Prognostication of DNA Damage Response Protein Expression Patterns in Chronic Lymphocytic Leukemia
Abstract
1. Introduction
2. Results
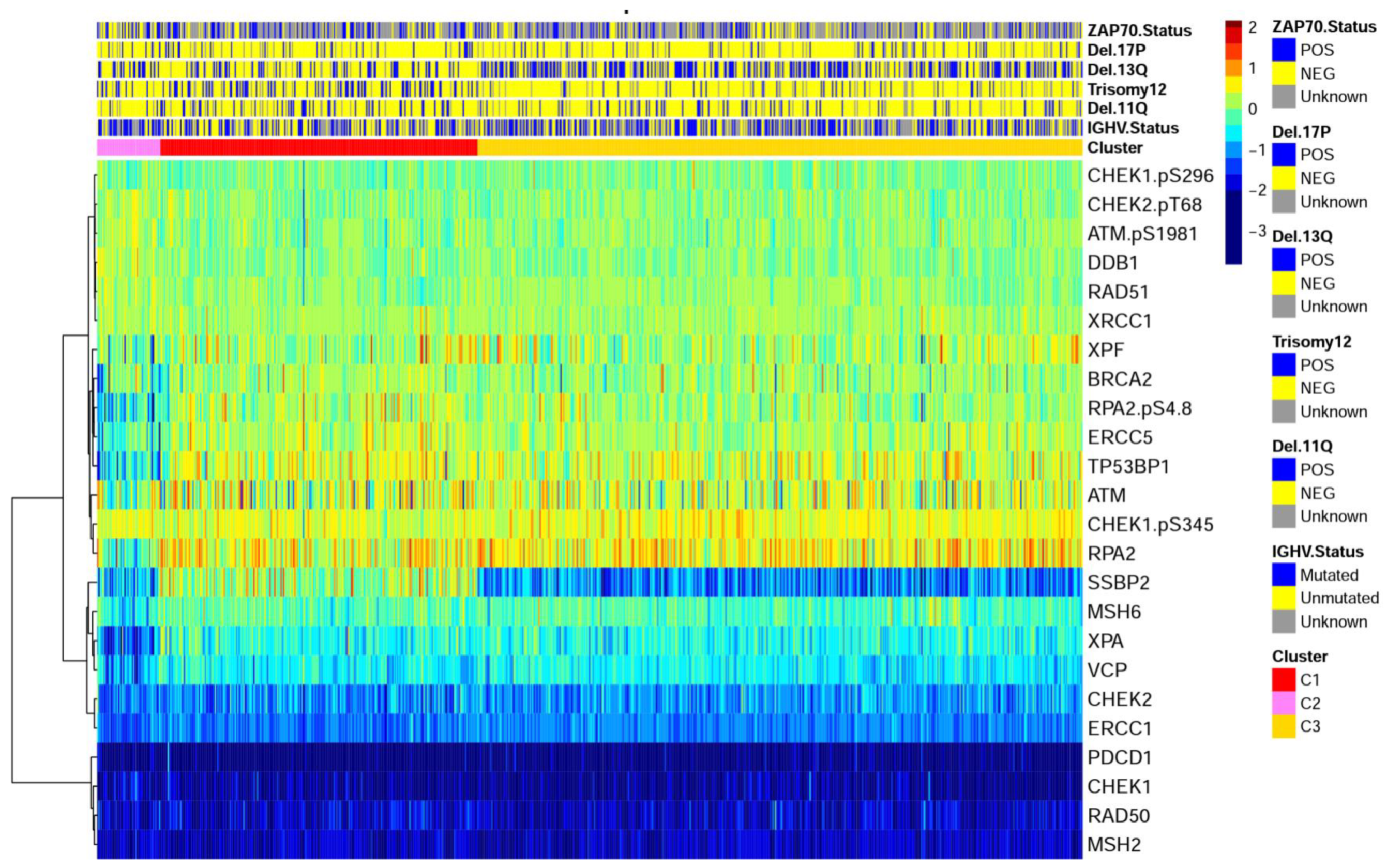
2.1. DDR Protein Expression in Chronic Lymphocytic Leukemia Is Different from Normal and Forms Recurrent Expression Patterns
2.2. DDR Cluster Membership Is Independent of Most Traditional Prognostic Features
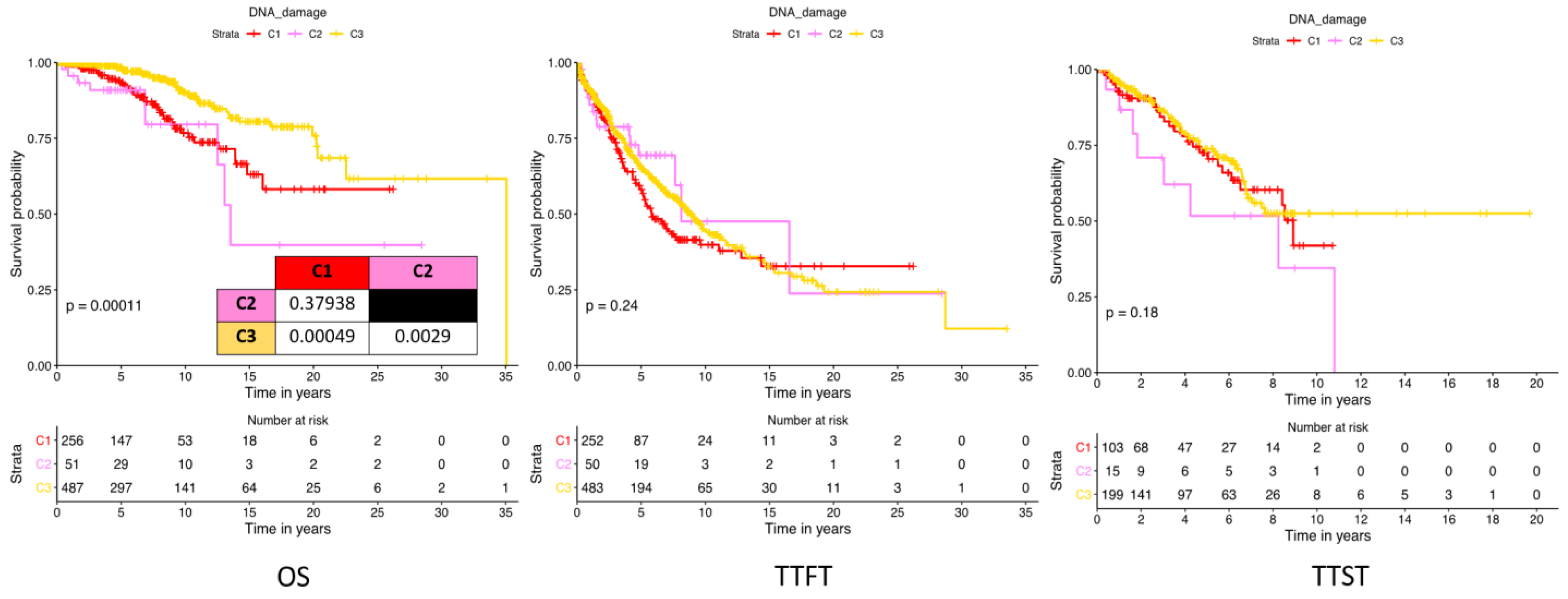
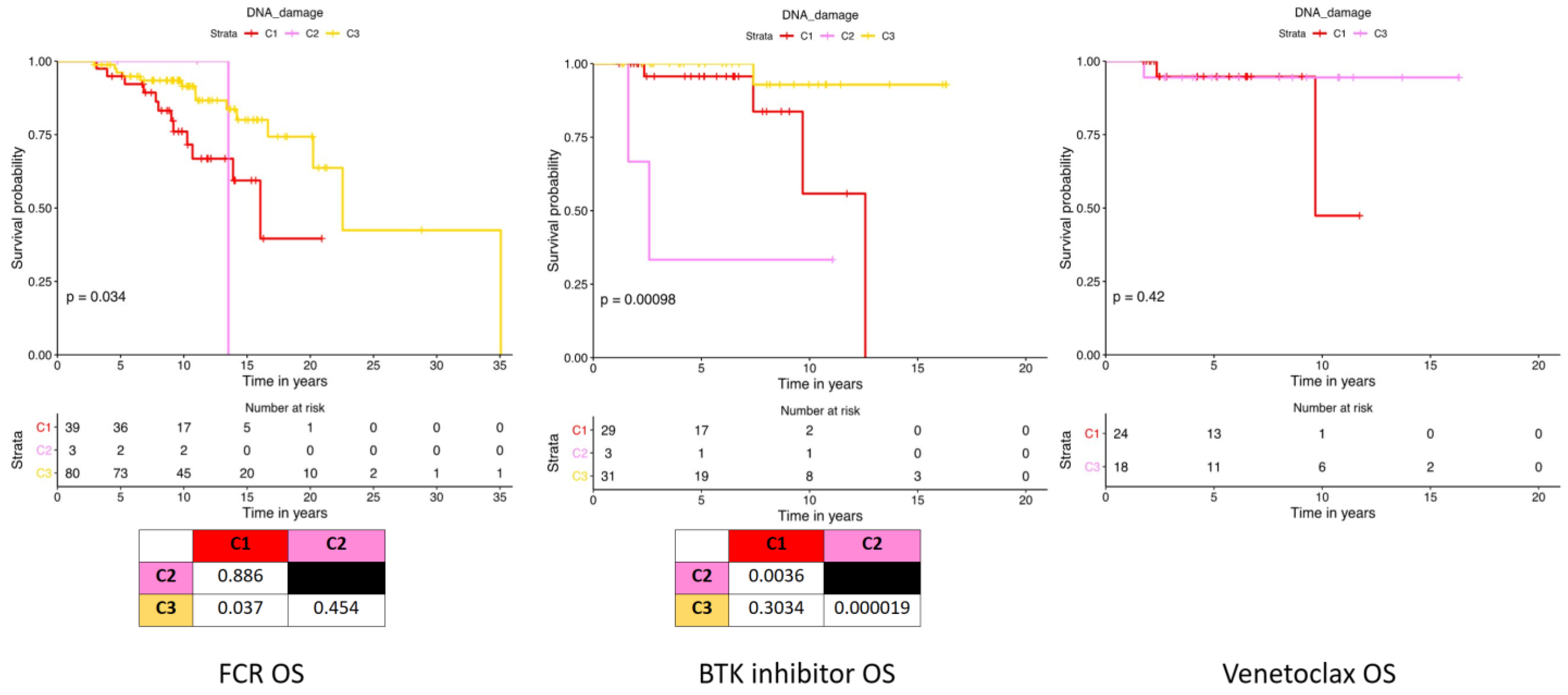
2.3. DNA Damage Expression Patterns Are Associated with Adverse Patient Outcomes, Chemotherapy Responses, and Prognostic Factors
2.4. Prognostication of Individual DNA Damage Member Proteins
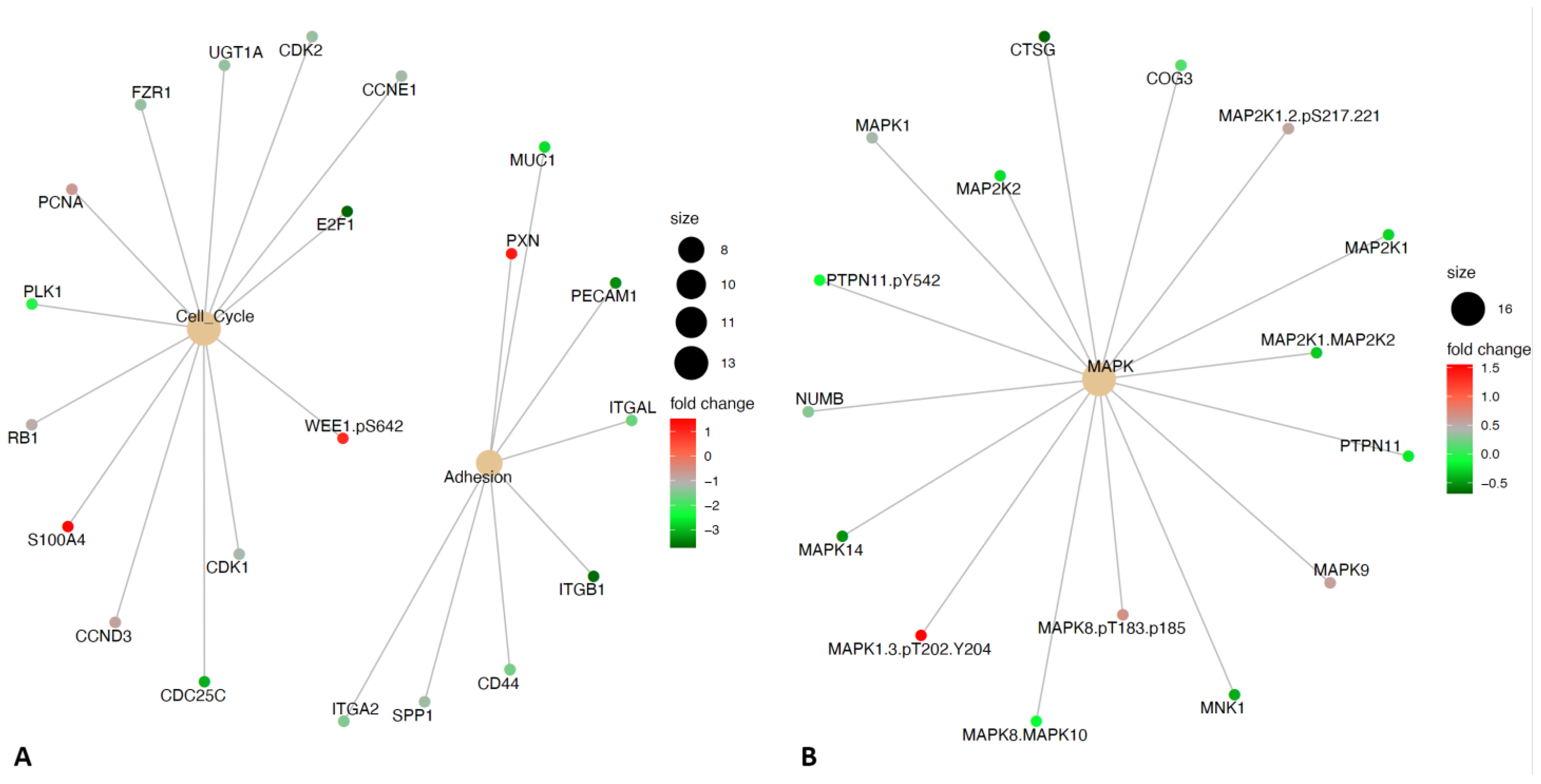
2.5. Differential Expression of DNA Damage Protein Expression Groups Reveals Altered Utilization of Adhesion, Cell Cycle, and MAPK Signaling
3. Discussion
4. Materials and Methods
4.1. RPPA Methodology
4.2. Data Processing, Normalization, and Quality Control
4.3. Statistics
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Popp, H.D.; Flach, J.; Brendel, S.; Ruppenthal, S.; Kleiner, H.; Seifarth, W.; Schneider, S.; Schulze, T.J.; Weiss, C.; Wenz, F.; et al. Accumulation of DNA damage and alteration of the DNA damage response in monoclonal B-cell lymphocytosis and chronic lymphocytic leukemia. Leuk. Lymphoma 2019, 60, 795–804. [Google Scholar] [CrossRef] [PubMed]
- Hernández-Sánchez, M.; Rodríguez-Vicente, A.E.; Gonzalez-Gascon, Y.M.I.; Quijada-Álamo, M.; Hernández-Sánchez, J.M.; Martín-Izquierdo, M.; Hernández-Rivas, J.; Benito, R.; Hernández-Rivas, J.M. DNA damage response-related alterations define the genetic background of patients with chronic lymphocytic leukemia and chromosomal gains. Exp. Hematol. 2019, 72, 9–13. [Google Scholar] [CrossRef]
