rev. Genet. 2004, 5, 101–113. [Google Scholar] [CrossRef] [PubMed]
- Vidal, M.; Cusick, M.E.; Barabasi, A.L. Interactome networks and human disease. Cell 2011, 144, 986–998. [Google Scholar] [CrossRef] [PubMed]
- Hahn, M.W.; Kern, A.D. Comparative genomics of centrality and essentiality in three eukaryotic protein-interaction networks. Mol. Biol. Evol. 2005, 22, 803–806. [Google Scholar] [CrossRef] [PubMed]
- Ravasz, E.; Somera, A.L.; Mongru, D.A.; Oltvai, Z.N.; Barabási, A.L. Hierarchical organization of modularity in metabolic networks. Science 2002, 297, 1551–1555. [Google Scholar] [CrossRef]
- Ravasz, E.; Barabasi, A.L. Hierarchical organization in complex networks. Phys. Rev. E Stat. Nonlin. Soft Matter. Phys. 2003, 67, 026112. [Google Scholar] [CrossRef]
- Valizadeh, R.; Bahadorimonfared, A.; Rezaei-Tavirani, M.; Norouzinia, M.; Ehsani Ardakani, M.I. Evaluation of involved proteins in colon adenocarcinoma: An interactome analysis. Gastroenterol. Hepatol. Bed Bench 2017, 10, S129–S138. [Google Scholar]
- Mansouri, V.; Rezaei Tavirani, M.; Rezaei Tavirani, S. Gene screening of colorectal cancers via network analysis. Gastroenterol. Hepatol. Bed Bench 2019, 12, 149–154. [Google Scholar]
- Arjmand, B.; Khodadoost, M.; Jahani Sherafat, S.; Rezaei Tavirani, M.; Ahmadi, N.; Hamzeloo Moghadam, M.; Rezaei Tavirani, S.; Khanabadi, B.; Iranshahi, M. Assessment of colon cancer molecular mechanism: A system biology approach. Gastroenterol. Hepatol. Bed Bench 2021, 14, S51–S57. [Google Scholar]
- Kanehisa, M.; Goto, S. KEGG: Kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 2000, 28, 29–34. [Google Scholar] [CrossRef]
- Von Mering, C.; Huynen, M.; Jaeggi, D.; Schmidt, S.; Bork, P.; Snel, B. STRING: A database of predicted functional associations between proteins. Nucleic Acids Res. 2003, 31, 258–261. [Google Scholar] [CrossRef]
- Assenov, Y.; Ramirez, F.; Schelhorn, S.E.; Lengauer, T.; Albrecht, M. Computing topological parameters of biological networks. Bioinformatics 2008, 24, 282–284. [Google Scholar] [CrossRef] [PubMed]
- Traag, V.A.; Krings, G.; Van Dooren, P. Significant scales in community structure. Sci. Rep. 2013, 3, 2930. [Google Scholar] [CrossRef] [PubMed]
- Borgatti, S.P.; Everett, M.G. A Graph-theoretic perspective on centrality. Soc. Netw. 2006, 28, 466–484. [Google Scholar] [CrossRef]
- Barabasi, A.L.; Gulbahce, N.; Loscalzo, J. Network medicine: A network-based approach to human disease. Nat. Rev. Genet. 2011, 12, 56–68. [Google Scholar] [CrossRef]
