Circulating Extracellular Vesicles microRNAs Are Altered in Women Undergoing Preterm Birth
Abstract
:1. Introduction
2. Results
2.1. Patients
2.2. sEV Characterization
2.3. miRNA Expression
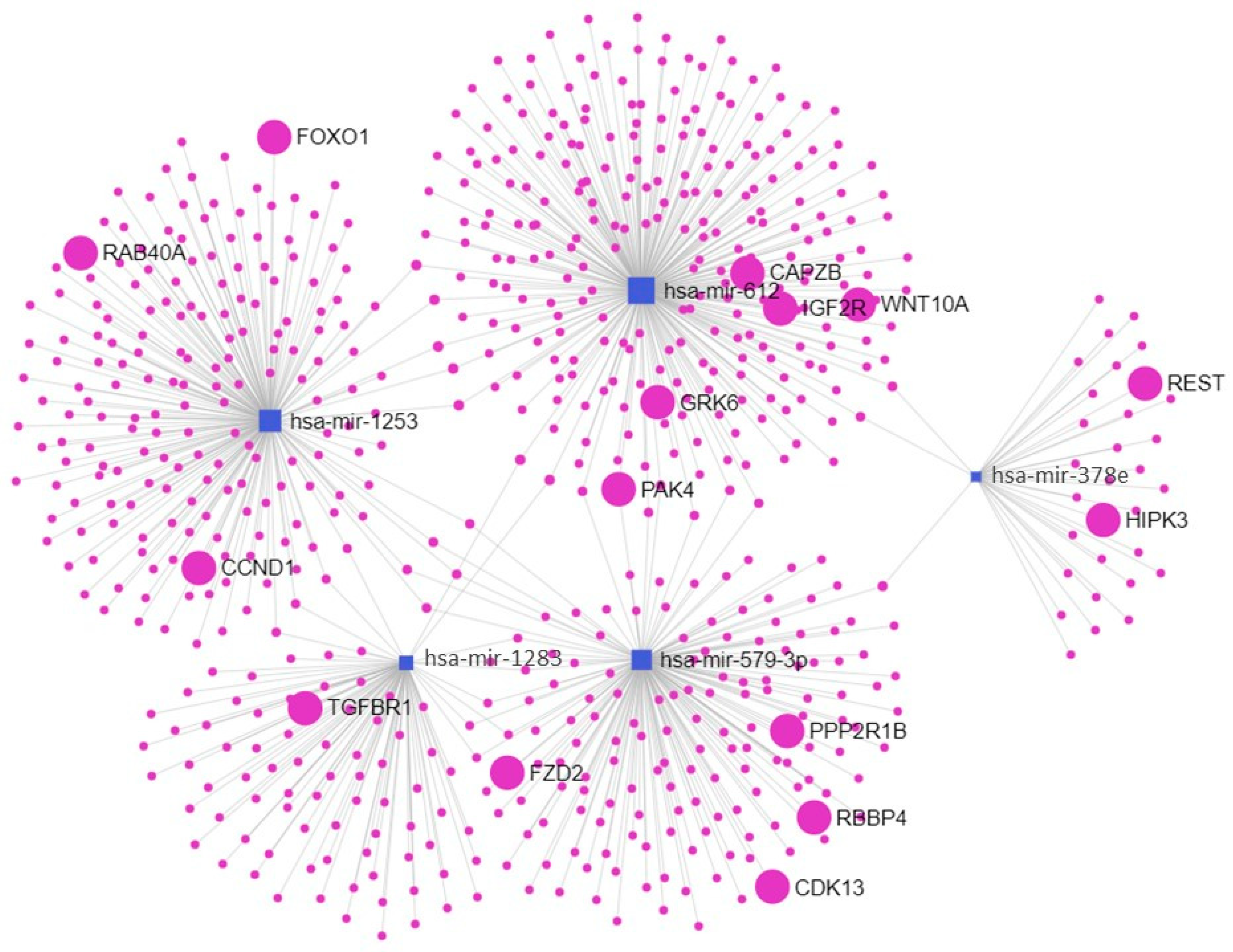
2.4. Enriched Pathways
3. Discussion
4. Materials and Methods
4.1. Patients
4.2. Sample Collection
4.3. Definition and Isolation of sEV
4.4. Characterization of sEV
4.5. Total RNA Extraction and Purification
4.6. NanoString nCounter Profiling Analysis
4.7. Pathway Enrichment and Network Analyses
4.8. Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Goldenberg, R.L.; Culhane, J.F.; Iams, J.D.; Romero, R. Epidemiology and causes of preterm birth. Lancet 2008, 371, 75–84. [Google Scholar] [CrossRef] [PubMed]
- Tracy, S.; Tracy, M.; Dean, J.; Laws, P.; Sullivan, E. Spontaneous preterm birth of liveborn infants in women at low risk in Australia over 10 years: A population-based study. BJOG Int. J. Obstet. Gynaecol. 2007, 114, 731–735. [Google Scholar] [CrossRef] [PubMed]
- McPheeters, M.L.; Miller, W.C.; Hartmann, K.E.; Savitz, D.A.; Kaufman, J.S.; Garrett, J.M.; Thorp, J.M. The epidemiology of threatened preterm labor: A prospective cohort study. Am. J. Obstet. Gynecol. 2005, 192, 1325–1329. [Google Scholar] [CrossRef] [PubMed]
- Arpino, C.; D’Argenzio, L.; Ticconi, C.; Di Paolo, A.; Stellin, V.; Lopez, L.; Curatolo, P. Brain damage in preterm infants: Etiological pathways. Ann. Dell’istituto Super. Sanitã 2005, 41, 229–237. [Google Scholar]
- Beck, S. The world-wide incidence of preterm birth: A systematic review of maternal mortality and morbidity. Bull. World Health Organ. 2010, 88, 31–88. [Google Scholar] [CrossRef] [PubMed]
