Heterologous Expression and Bioactivity Determination of Monochamus alternatus Antibacterial Peptide Gene in Komagataella phaffii (Pichia pastoris)
Abstract
:1. Introduction
2. Results
2.1. Phylogenetic Analysis of M. alternatus AMP Genes
2.2. Bioassays of AMPs
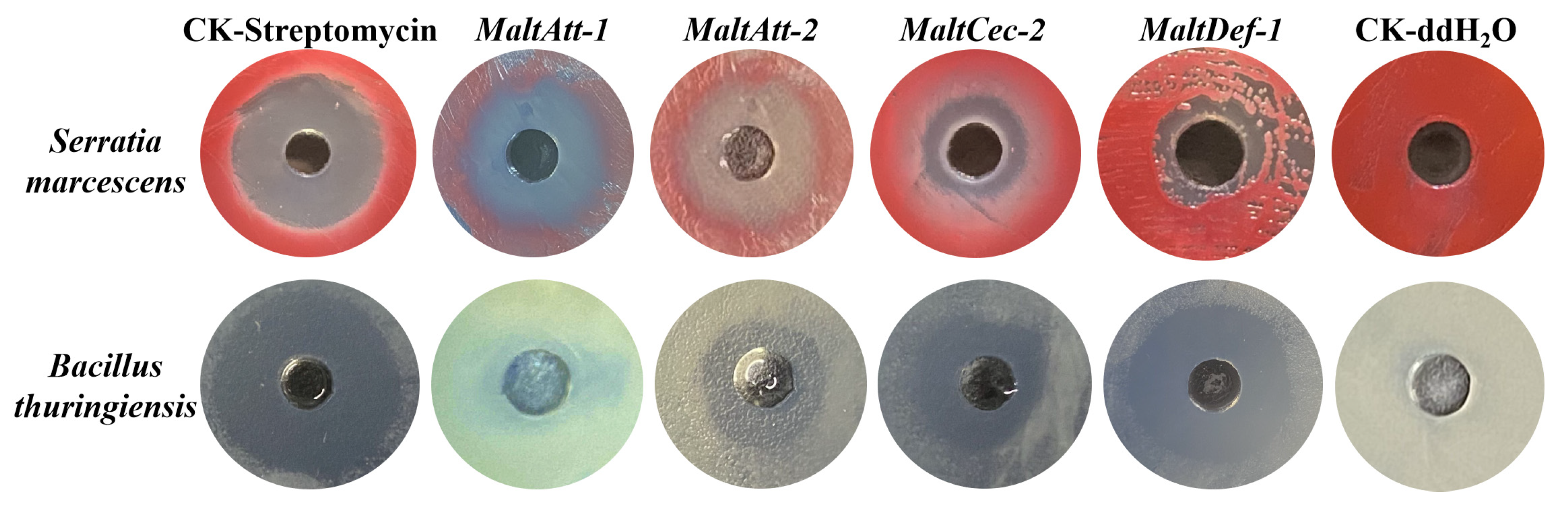
2.2.1. Antimicrobial Activity Detection of AMPs
2.2.2. Hemolytic Activity Detection of AMPs
2.2.3. Bioassays with B. xylophilus
2.3. Effects of M. alternatus AMPs on Morphological Structure of B. xylophilus
3. Discussion
4. Materials and Methods
4.1. M. alternatus Larvae Rearing
4.2. Phylogenetic Analysis of M. alternatus AMP Genes
4.3. Construction of M. alternatus AMP Genes Eukaryotic Expression Vector
4.4. Induction Expression and Purification of AMPs
4.5. Assays of AMPs
4.5.1. Antimicrobial Activity Detection of AMPs
4.5.2. Hemolytic Activity Detection of AMPs
4.5.3. Bioassays with B. xylophilus
4.6. Observation of B. xylophilus by Scanning Electron Microscope (SEM)
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Acknowledgments
Conflicts of Interest
References
- Hillyer, J.F. Insect immunology and hematopoiesis. Dev. Comp. Immunol. 2016, 58, 102–118. [Google Scholar] [CrossRef] [Green Version]
- Mylonakis, E.; Podsiadlowski, L.; Muhammed, M.; Vilcinskas, A. Diversity, evolution and medical applications of insect antimicrobial peptides. Philos. T. R. Soc. B 2016, 371, 20150290. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ali Mohammadie Kojour, M.; Han, Y.S.; Jo, Y.H. An overview of insect innate immunity. Entomol. Res. 2020, 50, 282–291. [Google Scholar] [CrossRef]
- Marieshwari, B.N.; Bhuvaragavan, S.; Sruthi, K.; Mullainadhan, P.; Janarthanan, S. Insect phenoloxidase and its diverse roles: Melanogenesis and beyond. J. Comp. Physiol. B 2022, 193, 1–23. [Google Scholar] [CrossRef] [PubMed]
- Choi, B.; Park, W.R.; Kim, Y.J.; Mun, S.; Park, S.J.; Jeong, J.H.; Choi, H.S.; Kim, D.K. Nuclear receptor estrogen-related receptor modulates antimicrobial peptide expression for host innate immunity in Tribolium castaneum. Insect. Biochem. Molec. 2022, 148, 103816. [Google Scholar] [CrossRef]
- Li, S.; Mandal, S.D.; Xu, X.; Jin, F.L. The tripartite interaction of host immunity-Bacillus thuringiensis infection-gut microbiota. Toxins 2020, 12, 514. [Google Scholar] [CrossRef]
- Manniello, M.D.; Moretta, A.; Salvia, R.; Scieuzo, C.; Lucchetti, D.; Vogel, H.; Sgambato, A.; Falabella, P. Insect antimicrobial peptides: Potential weapons to counteract the antibiotic resistance. Cell. Mol. Life Sci. 2021, 78, 4259–4282. [Google Scholar] [CrossRef]
- Zeng, T.; Jaffar, S.; Xu, Y.; Qi, Y.X. The intestinal immune defense system in insects. Int. J. Mol. Sci. 2022, 23, 15132. [Google Scholar] [CrossRef]