- Lim, J.; Hao, T.; Shaw, C.; Patel, A.J.; Szabo, G.; Rual, J.F.; Fisk, C.J.; Li, N.; Smolyar, A.; Hill, D.E.; et al. A protein-protein interaction network for human inherited ataxias and disorders of Purkinje cell degeneration. Cell 2006, 125, 801–814. [Google Scholar] [CrossRef]
- Stelzl, U.; Worm, U.; Lalowski, M.; Haenig, C.; Brembeck, F.H.; Goehler, H.; Stroedicke, M.; Zenkner, M.; Schoenherr, A.; Koeppen, S.; et al. A human protein-protein interaction network: A resource for annotating the proteome. Cell 2005, 122, 957–968. [Google Scholar] [CrossRef]
- Verma, R.N.; Malik, M.Z.; Subbarao, N.; Singh, G.P.; Sinha, D.N. Entamoeba histolytica HM-1: IMSS gene expression profiling identifies key hub genes, potential biomarkers, and pathways in Amoebiasis infection: A systematic network meta-analysis. BioSci. Rep. 2022, 42, BSR20220191. [Google Scholar] [CrossRef]
- Mangangcha, I.R.; Malik, M.Z.; Kucuk, O.; Ali, S.; Singh, R.K.B. Kinless hubs are potential target genes in prostate cancer network. Genomics 2020, 112, 5227–5239. [Google Scholar] [CrossRef]
- Bader, G.D.; Hogue, C.W. An automated method for finding molecular complexes in large protein interaction networks. BMC Bioinform. 2003, 4, 2. [Google Scholar] [CrossRef]
- Nafis, S.; Kalaiarasan, P.; Brojen Singh, R.K.; Husain, M.; Bamezai, R.N. Apoptosis regulatory protein-protein interaction demonstrates hierarchical scale-free fractal network. Brief Bioinform. 2015, 16, 675–699. [Google Scholar] [CrossRef]
- Reimand, J.; Arak, T.; Adler, P.; Kolberg, L.; Reisberg, S.; Peterson, H.; Vilo, J. g:Profiler-a web server for functional interpretation of gene lists (2016 update). Nucleic Acids Res. 2016, 44, W83–W89. [Google Scholar] [CrossRef] [PubMed]
- Xie, Z.; Bailey, A.; Kuleshov, M.V.; Clarke, D.J.B.; Evangelista, J.E.; Jenkins, S.L.; Lachmann, A.; Wojciechowicz, M.L.; Kropiwnicki, E.; Jagodnik, K.M.; et al. Gene Set Knowledge Discovery with Enrichr. Curr. Protoc. 2021, 1, e90. [Google Scholar] [CrossRef] [PubMed]
- Agarwal, V.; Bell, G.W.; Nam, J.W.; Bartel, D.P. Predicting effective microRNA target sites in mammalian mRNAs. Elife 2015, 4, e05005. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Wang, X. miRDB: An online database for prediction of functional microRNA targets. Nucleic Acids Res. 2020, 48, D127–D131. [Google Scholar] [CrossRef] [PubMed]
- Huang, H.Y.; Lin, Y.C.; Li, J.; Huang, K.Y.; Shrestha, S.; Hong, H.C.; Tang, Y.; Chen, Y.G.; Jin, C.N.; Yu, Y.; et al. miRTarBase 2020: Updates to the experimentally validated microRNA-target interaction database. Nucleic Acids Res. 2020, 48, D148–D154. [Google Scholar] [CrossRef]