- Schnaiter, A.; Stilgenbauer, S. 17p deletion in chronic lymphocytic leukemia: Risk stratification and therapeutic approach. Hematol. Oncol. Clin. N. Am. 2013, 27, 289–301. [Google Scholar] [CrossRef] [PubMed]
- Stankovic, T.; Skowronska, A. The role of ATM mutations and 11q deletions in disease progression in chronic lymphocytic leukemia. Leuk. Lymphoma 2014, 55, 1227–1239. [Google Scholar] [CrossRef]
- Campo, E.; Cymbalista, F.; Ghia, P.; Jager, U.; Pospisilova, S.; Rosenquist, R.; Schuh, A.; Stilgenbauer, S. TP53 aberrations in chronic lymphocytic leukemia: An overview of the clinical implications of improved diagnostics. Haematologica 2018, 103, 1956–1968. [Google Scholar] [CrossRef]
- Turgut, B.; Vural, O.; Pala, F.; Pamuk, G.; Tabakcioğlu, K.; Demir, M.; Ongören, S.; Soysal, T.; Algüneş, C. 17p Deletion is associated with resistance of B-cell chronic lymphocytic leukemia cells to in vitro fludarabine-induced apoptosis. Leuk. Lymphoma 2007, 48, 311–320. [Google Scholar] [CrossRef]
- Tahir, S.K.; Smith, M.L.; Hessler, P.; Rapp, L.R.; Idler, K.B.; Park, C.H.; Leverson, J.D.; Lam, L.T. Potential mechanisms of resistance to venetoclax and strategies to circumvent it. BMC Cancer 2017, 17, 399. [Google Scholar] [CrossRef] [PubMed]
- Landau, D.A.; Carter, S.L.; Stojanov, P.; McKenna, A.; Stevenson, K.; Lawrence, M.S.; Sougnez, C.; Stewart, C.; Sivachenko, A.; Wang, L.; et al. Evolution and impact of subclonal mutations in chronic lymphocytic leukemia. Cell 2013, 152, 714–726. [Google Scholar] [CrossRef]