- Azmiera, N.; Krasilnikova, A.; Sahudin, S.; Al-Talib, H.; Heo, C.C. Antimicrobial peptides isolated from insects and their potential applications. J. Asia-Pac. Entomol. 2022, 25, 101892. [Google Scholar] [CrossRef]
- Bang, K.; Park, S.; Yoo, J.Y.; Cho, S. Characterization and expression of attacin, an antibacterial protein-encoding gene, from the beet armyworm, Spodoptera exigua (Hübner) (Insecta: Lepidoptera: Noctuidae). Mol. Biol. Rep. 2012, 39, 5151–5159. [Google Scholar] [CrossRef]
- Lu, D.D.; Geng, T.; Hou, C.X.; Huang, Y.X.; Qin, G.X.; Guo, X.J. Bombyx mori cecropin A has a high antifungal activity to entomopathogenic fungus Beauveria bassiana. Gene 2016, 583, 29–35. [Google Scholar] [CrossRef]
- Browne, K.; Chakraborty, S.; Chen, R.; Willcox, M.D.; Black, D.S.; Walsh, W.R.; Kumar, N. A new era of antibiotics: The clinical potential of antimicrobial peptides. Int. J. Mol. Sci. 2020, 21, 7047. [Google Scholar] [CrossRef] [PubMed]
- Meng, F.; Li, Y.X.; Liu, Z.K.; Feng, Y.Q.; Wang, X.; Zhang, X.Y. Expression of the thaumatin-like protein-1 gene (Bx-tlp-1) from pine wood nematode Bursaphelenchus xylophilus affects terpene metabolism in pine trees. Phytopathology 2022, 112, 888–897. [Google Scholar] [CrossRef] [PubMed]
- Dou, G.M.; Yan, D.H. Research progress on biocontrol of pine wilt disease by microorganisms. Forests 2022, 13, 1047. [Google Scholar] [CrossRef]
- Akbulut, S.; Stamps, W.T. Insect vectors of the pinewood nematode: A review of the biology and ecology of Monochamus species. For. Pathol. 2012, 42, 89–99. [Google Scholar] [CrossRef]
- Wang, H.; Meng, X.L.; Xu, J.P.; Wang, J.; Wang, H.; Ma, C.W. Production, purification, and characterization of the cecropin from Plutella xylostella, pxCECA1, using an intein-induced self-cleavable system in Escherichia coli. App. Microbiol. Biot. 2012, 94, 1031–1039. [Google Scholar] [CrossRef] [PubMed]
- Xu, H.H.; Tie, K.Y.; Zhang, Y.; Feng, X.; Cao, Y.; Han, W.Y. Design, expression, and characterization of the hybrid antimicrobial peptide T-catesbeianin-1 based on FyuA. J. Pept. Sci. 2018, 24, e3059. [Google Scholar] [CrossRef]
- Li, J.B.; Zhu, W.Z.; Luo, M.R.; Ren, H.H.; Tang, L.; Liao, H.H.; Wang, Y. Molecular cloning, expression and purification of lactoferrin from Tibetan sheep mammary gland using a yeast expression system. Protein. Expres. Purif. 2015, 109, 35–49. [Google Scholar] [CrossRef]
- Sang, M.; Wei, H.; Zhang, J.X.; Wei, Z.H.; Wu, X.L.; Chen, Y.; Zhuge, Q. Expression and characterization of the antimicrobial peptide ABP-dHC-cecropin A in the methylotrophic yeast Pichia pastoris. Protein. Expres. Purif. 2017, 140, 44–51. [Google Scholar] [CrossRef]