- Han, H.; Shim, H.; Shin, D.; Shim, J.E.; Ko, Y.; Shin, J.; Kim, H.; Cho, A.; Kim, E.; Lee, T.; et al. TRRUST: A reference database of human transcriptional regulatory interactions. Sci. Rep. 2015, 5, 11432. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Lu, M.; Qiu, C.; Cui, Q. TransmiR: A transcription factor-microRNA regulation database. Nucleic Acids Res. 2010, 38, D119–D122. [Google Scholar] [CrossRef]
- Excoffier, L.; Gouy, A.; Daub, J.T.; Shannon, P.; Markiel, A.; Ozier, O.; Baliga, N.S.; Wang, J.T.; Ramage, D.; Amin, N.; et al. Cytoscape: A Software Environment for Integrated Models of Biomolecular Interaction Networks. Nucleic Acids Res. 2017, 13, 2498–2504. [Google Scholar]
- Le, D.H.; Kwon, Y.K. A coherent feedforward loop design principle to sustain robustness of biological networks. Bioinformatics 2013, 29, 630–637. [Google Scholar] [CrossRef]
- Mangan, S.; Alon, U. Structure and function of the feed-forward loop network motif. Proc. Natl. Acad. Sci. USA 2003, 100, 11980–11985. [Google Scholar] [CrossRef]
- Su, L.N.; Song, X.Q.; Xue, Z.X.; Zheng, C.Q.; Yin, H.F.; Wei, H.P. Network analysis of microRNAs, transcription factors, and target genes involved in axon regeneration. J. Zhejiang Univ. Sci. B 2018, 19, 293–304. [Google Scholar] [CrossRef] [PubMed]
- Kalir, S.; Mangan, S.; Alon, U. A coherent feed-forward loop with a SUM input function prolongs flagella expression in Escherichia coli. Mol. Syst. Biol. 2005, 1, 2005-0006. [Google Scholar] [CrossRef] [PubMed]
- Dong, J.; Horvath, S. Understanding network concepts in modules. BMC Syst. Biol. 2007, 1, 24. [Google Scholar] [CrossRef] [PubMed]
- Koveitypour, Z.; Panahi, F.; Vakilian, M.; Peymani, M.; Seyed Forootan, F.; Nasr Esfahani, M.H.; Ghaedi, K. Signaling pathways involved in colorectal cancer progression. Cell Biosci. 2019, 9, 97. [Google Scholar] [CrossRef]
- Bhullar, K.S.; Lagarón, N.O.; McGowan, E.M.; Parmar, I.; Jha, A.; Hubbard, B.P.; Rupasinghe, H.P.V. Kinase-targeted cancer therapies: Progress, challenges and future directions. Mol. Cancer 2018, 17, 48. [Google Scholar] [CrossRef]
- Morris, V.K.; Kennedy, E.B.; Baxter, N.N.; III, A.B.B.; Cercek, A.; Cho, M.; Ciombor, K.K.; Cremolini, C.; Davis, A.; Deming, D.A.; et al. Treatment of Metastatic Colorectal Cancer: ASCO Guideline. J. Clin. Oncol. 2023, 41, 678–700. [Google Scholar] [CrossRef]
- Pinero, J.; Ramirez-Anguita, J.M.; Sauch-Pitarch, J.; Ronzano, F.; Centeno, E.; Sanz, F.; Furlong, L.I. The DisGeNET knowledge platform for disease genomics: 2019 update. Nucleic Acids Res. 2020, 48, D845–D855. [Google Scholar] [CrossRef]