- Puente, X.S.; Pinyol, M.; Quesada, V.; Conde, L.; Ordóñez, G.R.; Villamor, N.; Escaramis, G.; Jares, P.; Beà, S.; González-Díaz, M.; et al. Whole-genome sequencing identifies recurrent mutations in chronic lymphocytic leukaemia. Nature 2011, 475, 101–105. [Google Scholar] [CrossRef]
- Austen, B.; Skowronska, A.; Baker, C.; Powell, J.E.; Gardiner, A.; Oscier, D.; Majid, A.; Dyer, M.; Siebert, R.; Taylor, A.M.; et al. Mutation status of the residual ATM allele is an important determinant of the cellular response to chemotherapy and survival in patients with chronic lymphocytic leukemia containing an 11q deletion. J. Clin. Oncol. 2007, 25, 5448–5457. [Google Scholar] [CrossRef]
- Seiffert, M.; Dietrich, S.; Jethwa, A.; Glimm, H.; Lichter, P.; Zenz, T. Exploiting biological diversity and genomic aberrations in chronic lymphocytic leukemia. Leuk. Lymphoma 2012, 53, 1023–1031. [Google Scholar] [CrossRef]
- Reiners, K.S.; Topolar, D.; Henke, A.; Simhadri, V.R.; Kessler, J.; Sauer, M.; Bessler, M.; Hansen, H.P.; Tawadros, S.; Herling, M.; et al. Soluble ligands for NK cell receptors promote evasion of chronic lymphocytic leukemia cells from NK cell anti-tumor activity. Blood 2013, 121, 3658–3665. [Google Scholar] [CrossRef]
- Shatnyeva, O.M.; Hansen, H.P.; Reiners, K.S.; Sauer, M.; Vyas, M.; von Strandmann, E.P. DNA damage response and evasion from immunosurveillance in CLL: New options for NK cell-based immunotherapies. Front. Genet. 2015, 6, 11. [Google Scholar] [CrossRef]
- Bloehdorn, J.; Braun, A.; Taylor-Weiner, A.; Jebaraj, B.M.C.; Robrecht, S.; Krzykalla, J.; Pan, H.; Giza, A.; Akylzhanova, G.; Holzmann, K.; et al. Multi-platform profiling characterizes molecular subgroups and resistance networks in chronic lymphocytic leukemia. Nat. Commun. 2021, 12, 5395. [Google Scholar] [CrossRef]