- Zhan, N.; Wang, T.Y.; Zhang, L.C.; Shan, A.S. A eukaryotic expression strategy for producing the novel antimicrobial peptide PRW4. Braz. J. Microbiol. 2020, 51, 999–1008. [Google Scholar] [CrossRef]
- Robertson, L.; Arcos, S.C.; Escuer, M.; Merino, R.S.; Esparrago, G.; Abelleira, A.; Navas, A. Incidence of the pinewood nematode Bursaphelenchus xylophlius Steiner & Buhrer, 1934 (Nickle, 1970) in Spain. Nematology 2011, 13, 755–757. [Google Scholar]
- Martens, E.; Demain, A. The antibiotic resistance crisis, with a focus on the United States. J. Antibiot. 2017, 70, 520–526. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kanost, M.R.; Jiang, H.B.; Yu, X.Q. Innate immune responses of a lepidopteran insect, Manduca sexta. Immunol. Rev. 2004, 198, 97–105. [Google Scholar] [CrossRef] [PubMed]
- Lemaitre, B.; Hoffmann, J. The host defense of Drosophila melanogaster. Annu. Rev. Immunol. 2007, 25, 697–743. [Google Scholar] [CrossRef] [Green Version]
- Tsakas, S.; Marmaras, V.J. Insect immunity and its signalling: An overview. Invertebr. Surviv. J. 2010, 7, 228–238. [Google Scholar]
- Zhang, K.Y.; Li, X.; Yu, C.; Wang, Y. Promising Therapeutic strategies against microbial biofilm challenges. Front. Cell. Infect. Microbiol. 2020, 10, 359. [Google Scholar] [CrossRef]
- Sheehan, G.; Garvey, A.; Croke, M.; Kavanagh, K. Innate humoral immune defences in mammals and insects: The same, with differences? Virulence 2018, 9, 1625–1639. [Google Scholar] [CrossRef] [Green Version]
- Andoh, M.; Ueno, T.; Kawasaki, K. Tissue-dependent induction of antimicrobial peptide genes after body wall injury in house fly (Musca domestica) larvae. Drug. Discov. Ther. 2018, 12, 355–362. [Google Scholar] [CrossRef] [Green Version]
- Moghaddam, M.R.B.; Tonk, M.; Schreiber, C.; Salzig, D.; Czermak, P.; Vilcinskas, A.; Rahnamaeian, M. The potential of the Galleria mellonella innate immune system is maximized by the co-presentation of diverse antimicrobial peptides. Biol. Chem. 2016, 397, 939–945. [Google Scholar] [CrossRef]
- Wu, T.Q.; Chen, Y.F.; Chen, W.D.; Zou, S.H.; Zhang, Y.Q.; Lin, Y.; Liang, Z.J.; Tang, D.Z. Transgenic expression of an insect diapause-specific peptide (DSP) in Arabidopsis resists phytopathogenic fungal attacks. Eur. J. Plant. Pathol. 2013, 137, 93–101. [Google Scholar] [CrossRef]
- Zhang, W.; Tettamanti, G.; Bassal, T.; Heryanto, C.; Eleftherianos, L.; Mohamed, A. Regulators and signalling in insect antimicrobial innate immunity: Functional molecules and cellular pathways. Cell Signal. 2021, 83, 110003. [Google Scholar] [CrossRef] [PubMed]
- Pretzel, J.; Mohring, F.; Rahlfs, S.; Becker, K. Antiparasitic peptides. In Yellow Biotechnology I; Springer: Berlin/Heidelberg, Germany, 2013; pp. 157–192. [Google Scholar]
- Kurz, C.L.; Tan, M.W. Regulation of aging and innate immunity in C. elegans. Aging Cell 2004, 3, 185–193. [Google Scholar] [CrossRef] [PubMed]