- Li, V.D.; Li, K.H.; Li, J.T. TP53 mutations as potential prognostic markers for specific cancers: Analysis of data from The Cancer Genome Atlas and the International Agency for Research on Cancer TP53 Database. J. Cancer Res. Clin. Oncol. 2019, 145, 625–636. [Google Scholar] [CrossRef]
- Chen, S.L.; Liu, L.L.; Wang, C.H.; Lu, S.X.; Yang, X.; He, Y.F.; Zhang, C.Z.; Yun, J.P. Loss of RDM1 enhances hepatocellular carcinoma progression via p53 and Ras/Raf/ERK pathways. Mol. Oncol. 2020, 14, 373–386. [Google Scholar] [CrossRef]
- Liebl, M.C.; Hofmann, T.G. The Role of p53 Signaling in Colorectal Cancer. Cancers 2021, 13, 2125. [Google Scholar] [CrossRef]
- Baugh, E.H.; Ke, H.; Levine, A.J.; Bonneau, R.A.; Chan, C.S. Why are there hotspot mutations in the TP53 gene in human cancers? Cell Death Differ. 2018, 25, 154–160. [Google Scholar] [CrossRef] [PubMed]
- Roszkowska, K.A.; Piecuch, A.; Sady, M.; Gajewski, Z.; Flis, S. Gain of Function (GOF) Mutant p53 in Cancer-Current Therapeutic Approaches. Int. J. Mol. Sci. 2022, 23, 13287. [Google Scholar] [CrossRef] [PubMed]
- Pham, T.H.; Park, H.M.; Kim, J.; Hong, J.T.; Yoon, D.Y. STAT3 and p53: Dual Target for Cancer Therapy. Biomedicines 2020, 8, 637. [Google Scholar] [CrossRef] [PubMed]
- Levine, A.J.; Oren, M. The first 30 years of p53: Growing ever more complex. Nat. Rev. Cancer 2009, 9, 749–758. [Google Scholar] [CrossRef] [PubMed]
- Sakai, H.; Tsukamoto, T.; Yamamoto, M.; Shirai, N.; Iidaka, T.; Hirata, A.; Yanai, T.; Masegi, T.; Donehower, L.A.; Tatematsu, M. High susceptibility of nullizygous p53 knockout mice to colorectal tumor induction by 1,2-dimethylhydrazine. J. Cancer Res. Clin. Oncol. 2003, 129, 335–340. [Google Scholar] [CrossRef] [PubMed]
- Hu, Y.; Le Leu, R.K.; Belobrajdic, D.; Young, G.P. The potential of sphingomyelin as a chemopreventive agent in AOM-induced colon cancer model: Wild-type and p53+/– mice. Mol. Nutr. Food Res. 2008, 52, 558–566. [Google Scholar] [CrossRef] [PubMed]
- Mundhenk, J.; Hennenlotter, J.; Zug, L.; Alloussi, S.H.; Todenhoefer, T.; Gakis, G.; Aufderklamm, S.; Scharpf, M.; Kuehs, U.; Stenzl, A.; et al. Evidence for PTEN-independent Akt activation and Akt-independent p27(Kip1) expression in advanced bladder cancer. Oncol. Lett. 2011, 2, 1089–1093. [Google Scholar] [CrossRef]
- Zhang, X.; Chen, X.; Zhai, Y.; Cui, Y.; Cao, P.; Zhang, H.; Wu, Z.; Li, P.; Yu, L.; Xia, X.; et al. Combined effects of genetic variants of the PTEN, AKT1, MDM2 and p53 genes on the risk of nasopharyngeal carcinoma. PLoS ONE 2014, 9, e92135. [Google Scholar] [CrossRef]