- Boudny, M.; Zemanova, J.; Khirsariya, P.; Borsky, M.; Verner, J.; Cerna, J.; Oltova, A.; Seda, V.; Mraz, M.; Jaros, J.; et al. Novel CHK1 inhibitor MU380 exhibits significant single-agent activity in TP53-mutated chronic lymphocytic leukemia cells. Haematologica 2019, 104, 2443–2455. [Google Scholar] [CrossRef]
- Zemanova, J.; Hylse, O.; Collakova, J.; Vesely, P.; Oltova, A.; Borsky, M.; Zaprazna, K.; Kasparkova, M.; Janovska, P.; Verner, J.; et al. Chk1 inhibition significantly potentiates activity of nucleoside analogs in TP53-mutated B-lymphoid cells. Oncotarget 2016, 7, 62091–62106. [Google Scholar] [CrossRef]
- Boudny, M.; Trbusek, M. ATR-CHK1 pathway as a therapeutic target for acute and chronic leukemias. Cancer Treat. Rev. 2020, 88, 102026. [Google Scholar] [CrossRef] [PubMed]
- Pepper, C.; Lowe, H.; Fegan, C.; Thurieau, C.; Thurston, D.E.; Hartley, J.A.; Delavault, P. Fludarabine-mediated suppression of the excision repair enzyme ERCC1 contributes to the cytotoxic synergy with the DNA minor groove crosslinking agent SJG-136 (NSC 694501) in chronic lymphocytic leukaemia cells. Br. J. Cancer 2007, 97, 253–259. [Google Scholar] [CrossRef]
- McNeil, E.M.; Melton, D.W. DNA repair endonuclease ERCC1-XPF as a novel therapeutic target to overcome chemoresistance in cancer therapy. Nucleic Acids Res. 2012, 40, 9990–10004. [Google Scholar] [CrossRef] [PubMed]
- Usanova, S.; Piee-Staffa, A.; Sied, U.; Thomale, J.; Schneider, A.; Kaina, B.; Köberle, B. Cisplatin sensitivity of testis tumour cells is due to deficiency in interstrand-crosslink repair and low ERCC1-XPF expression. Mol. Cancer 2010, 9, 248. [Google Scholar] [CrossRef] [PubMed]