- Mercuro, G.; Bassareo, P.P.; Flore, G.; Fanos, V.; Dentamaro, I.; Scicchitano, P.; Laforgia, N.; Ciccone, M. Prematurity and low weight at birth as new conditions predisposing to an increased cardiovascular risk. Eur. J. Prev. Cardiol. 2012, 20, 357–367. [Google Scholar] [CrossRef] [PubMed]
- Venkatesh, K.K.; Leviton, A.; Hecht, J.L.; Joseph, R.M.; Douglass, L.M.; Frazier, J.A.; Daniels, J.L.; Fry, R.C.; O’Shea, T.M.; Kuban, K.C. Histologic chorioamnionitis and risk of neurodevelopmental impairment at age 10 years among extremely preterm infants born before 28 weeks of gestation. Am. J. Obstet. Gynecol. 2020, 223, 745.e1–745.e10. [Google Scholar] [CrossRef] [PubMed]
- Nour, N. Preterm delivery and the millennium development goal. Rev. Obstet. Gynecol. 2012, 5, 100–105. [Google Scholar] [PubMed]
- Lo, C.C.; Hsu, J.J.; Hsieh, C.C.; Hsieh, T.T.; Hung, T.H. Risk factors for spontaneous preterm delivery before 34 weeks of gestation among Taiwanese women. Taiwan J. Obstet. Gyneco. 2007, 46, 389–394. [Google Scholar] [CrossRef] [Green Version]
- Varma, R.; Gupta, J.K.; James, D.K.; Kilby, M.D. Do screening-preventative interventions in asymptomatic pregnancies reduce the risk of preterm delivery--a critical appraisal of the literature. Eur. J. Obstet. Gynecol. Reprod. Biol. 2006, 127, 145–159. [Google Scholar] [CrossRef]
- Ancel, P.Y. Perspectives in the prevention of premature birth. Eur. J. Obstet. Gynecol. Reprod. Biol. 2004, 117, S2–S5. [Google Scholar] [CrossRef] [PubMed]
- Ouyang, Y.; Mouillet, J.-F.; Coyne, C.; Sadovsky, Y. Review: Placenta-specific microRNAs in exosomes—Good things come in nano-packages. Placenta 2014, 35, S69–S73. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Available online: http://www.mirbase.org/index.shtml (accessed on 18 August 2022).