- Ying, J.; Xu, Q.; Liu, B.; Zhang, G.; Chen, L.; Pan, H. The expression of the PI3K/AKT/mTOR pathway in gastric cancer and its role in gastric cancer prognosis. Onco Targets Ther. 2015, 8, 2427–2433. [Google Scholar] [CrossRef]
- Wang, M.Y.; He, J.; Zhu, M.L.; Teng, X.Y.; Li, Q.X.; Sun, M.H.; Wang, X.F.; Yang, Y.J.; Wang, J.C.; Jin, L.; et al. A Functional Polymorphism (rs2494752) in the AKT1 Promoter Region and Gastric Adenocarcinoma Risk in an Eastern Chinese Population. Sci. Rep. 2016, 6, 20008. [Google Scholar] [CrossRef]
- Dang, K.; Davidson, N.E.; Del Vecchio Fitz, C.; Dogan, S.; DuBois, R.N.; Ducar, M.D.; Futreal, P.A.; Gao, J.; Garcia, F.; Gardos, S.; et al. AACR Project GENIE: Powering Precision Medicine through an International Consortium. Cancer Discov. 2017, 7, 818–831. [Google Scholar]
- Arnold, A.A.-O.; Tronser, M.; Sers, C.; Ahadova, A.; Endris, V.; Mamlouk, S.; Horst, D.; Möbs, M.; Bischoff, P.; Kloor, M.; et al. The majority of β-catenin mutations in colorectal cancer is homozygous. BMC Cancer 2020, 20, 1–10. [Google Scholar]
- Korinek, V.; Barker, N.; Morin, P.J.; van Wichen, D.; de Weger, R.; Kinzler, K.W.; Vogelstein, B.; Clevers, H. Constitutive transcriptional activation by a beta-catenin-Tcf complex in APC-/- colon carcinoma. Science 1997, 275, 1784–1787. [Google Scholar] [CrossRef] [PubMed]
- Morin, P.J.; Sparks, A.B.; Korinek, V.; Barker, N.; Clevers, H.; Vogelstein, B.; Kinzler, K.W. Activation of beta-catenin-Tcf signaling in colon cancer by mutations in beta-catenin or APC. Science 1997, 275, 1787–1790. [Google Scholar] [CrossRef]
- Polakis, P. Wnt signaling in cancer. Cold Spring Harb. Perspect. Biol. 2012, 4, a008052. [Google Scholar] [CrossRef]
- Murugan, A.K.; Grieco, M.; Tsuchida, N. RAS mutations in human cancers: Roles in precision medicine. Semin. Cancer Biol. 2019, 59, 23–35. [Google Scholar] [CrossRef] [PubMed]
- Zenonos, K.; Kyprianou, K. RAS signaling pathways, mutations and their role in colorectal cancer. World J. Gastrointest. Oncol. 2013, 5, 97–101. [Google Scholar] [CrossRef]
- Luo, F.; Brooks, D.G.; Ye, H.; Hamoudi, R.; Poulogiannis, G.; Patek, C.E.; Winton, D.J.; Arends, M.J. Conditional expression of mutated K-ras accelerates intestinal tumorigenesis in Msh2-deficient mice. Oncogene 2007, 26, 4415–4427. [Google Scholar] [CrossRef]
- Haigis, K.M.; Kendall, K.R.; Wang, Y.; Cheung, A.; Haigis, M.C.; Glickman, J.N.; Niwa-Kawakita, M.; Sweet-Cordero, A.; Sebolt-Leopold, J.; Shannon, K.M.; et al. Differential effects of oncogenic K-Ras and N-Ras on proliferation, differentiation and tumor progression in the colon. Nat. Genet. 2008, 40, 600–608. [Google Scholar] [CrossRef]