- Kirschner, K.; Melton, D.W. Multiple roles of the ERCC1-XPF endonuclease in DNA repair and resistance to anticancer drugs. Anticancer Res. 2010, 30, 3223–3232. [Google Scholar]
- Schuhwerk, H.; Kleemann, J.; Gupta, P.; van Roey, R.; Armstark, I.; Kreileder, M.; Feldker, N.; Ramesh, V.; Hajjaj, Y.; Fuchs, K.; et al. The EMT transcription factor ZEB1 governs a fitness-promoting but vulnerable DNA replication stress response. Cell Rep. 2022, 41, 111819. [Google Scholar] [CrossRef] [PubMed]
- Shukla, A.; Shukla, V.; Joshi, S.S. Regulation of MAPK signaling and implications in chronic lymphocytic leukemia. Leuk. Lymphoma 2018, 59, 1565–1573. [Google Scholar] [CrossRef] [PubMed]
- Li, W.; Melton, D.W. Cisplatin regulates the MAPK kinase pathway to induce increased expression of DNA repair gene ERCC1 and increase melanoma chemoresistance. Oncogene 2012, 31, 2412–2422. [Google Scholar] [CrossRef]
- Griffen, T.L.; Hoff, F.W.; Qiu, Y.; Lillard, J.W.; Ferrajoli, A.; Thompson, P.; Toro, E.; Ruiz, K.; Burger, J.; Wierda, W.; et al. Proteomic Profiling Based Classification of CLL Provides Prognostication for Modern Therapy and Identifies Novel Therapeutic Targets. Blood 2021, 138, 1538. [Google Scholar] [CrossRef]


| TOTAL | C1 | C2 | C3 | p-Value | ||
|---|---|---|---|---|---|---|
| Number | 795 | 256 | 51 | 488 | ||
| Age (Mean +/- STD) | 65 (±9.8) | 65 (±11) | 65 (±11) | 65 ± 11 | 0.91 | |
| Vital Status (Dead) | 88 | 11.1% | 14.8% | 17.6% | 8.4% | 0.02 |
| Race | Number | Percentage | 0.97 | |||
| Asian | 7 | 0.9% | 0.4% | 2.1% | 1.1% | |