- Nicholas, H.R.; Hodgkin, J. Responses to infection and possible recognition strategies in the innate immune system of Caenorhabditis elegans. Mol. Immunol. 2004, 41, 479–493. [Google Scholar] [CrossRef] [PubMed]
- Castillo, J.C.; Reynolds, S.E.; Eleftherianos, I. Insect immune responses to nematode parasites. Trends Parasitol. 2011, 27, 537–547. [Google Scholar] [CrossRef] [PubMed]
- Eleftherianos, I.; Shokal, U.; Yadav, S.; Kenney, E.; Maldonado, T. Insect immunity to entomopathogenic nematodes and their mutualistic bacteria. Curr. Top. Microbiol. Immunol. 2017, 402, 123–156. [Google Scholar]
- Gillespie, J.P.; Kanost, M.R.; Trenczek, T. Biological mediators of insect immunity. Annual. Rev. Entomol. 1997, 42, 611–643. [Google Scholar] [CrossRef]
- Kim, C.H.; Park, J.W.; Ha, N.C.; Kang, H.J.; Lee, B.L. Innate immune response in insects: Recognition of bacterial peptidoglycan and amplification of its recognition signal. BMB Rep. 2008, 41, 93–101. [Google Scholar] [CrossRef] [Green Version]
- Kumar, S.; Christophides, G.K.; Cantera, R.; Barillas-Mury, S. The role of reactive oxygen species on Plasmodium melanotic encapsulation in Anopheles gambiae. Proc. Natl. Acad. Sci. USA 2003, 100, 14139–14144. [Google Scholar] [CrossRef] [Green Version]
- Peña, J.M.; Carrillo, M.A.; Hallem, E.A. Variation in the susceptibility of Drosophila to different entomopathogenic nematodes. Infect. Immun. 2015, 83, 1130–1138. [Google Scholar] [CrossRef] [Green Version]
- Garriga, A.; Mastore, M.; Morton, A.; Garcia del Pino, F.; Brivio, M.F. Immune response of Drosophila suzukii larvae to infection with the nematobacterial complex Steinernema carpocapsae-Xenorhabdus nematophila. Insects 2020, 11, 210. [Google Scholar] [CrossRef] [Green Version]
- Cabezas-Cruz, A.; Tonk, M.; Bouchut, A.; Pierrot, C.; Pierce, R.J.; Kotsyfakis, M.; Rahnamaeian, M.; Vilcinskas, A.; Khalife, J.; Valdés, J.J. Antiplasmodial activity is an ancient and conserved feature of tick defensins. Front. Microbiol. 2016, 7, 1682. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pitale, D.M.; Kaur, G.; Baghel, M.; Kaur, K.J.; Shaha, C. Halictine-2 antimicrobial peptide shows promising anti-parasitic activity against Leishmania spp. Exp. Parasitol. 2020, 218, 107987. [Google Scholar] [CrossRef] [PubMed]
- Fieck, A.; Hurwitz, I.; Kang, A.S.; Durvasula, R. Trypanosoma cruzi: Synergistic cytotoxicity of multiple amphipathic anti-microbial peptides to T. cruzi and potential bacterial hosts. Exp. Parasitol. 2010, 125, 342–347. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, Q.Y.; Yan, Z.B.; Meng, Y.M.; Hong, X.Y.; Shao, G.; Ma, J.J.; Chen, X.R.; Liu, J.; Kang, J.; Fu, C.Y. Antimicrobial peptides: Mechanism of action, activity and clinical potential. Mil. Med. Res. 2021, 8, 48. [Google Scholar] [CrossRef]