- Wang, H.; Birkenbach, M.; Hart, J. Expression of Jun family members in human colorectal adenocarcinoma. Carcinogenesis 2000, 21, 1313–1317. [Google Scholar] [CrossRef]
- Hughes, M.; Sehgal, A.; Hadman, M.; Hadman, M.; Bos, T.; Bos, T. Heterodimerization with c-Fos is not required for cell transformation of chicken embryo fibroblasts by Jun. Cell Growth Differ. 1992, 3, 889–897. [Google Scholar]
- Castellazzi, M.; Spyrou, G.; La Vista, N.; Dangy, J.P.; Piu, F.; Yaniv, M.; Brun, G. Overexpression of c-jun, junB, or junD affects cell growth differently. Proc. Natl. Acad. Sci. USA 1991, 88, 8890–8894. [Google Scholar] [CrossRef] [PubMed]
- Jin, H.; Tan, X.; Liu, X.; Ding, Y. Downregulation of AP-1 gene expression is an initial event in the oridonin-mediated inhibition of colorectal cancer: Studies in vitro and in vivo. J. Gastroenterol. Hepatol. 2011, 26, 706–715. [Google Scholar] [CrossRef] [PubMed]
- Wang, T.; Rao, D.; Yu, C.; Sheng, J.; Luo, Y.; Xia, L.; Huang, W. RHO GTPase family in hepatocellular carcinoma. Exp. Hematol. Oncol. 2022, 11, 91. [Google Scholar] [CrossRef] [PubMed]
- Takami, Y.; Higashi, M.; Kumagai, S.; Kuo, P.C.; Kawana, H.; Koda, K.; Miyazaki, M.; Harigaya, K. The activity of RhoA is correlated with lymph node metastasis in human colorectal cancer. Dig. Dis. Sci. 2008, 53, 467–473. [Google Scholar] [CrossRef] [PubMed]
- Wee, P.; Wang, Z. Epidermal Growth Factor Receptor Cell Proliferation Signaling Pathways. Cancers 2017, 9, 52. [Google Scholar] [CrossRef] [PubMed]
- Gazdar, A.F. Activating and resistance mutations of EGFR in non-small-cell lung cancer: Role in clinical response to EGFR tyrosine kinase inhibitors. Oncogene 2009, 28 (Suppl. S1), S24–S31. [Google Scholar] [CrossRef]
- Sigismund, S.; Avanzato, D.; Lanzetti, L. Emerging functions of the EGFR in cancer. Mol. Oncol. 2018, 12, 3–20. [Google Scholar] [CrossRef]
- Souza, A.; Costa-Casagrande, T.A. Animal Models for Colorectal Cancer. Arq. Bras. Cir. Dig. 2018, 31, e1369. [Google Scholar] [CrossRef]
- Bai, J.; Hu, S. Transcriptome network analysis reveals potential candidate genes for squamous lung cancer. Int. J. Mol. Med. 2012, 29, 95–101. [Google Scholar] [CrossRef]
- Rowland, B.; Bernards, R.; Peeper, D. The KLF4 tumour suppressor is a transcriptional repressor of p53 that acts as a context-dependent oncogene. Nat. Cell Biol. 2005, 7, 1074–1082. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Zhao, X.; Shi, D.; Chen, P.; Yu, Y.; Yang, L.; Xie, L. Overexpression of SIRT1 promotes high glucose-attenuated corneal epithelial wound healing via p53 regulation of the IGFBP3/IGF-1R/AKT pathway. Investig. Ophthalmol. Vis. Sci. 2013, 54, 3806–3814. [Google Scholar] [CrossRef] [PubMed]