| Black | 33 | 4.2% | 5.2% | 2.1% | 4.0% | |
| Hispanic | 22 | 2.9% | 2.8% | 2.1% | 2.9% | |
| White | 710 | 92.0% | 91.5% | 93.6% | 92.0% | |
| Gender | 0.12 | |||||
| Female | 310 | 39.0% | 35.2% | 52.9% | 39.5% | |
| Male | 485 | 61.0% | 64.8% | 47.1% | 60.5% | |
| Binet Stage | 0.54 | |||||
| A | 478 | 478 (61.0%) | 59.5% | 29 (58.0%) | 62.0% | |
| B | 71 | 71 (9.06%) | 7.9% | 2 (4.00%) | 10.2% | |
| C | 235 | 235 (30.0%) | 32.5% | 19 (38.0%) | 27.8% | |
| Rai Stage | 0.61 | |||||
| 0 | 268 | 34.2% | 34.5% | 38.0% | 33.6% | |
| I | 234 | 29.8% | 27.8% | 22.0% | 31.7% | |
| II | 47 | 6.0% | 5.1% | 2.0% | 6.9% | |
| III | 132 | 16.8% | 17.9% | 28.0% | 15.1% | |
| IV | 103 | 13.1% | 14.7% | 10.0% | 12.7% | |
| Biomarkers | ||||||
| IGHV Status (Unmutated) | 280 | 48.6% | 58.3% | 25.0% | 45.7% | 0.002 |
| ZAP70 | 189 | 50.3% | 59.3% | 40.5% | 47.1% | 0.10 |
| SF3B1 | 34 | 16.1% | 19.8% | 14.3% | 13.8% | 0.73 |
| Cytogenetic abberations | ||||||
| Deletion 11Q | 100 | 14.1% | 19.0% | 6.5% | 12.2% | 0.05 |
| Deletion 13Q | 273 | 38.4% | 22.0% | 37.0% | 47.3% | <0.001 |
| Trisomy 12 | 109 | 15.3% | 30.0% | 17.4% | 7.2% | <0.001 |
| Deletion 17P | 68 | 95.6% | 10.8% | 13.0% | 8.6% | 0.67 |
| TP53 | 34 | 4.3% | 4.3% | 2.0% | 4.5% | 0.866 |
| No Abberations | 165 | 23.2% | 20.7% | 28.3% | 24.0% | 0.65 |
| Lab Tests | Units | |||||
| PB Platelets | K/uL | 190 (±72) | 190 (±75) | 220 (±77) | 190 (±70) | 0.03 |
| Hemoglobin | g/dL | 13 (±1.8) | 13 (±2.0) | 14 (±1.5) | 14 (±1.7) | 0.90 |
| Serum B2M | mg/L | 2.8 (±1.8) | 2.7 (±1.4) | 2.2 (±1.0) | 2.8 (±2.0) | 0.07 |
| Serum LDH | IU | 480 (±240) | 490 (±300) | 520 (±210) | 460 (±200) | 0.27 |