- Ji, L.; Brkić, J.; Liu, M.; Fu, G.; Peng, C.; Wang, Y.-L. Placental trophoblast cell differentiation: Physiological regulation and pathological relevance to preeclampsia. Mol. Asp. Med. 2013, 34, 981–1023. [Google Scholar] [CrossRef]
- Seitz, H.; Royo, H.; Bortolin, M.-L.; Lin, S.-P.; Ferguson-Smith, A.C.; Cavaillé, J. A Large Imprinted microRNA Gene Cluster at the Mouse Dlk1-Gtl2 Domain. Genome Res. 2004, 14, 1741–1748. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Luo, L.; Ye, G.; Nadeem, L.; Fu, G.; Yang, B.B.; Dunk, C.; Lye, S.; Peng, C. MicroRNA-378a-5p promotes trophoblast cell survival, migration and invasion by targeting Nodal. J. Cell Sci. 2012, 125, 3124–3132. [Google Scholar] [CrossRef] [Green Version]
- Forbes, K.; Farrokhnia, F.; Aplin, J.; Westwood, M. Dicer-dependent miRNAs provide an endogenous restraint on cytotrophoblast proliferation. Placenta 2012, 33, 581–585. [Google Scholar] [CrossRef]
- Williams, Z.; Ben-Dov, I.Z.; Elias, R.; Mihailovic, A.; Brown, M.; Rosenwaks, Z.; Tuschl, T. Comprehensive profiling of circulating microRNA via small RNA sequencing of cDNA libraries reveals biomarker potential and limitations. Proc. Natl. Acad. Sci. USA 2013, 110, 4255–4260. [Google Scholar] [CrossRef] [Green Version]
- Renthal, N.E.; Williams, K.C.; Mendelson, C.R. MicroRNAs—Mediators of myometrial contractility during pregnancy and labour. Nat. Rev. Endocrinol. 2013, 9, 391–401. [Google Scholar] [CrossRef]
- Hromadnikova, I.; Kotlabova, K.; Ondrackova, M.; Pirkova, P.; Kestlerova, A.; Novotna, V.; Hympanova, L.; Krofta, L. Expression Profile of C19MC microRNAs in Placental Tissue in Pregnancy-Related Complications. DNA Cell Biol. 2015, 34, 437–457. [Google Scholar] [CrossRef]
- Elovitz, M.A.; Brown, A.G.; Anton, L.; Gilstrop, M.; Heiser, L.; Bastek, J. Distinct cervical microRNA profiles are present in women destined to have a preterm birth. Am. J. Obstet. Gynecol. 2014, 210, 221.e1–221.e11. [Google Scholar] [CrossRef]
- Paquette, A.G.; Shynlova, O.; Wu, X.; Kibschull, M.; Wang, K.; Price, N.D.; Lye, S.J. MicroRNA-transcriptome networks in whole blood and monocytes of women undergoing preterm labour. J. Cell Mol. Med. 2019, 23, 6835–6845. [Google Scholar] [CrossRef] [PubMed]
- Winger, E.E.; Reed, J.L.; Ji, X.; Gomez-Lopez, N.; Pacora, P.; Romero, R. MicroRNAs isolated from peripheral blood in the first trimester predict spontaneous preterm birth. PLoS ONE 2020, 15, e0236805. [Google Scholar] [CrossRef]
- Sarker, S.; Scholz-Romero, K.; Perez, A.; Illanes, S.E.; Mitchell, M.D.; Rice, G.E.; Salomon, C. Placenta-derived exosomes continuously increase in maternal circulation over the first trimester of pregnancy. J. Transl. Med. 2014, 12, 204. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Valadi, H.; Ekström, K.; Bossios, A.; Sjöstrand, M.; Lee, J.J.; Lötvall, J.O. Exosome-mediated transfer of mRNAs and microRNAs is a novel mechanism of genetic exchange between cells. Nat. Cell Biol. 2007, 9, 654–659. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Record, M. Intercellular communication by exosomes in placenta: A possible role in cell fusion? Placenta 2014, 35, 297–302. [Google Scholar] [CrossRef]
- Radnaa, E.; Richardson, L.S.; Sheller-Miller, S.; Baljinnyam, T.; Silva, M.D.C.; Kammala, A.K.; Urrabaz-Garza, R.; Kechichian, T.; Kim, S.; Han, A.; et al. Extracellular vesicle mediated feto-maternal HMGB1 signaling induces preterm birth. Lab Chip 2021, 21, 1956–1973. [Google Scholar] [CrossRef]
- Ghafourian, M.; Mahdavi, R.; Jonoush, Z.A.; Sadeghi, M.; Ghadiri, N.; Farzaneh, M.; Salehi, A.M. The implications of exosomes in pregnancy: Emerging as new diagnostic markers and therapeutics targets. Cell Commun. Signal. 2022, 20, 51. [Google Scholar] [CrossRef]