- Li, P.; Nicosia, S.V.; Bai, W. Antagonism between PTEN/MMAC1/TEP-1 and androgen receptor in growth and apoptosis of prostatic cancer cells. J. Biol. Chem. 2001, 276, 20444–20450. [Google Scholar] [CrossRef] [PubMed]
- Wang, A.H.; Bertos, N.R.; Vezmar, M.; Pelletier, N.; Crosato, M.; Heng, H.H.; Th’ng, J.; Han, J.; Yang, X.-J. HDAC4, a Human Histone Deacetylase Related to Yeast HDA1, Is a Transcriptional Corepressor. Mol. Cell. Biol. 1999, 19, 7816–7827. [Google Scholar] [CrossRef]
- McGregor, N.; Patel, L.; Craig, M.; Weidner, S.; Wang, S.; Pienta, K.J. AT-101 (R-(-)-gossypol acetic acid) enhances the effectiveness of androgen deprivation therapy in the VCaP prostate cancer model. J. Cell Biochem. 2010, 110, 1187–1194. [Google Scholar] [CrossRef]
- Chen, M.; Cai, H.; Yang, J.-L.; Lu, C.-L.; Liu, T.; Yang, W.; Guo, J.; Hu, X.-Q.; Fan, C.-H.; Hu, Z.-Y.; et al. Effect of heat stress on expression of junction-associated molecules and upstream factors androgen receptor and Wilms’ tumor 1 in monkey sertoli cells. Endocrinology 2008, 149, 4871–4882. [Google Scholar] [CrossRef]
- Yuan, H.; Young, C.Y.; Tian, Y.; Liu, Z.; Zhang, M.; Lou, H. Suppression of the androgen receptor function by quercetin through protein-protein interactions of Sp1, c-Jun, and the androgen receptor in human prostate cancer cells. Mol. Cell Biochem. 2010, 339, 253–262. [Google Scholar] [CrossRef]
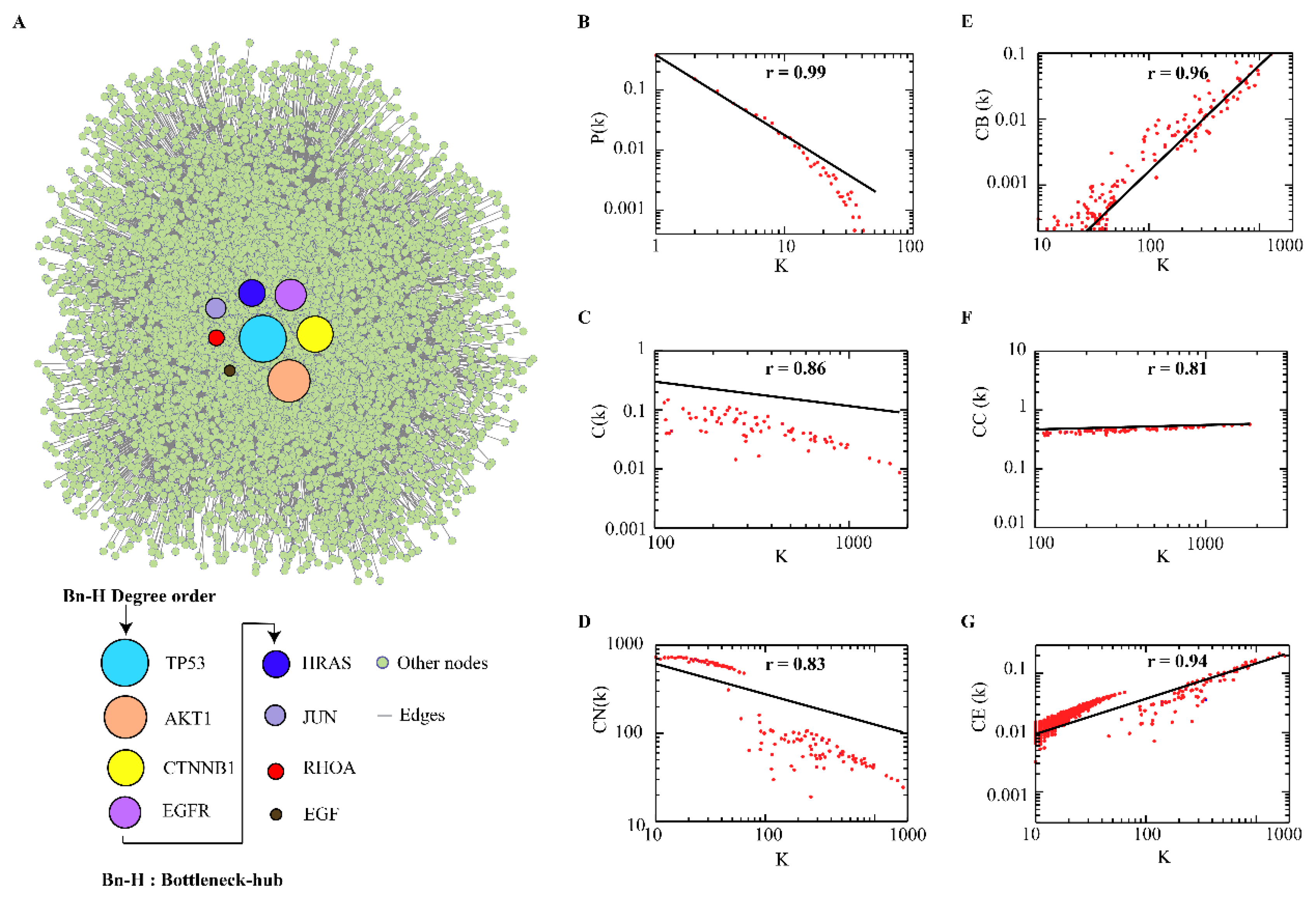
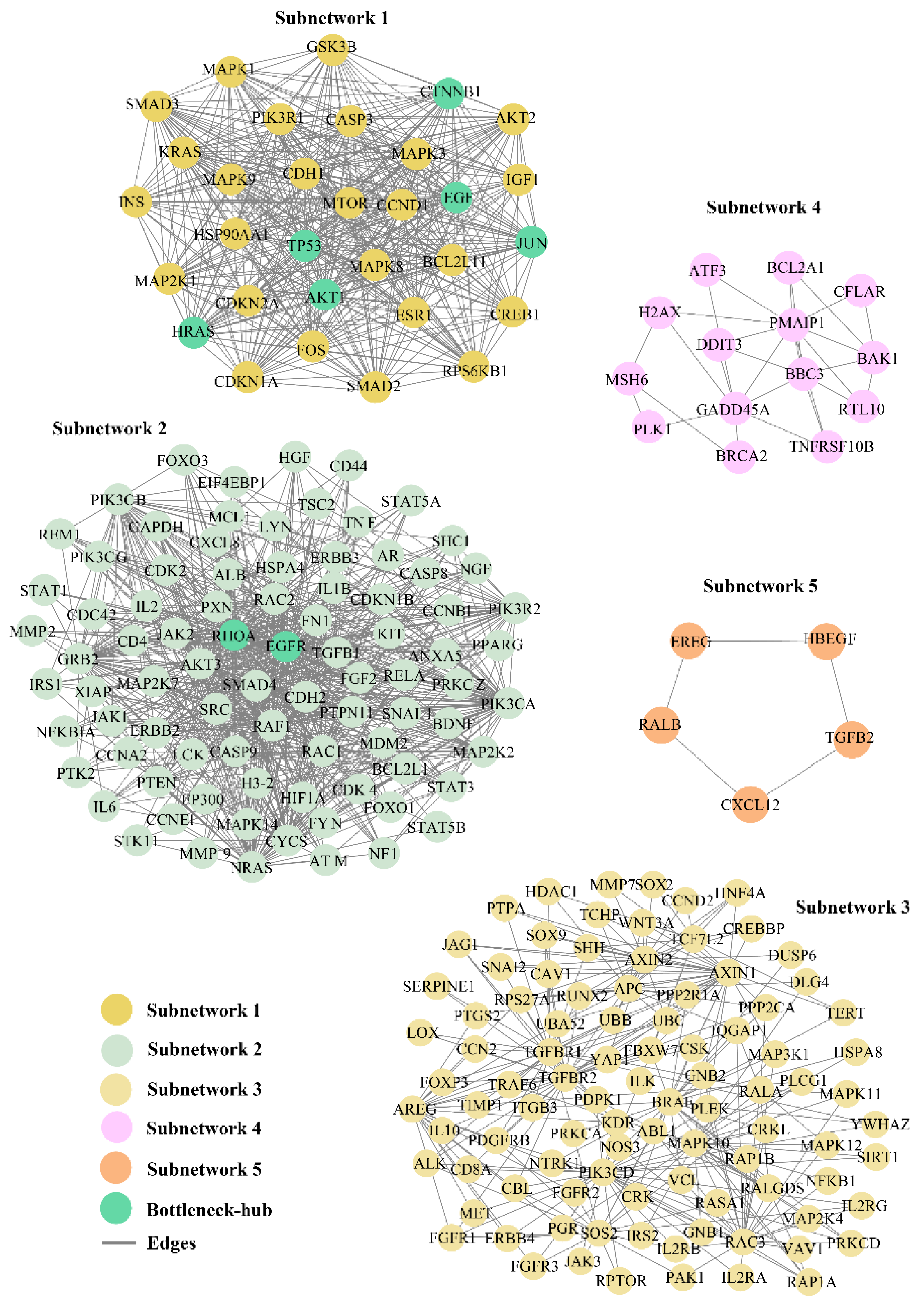

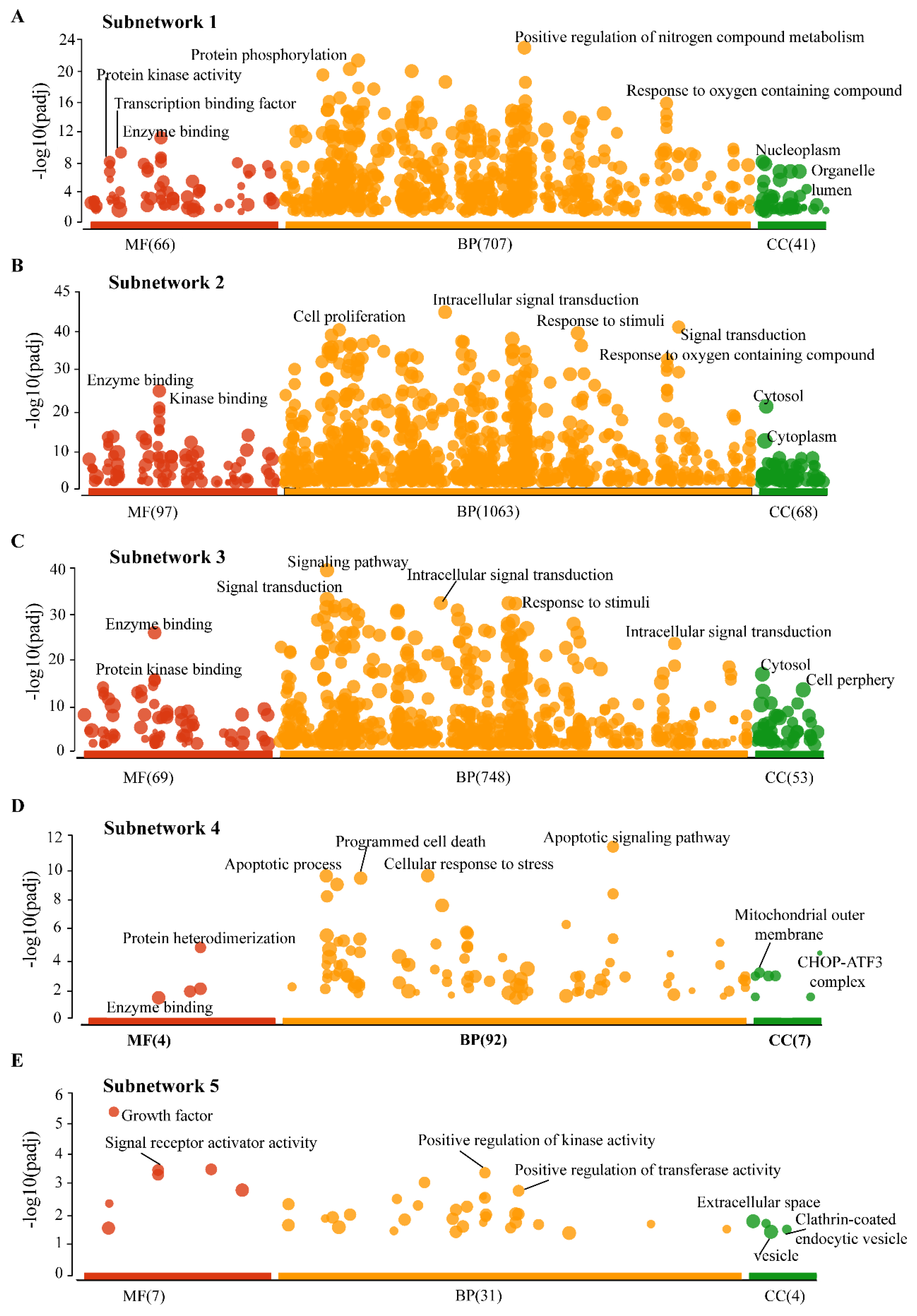
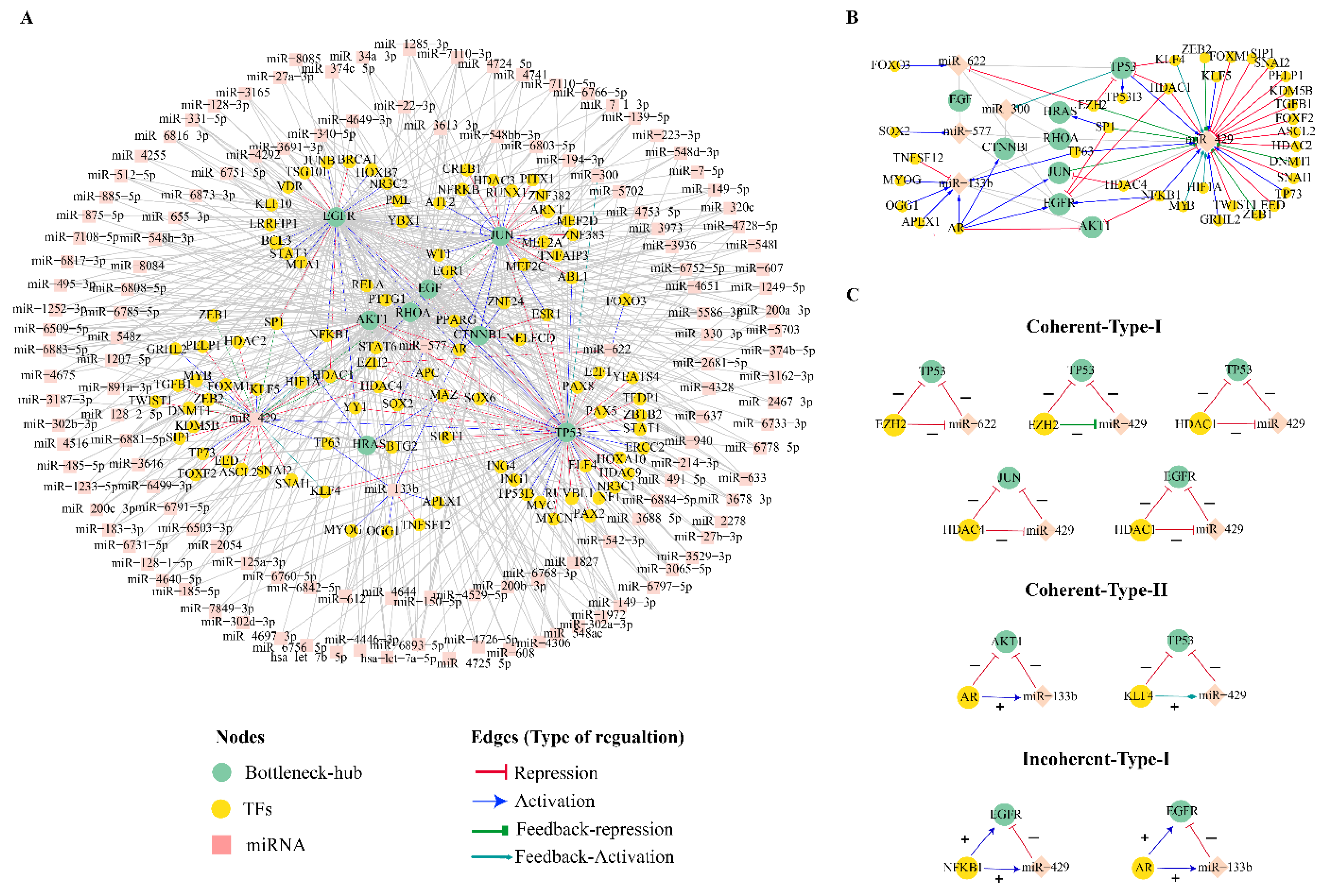
| S.NO | Name | Degree (K) | Name | Betweeness (CB) |
|---|---|---|---|---|
| 1 | TP53 * | 1817 | TP53 | 0.198765 |
| 2 | AKT1 * | 1623 | CTNNB1 | 0.11618 |
| 3 | CTNNB1 * | 1426 | AKT1 | 0.11499 |
| 4 | EGFR * | 1276 | EGFR | 0.096395 |
| 5 | HRAS * | 980 | CYCS | 0.072343 |
| 6 | JUN * | 966 | RHOA | 0.063284 |
| 7 | MAPK3 | 908 | JUN | 0.054568 |
| 8 | RHOA * | 838 | HRAS | 0.0481 |
| 9 | EGF * | 814 | EGF | 0.047273 |
| 10 | KRAS | 801 | FOS | 0.041709 |
| Name of Bn-H | SN-1 | SN-2 | SN-3 | SN-4 | SN-5 | Total |
|---|---|---|---|---|---|---|
| HRAS* | 30 | 85 | 92 | 5 | 4 | 216 |
| TP53 | 30 | 83 | 72 | 14 | 3 | 202 |
| EGFR | 29 | 80 | 77 | 7 | 5 | 198 |
| JUN | 30 | 83 | 70 | 10 | 3 | 196 |
| AKT1 | 27 | 78 | 70 | 12 | 5 | 192 |
| CTNNB1 | 27 | 77 | 73 | 6 | 3 | 186 |
| EGF | 29 | 75 | 59 | 1 | 4 | 168 |