| Lymphocytes | K/uL | 38 (±54) | 42 (±61) | 18 (±19) | 38 (±51) | 0.02 |
| Immunophenotypic Markers | ||||||
| CD5 | % cells positive+ | 94 (±11) | 93 (±9.5) | 94 (±4.0) | 94 (±12) | 0.15 |
| CD19 | 81 (±15) | 82 (±16) | 76 (±16) | 82 (±14) | 0.09 | |
| CD20 | 78 (±20) | 78 (±21) | 79 (±22) | 78 (±19) | 0.69 | |
| CD22 | 63 (±39) | 68 (±38) | 74 (±37) | 59 (±40) | <0.001 | |
| CD23 | 87 (±18) | 86 (±19) | 85 (±20) | 88 (±17) | 0.99 | |
| CD38 | 24 (±27) | 33 (±31) | 23 (±29) | 19 (±23) | <0.001 | |
| CD79b | 43 (±38) | 48 (±33) | 40 (±36) | 40 (±40) | 0.02 | |
| Univariate Overall Survival | Multivariate Overall Survival | |||||||
|---|---|---|---|---|---|---|---|---|
| Variable | Est. | 2.50% | 97.50% | p-Value | Est. | 2.50% | 97.50% | p-Value |
| DDR Cluster 1 | 6.53 | 5.82 | 7.24 | p < 0.01 | 8.32 | 4.01 | 12.62 | p < 0.01 |
| DDR Cluster 2 | 6.37 | 4.77 | 7.96 | p < 0.01 | 7.04 | 2.39 | 11.7 | p < 0.01 |
| DDR Cluster 3 | 7.73 | 7.22 | 8.25 | p < 0.01 | 9.3 | 5.28 | 13.32 | p < 0.01 |
| Gender Male | 6.98 | 6.46 | 7.5 | p < 0.01 | −0.69 | −2.07 | 0.69 | 0.33 |
| Binet Stage B | 9.62 | 8.28 | 10.96 | p < 0.01 | 2.17 | −0.72 | 5.05 | 0.14 |
| Binet Stage C | 5.97 | 5.23 | 6.71 | p < 0.01 | −0.52 | −3.59 | 2.55 | 0.74 |
| Rai Stage I | 8.91 | 8.17 | 9.64 | p < 0.01 | 1.7 | −0.2 | 3.59 | 0.08 |
| Rai Stage II | 8.49 | 6.85 | 10.13 | p < 0.01 | 1.46 | −1.98 | 4.89 | 0.4 |
| Rai Stage III | 5.58 | 4.6 | 6.56 | p < 0.01 | −1.01 | −2.83 | 0.81 | 0.27 |
| Rai Stage IV | 6.47 | 5.37 | 7.58 | p < 0.01 | 0.01 | −1.83 | 1.86 | 0.99 |
| Del_11QPOS | 6.47 | 5.38 | 7.57 | p < 0.01 | −1.43 | −5.2 | 2.34 | 0.46 |