- Nair, S.; Salomon, C. Extracellular vesicles and their immunomodulatory functions in pregnancy. Semin. Immunopathol. 2018, 40, 425–437. [Google Scholar] [CrossRef] [Green Version]
- Liu, M.; Chen, Y.; Huang, B.; Mao, S.; Cai, K.; Wang, L.; Yao, X. Tumor-suppressing effects of microRNA-612 in bladder cancer cells by targeting malic enzyme 1 expression. Int. J. Oncol. 2018, 52, 1923–1933. [Google Scholar] [CrossRef]
- Zhu, Y.; Zhang, H.L.; Wang, Q.Y.; Chen, M.J.; Liu, L.B. Overexpression of microrna-612 restrains the growth, invasion, and tumorigenesis of melanoma cells by targeting espin. PLoS ONE 2018, 13, e0201217. [Google Scholar] [CrossRef]
- Wang, M.; Wang, Z.; Zhu, X.; Guan, S.; Liu, Z. NFKB1-miR-612-FAIM2 pathway regulates tumorigenesis in neurofibromatosis type 1. Endocrinology 2019, 160, 249–275. [Google Scholar] [CrossRef] [PubMed]
- Garcia-Lacarte, M.; Martinez, J.A.; Zulet, M.A.; Milagro, F.I. Implication of miR-612 and miR-1976 in the regulation of TP53 and CD40 and their relationship in the response to specific weight-loss diets. In Vitro Cell Dev. Biol. Anim. 2019, 55, 491–500. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gomez-Lopez, N.; Romero, R.; Garcia-Flores, V.; Xu, Y.; Leng, Y.; Alhousseini, A.; Hassan, S.S.; Panaitescu, B. Amniotic fluid neutrophils can phagocytize bacteria: A mechanism for microbial killing in the amniotic cavity. Am. J. Reprod. Immunol. 2017, 78, e12723. [Google Scholar] [CrossRef] [PubMed]
- Available online: https://maayanlab.cloud/Enrichr/enrich?dataset=ca36541ba817bd95fa9ce6f6ead9dd48# (accessed on 18 August 2022).
- Wang, D.; Song, W.; Na, Q. The emerging roles of placenta-specific microRNAs in regulating trophoblast proliferation during the first trimester. Aust. N. Z. J. Obstet. Gynaecol. 2012, 52, 565–570. [Google Scholar] [CrossRef] [PubMed]
- Park, S.; Lim, W.; Bazer, F.W.; Whang, K.-Y.; Song, G. Quercetin inhibits proliferation of endometriosis regulating cyclin D1 and its target microRNAs in vitro and in vivo. J. Nutr. Biochem. 2018, 63, 87–100. [Google Scholar] [CrossRef]
- Available online: https://maayanlab.cloud/Enrichr/enrich?dataset=a4c309d2b4db51232173445e28b646b3# (accessed on 18 August 2022).
- Polettini, J.; da Silva, M.G. Telomere-related disorders in fetal membranes associated with birth and adverse pregnancy outcomes. Front. Physiol. 2020, 11, 561771. [Google Scholar] [CrossRef] [PubMed]
- Menon, R.; Richardson, L.S.; Lappas, M. Fetal membrane architecture, aging and inflammation in pregnancy and parturition. Placenta 2019, 79, 40–45. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.; Jia, J.; Gou, J.; Zhao, X.; Yi, T. MicroRNA-451 plays a role in murine embryo implantation through targeting Ankrd46, as implicated by a microarray-based analysis. Fertil. Steril. 2014, 103, 834–844.e4. [Google Scholar] [CrossRef]
- Dominguez, F.; Moreno-Moya, J.M.; Lozoya, T.; Romero, A.; Martínez, S.; Monterde, M.; Gurrea, M.; Ferri, B.; Núñez, M.J.; Simón, C.; et al. Embryonic miRNA Profiles of Normal and Ectopic Pregnancies. PLoS ONE 2014, 9, e102185. [Google Scholar] [CrossRef] [Green Version]