| RHOA | 30 | 68 | 63 | 1 | 5 | 167 |
| S.no | Term | p-Value | Adjusted p-Value | Odds Ratio | Combined Score | Genes |
|---|---|---|---|---|---|---|
| 1 | Signaling by ERBB2 R-HSA-1227986 | 3.98 × 1012 | 9.44 × 1010 | 755.6061 | 19,833.86 | EGF, AKT1, HRAS, EGFR, RHOA |
| 2 | Signaling by Non-Receptor Tyrosine Kinases R-HSA-9006927 | 5.43 × 1012 | 9.44 × 1010 | 707.2695 | 18,346.34 | EGF, AKT1, HRAS, EGFR, RHOA |
| 3 | ESR-mediated signaling R-HSA-8939211 | 3.81 × 109 | 4.42 × 107 | 180.4098 | 3497.451 | JUN, EGF, AKT1, HRAS, EGFR |
| 4 | Signaling by NOTCH R-HSA-157118 | 5.60 × 109 | 4.87 × 107 | 166.6162 | 3165.742 | JUN, EGF, AKT1, TP53, EGFR |
| 5 | GRB2 events in EGFR signaling R-HSA-179812 | 9.22 × 1012 | 5.62 × 107 | 1332.2 | 24,647.6 | EGF, HRAS, EGFR |
| 6 | Extra-nuclear estrogen signaling R-HSA-9009391 | 1.13 × 108 | 5.62 × 107 | 288.7391 | 5283.369 | EGF, AKT1, HRAS, EGFR |
| 7 | SHC1 events in EGFR signaling R-HSA-180336 | 1.20 × 108 | 5.62 × 107 | 1198.92 | 21,867.39 | EGF, HRAS, EGFR |
| 8 | Constitutive signaling by EGFRvIII R-HSA-5637810 | 1.53 × 108 | 5.62 × 107 | 1089.873 | 19,615.82 | EGF, HRAS, EGFR |
| 9 | GRB2 events in ERBB2 signaling R-HSA-1963640 | 1.91 × 108 | 5.62 × 107 | 999 | 17,757.53 | EGF, HRAS, EGFR |
| 10 | Signaling by ERBB2 ECD mutants R-HSA-9665348 | 1.91 × 108 | 5.62 × 107 | 999 | 17,757.53 | EGF, HRAS, EGFR |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kumar, R.; Mahmoud, M.M.; Tashkandi, H.M.; Haque, S.; Harakeh, S.; Ponnusamy, K.; Haider, S. Combinatorial Network of Transcriptional and miRNA Regulation in Colorectal Cancer. Int. J. Mol. Sci. 2023, 24, 5356. https://doi.org/10.3390/ijms24065356
Kumar R, Mahmoud MM, Tashkandi HM, Haque S, Harakeh S, Ponnusamy K, Haider S. Combinatorial Network of Transcriptional and miRNA Regulation in Colorectal Cancer. International Journal of Molecular Sciences. 2023; 24(6):5356. https://doi.org/10.3390/ijms24065356
Chicago/Turabian StyleKumar, Rupesh, Maged Mostafa Mahmoud, Hanaa M. Tashkandi, Shafiul Haque, Steve Harakeh, Kalaiarasan Ponnusamy, and Shazia Haider. 2023. "Combinatorial Network of Transcriptional and miRNA Regulation in Colorectal Cancer" International Journal of Molecular Sciences 24, no. 6: 5356. https://doi.org/10.3390/ijms24065356
APA StyleKumar, R., Mahmoud, M. M., Tashkandi, H. M., Haque, S., Harakeh, S., Ponnusamy, K., & Haider, S. (2023). Combinatorial Network of Transcriptional and miRNA Regulation in Colorectal Cancer. International Journal of Molecular Sciences, 24(6), 5356. https://doi.org/10.3390/ijms24065356