| Del_13QPOS | 6.79 | 6.13 | 7.46 | p < 0.01 | −1.47 | −5.18 | 2.23 | 0.44 |
| Del_17PPOS | 7.41 | 6.08 | 8.74 | p < 0.01 | −0.52 | −4.44 | 3.41 | 0.8 |
| T12POS | 5.67 | 4.63 | 6.71 | p < 0.01 | −2 | −5.78 | 1.79 | 0.3 |
| No Major Mutation | 6.58 | 6.12 | 7.05 | p < 0.01 | −0.9 | −4.79 | 3 | 0.65 |
| IGHV Unmutated | 6.32 | 5.66 | 6.99 | p < 0.01 | 0.31 | −1.18 | 1.79 | 0.68 |
| Zap70POS | 6.28 | 5.53 | 7.03 | p < 0.01 | −1.56 | −2.99 | −0.13 | 0.03 |
| Binet Stage A and Rai Stage 0 used as comparator | ||||||||
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Griffen, T.L.; Hoff, F.W.; Qiu, Y.; Burger, J.; Wierda, W.; Kornblau, S.M. Prognostication of DNA Damage Response Protein Expression Patterns in Chronic Lymphocytic Leukemia. Int. J. Mol. Sci. 2023, 24, 5481. https://doi.org/10.3390/ijms24065481
Griffen TL, Hoff FW, Qiu Y, Burger J, Wierda W, Kornblau SM. Prognostication of DNA Damage Response Protein Expression Patterns in Chronic Lymphocytic Leukemia. International Journal of Molecular Sciences. 2023; 24(6):5481. https://doi.org/10.3390/ijms24065481
Chicago/Turabian StyleGriffen, Ti’ara L., Fieke W. Hoff, Yihua Qiu, Jan Burger, William Wierda, and Steven M. Kornblau. 2023. "Prognostication of DNA Damage Response Protein Expression Patterns in Chronic Lymphocytic Leukemia" International Journal of Molecular Sciences 24, no. 6: 5481. https://doi.org/10.3390/ijms24065481
APA StyleGriffen, T. L., Hoff, F. W., Qiu, Y., Burger, J., Wierda, W., & Kornblau, S. M. (2023). Prognostication of DNA Damage Response Protein Expression Patterns in Chronic Lymphocytic Leukemia. International Journal of Molecular Sciences, 24(6), 5481. https://doi.org/10.3390/ijms24065481