- Kumar, P.; Kizhakkedathu, J.N.; Straus, S.K. Antimicrobial peptides: Diversity, Mechanism of action and strategies to improve the activity and biocompatibility in vivo. Biomolecules 2018, 8, 4. [Google Scholar] [CrossRef] [Green Version]
- Price, N.P.; Hartman, T.M.; Li, J.; Velpula, K.K.; Naumann, T.A.; Guda, M.R.; Yu, B.; Bischoff, K.M. Modified tunicamycins with reduced eukaryotic toxicity that enhance the antibacterial activity of Î2-lactams. J. Antibiot. 2017, 70, 50–53. [Google Scholar] [CrossRef]
- Rathinakumar, R.; Walkenhorst, W.F.; Wimley, W.C. Broad-Spectrum antimicrobial peptides by aational combinatorial design and High-Throughput screening: The importance of interfacial activity. J. Am. Chem. Soc. 2009, 131, 7609–7617. [Google Scholar] [CrossRef] [Green Version]
- Guo, X.; Chen, J.; Sun, H.; Luo, L.L.; Gu, Y.X.; Yi, Y.L.; Wang, X.; Shan, Y.Y.; Liu, B.F.; Zhou, Y.; et al. Mining, heterologous expression, purification and characterization of 14 novel bacteriocins from Lactobacillus rhamnosus LS-8. Int. J. Biol. Macromol. 2020, 164, 2162–2176. [Google Scholar] [CrossRef]
- Li, Z.X.; Cheng, Q.; Guo, H.N.; Zhang, R.J.; Si, D.Y. Expression of hybrid peptide EF-1 in Pichia pastoris, its purification, and antimicrobial characterization. Molecules 2020, 25, 5538. [Google Scholar] [CrossRef]





Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chu, X.; Jiang, D.; Yu, L.; Li, M.; Wu, S.; Zhang, F.; Hu, X. Heterologous Expression and Bioactivity Determination of Monochamus alternatus Antibacterial Peptide Gene in Komagataella phaffii (Pichia pastoris). Int. J. Mol. Sci. 2023, 24, 5421. https://doi.org/10.3390/ijms24065421
Chu X, Jiang D, Yu L, Li M, Wu S, Zhang F, Hu X. Heterologous Expression and Bioactivity Determination of Monochamus alternatus Antibacterial Peptide Gene in Komagataella phaffii (Pichia pastoris). International Journal of Molecular Sciences. 2023; 24(6):5421. https://doi.org/10.3390/ijms24065421
Chicago/Turabian StyleChu, Xu, Di Jiang, Lu Yu, Ming Li, Songqing Wu, Feiping Zhang, and Xia Hu. 2023. "Heterologous Expression and Bioactivity Determination of Monochamus alternatus Antibacterial Peptide Gene in Komagataella phaffii (Pichia pastoris)" International Journal of Molecular Sciences 24, no. 6: 5421. https://doi.org/10.3390/ijms24065421
APA StyleChu, X., Jiang, D., Yu, L., Li, M., Wu, S., Zhang, F., & Hu, X. (2023). Heterologous Expression and Bioactivity Determination of Monochamus alternatus Antibacterial Peptide Gene in Komagataella phaffii (Pichia pastoris). International Journal of Molecular Sciences, 24(6), 5421. https://doi.org/10.3390/ijms24065421