- Dixon, C.L.; Sheller-Miller, S.; Saade, G.R.; Fortunato, S.J.; Lai, A.; Palma, C.; Guanzon, D.; Salomon, C.; Menon, R. Amniotic Fluid Exosome Proteomic Profile Exhibits Unique Pathways of Term and Preterm Labor. Endocrinology 2018, 159, 2229–2240. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ramos, B.R.D.; Mendes, N.D.; Tanikawa, A.A.; Amador, M.A.T.; dos Santos, N.P.C.; dos Santos, S.E.B.; Castelli, E.C.; Witkin, S.S.; da Silva, M.G. Ancestry informative markers and selected single nucleotide polymorphisms in immunoregulatory genes on adverse gestational outcomes: A case control study. BMC Pregnancy Childbirth 2016, 16, 30. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lilliecreutz, C.; Larén, J.; Sydsjö, G.; Josefsson, A. Effect of maternal stress during pregnancy on the risk for preterm birth. BMC Pregnancy Childbirth 2016, 16, 5. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Menon, R.; Debnath, C.; Lai, A.; Guanzon, D.; Bhatnagar, S.; Kshetrapal, P.K.; Sheller-Miller, S.; Salomon, C.; The Garbhini Study Team. Circulating Exosomal miRNA Profile During Term and Preterm Birth Pregnancies: A Longitudinal Study. Endocrinology 2019, 160, 249–275. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Elovitz, M.A.; Anton, L.; Bastek, J.; Brown, A.G. Can microRNA profiling in maternal blood identify women at risk for preterm birth? Am. J. Obstet. Gynecol. 2015, 212, 782.e1–782.e5. [Google Scholar] [CrossRef] [PubMed]
- Goytain, A.; Ng, T. NanoString nCounter Technology: High-Throughput RNA Validation. Chimeric RNA Methods Protoc. 2020, 2079, 125–139. [Google Scholar] [CrossRef]
- Veldman-Jones, M.H.; Brant, R.; Rooney, C.; Geh, C.; Emery, H.; Harbron, C.G.; Wappett, M.; Sharpe, A.; Dymond, M.; Barrett, J.C.; et al. Evaluating Robustness and Sensitivity of the NanoString Technologies nCounter Platform to Enable Multiplexed Gene Expression Analysis of Clinical Samples. Cancer Res. 2015, 75, 2587–2593. [Google Scholar] [CrossRef] [Green Version]
- Jacinto, S.O.S.; Pamplona, K.; Soares, M. Manual Técnico de Gestação de Alto Risco; Editora MS: Brasília, Brazil, 2012; 302p. [Google Scholar]
- Théry, C.; Witwer, K.W.; Aikawa, E.; Alcaraz, M.J.; Anderson, J.D.; Andriantsitohaina, R.; Antoniou, A.; Arab, T.; Archer, F.; Atkin-Smith, G.K.; et al. Minimal information for studies of extracellular vesicles 2018 (MISEV2018): A position statement of the International Society for Extracellular Vesicles and update of the MISEV2014 guidelines. J. Extracell. Vesicles 2018, 7, 1535750. [Google Scholar] [CrossRef] [Green Version]
- Tronco, J.A.; Ramos, B.R.A.; Bastos, N.M.; Alcântara, S.A.; da Silveira, J.C.; da Silva, M.G. Alpha-2-macroglobulin from circulating exosome-like vesicles is increased in women with preterm pregnancies. Sci. Rep. 2020, 10, 16961. [Google Scholar] [CrossRef]
- Reis, P.P.; Tokar, T.; Goswami, R.S.; Xuan, Y.; Sukhai, M.; Seneda, A.L.; Móz, L.E.S.; Perez-Ordonez, B.; Simpson, C.; Goldstein, D.; et al. A 4-gene signature from histologically normal surgical margins predicts local recurrence in patients with oral carcinoma: Clinical validation. Sci. Rep. 2020, 10, 1713. [Google Scholar] [CrossRef] [Green Version]
- Reis, P.; Drigo, S.; Carvalho, R.; Lapa, R.L.; Felix, T.; Patel, D.; Cheng, D.; Pintilie, M.; Liu, G.; Tsao, M.-S. Circulating mir-16-5p, mir-92a-3p, and mir-451a in plasma from lung cancer patients: Potential application in early detection and a regulatory role in tumorigenesis pathways. Cancers 2020, 12, 2071. [Google Scholar] [CrossRef]
- Tokar, T.; Pastrello, C.; Rossos, A.E.M.; Abovsky, M.; Hauschild, A.-C.; Tsay, M.; Lu, R.; Jurisica, I. mirDIP 4.1-integrative database of human microRNA target predictions. Nucleic Acids Res. 2018, 46, 360–370. [Google Scholar] [CrossRef] [Green Version]
- Chen, E.Y.; Tan, C.M.; Kou, Y.; Duan, Q.; Wang, Z.; Meirelles, G.V.; Clark, N.R.; Ma’Ayan, A. Enrichr: Interactive and collaborative HTML5 gene list enrichment analysis tool. BMC Bioinform. 2013, 14, 128. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kuleshov, M.V.; Jones, M.R.; Rouillard, A.D.; Fernandez, N.F.; Duan, Q.; Wang, Z.; Koplev, S.; Jenkins, S.L.; Jagodnik, K.M.; Lachmann, A.; et al. Enrichr: A comprehensive gene set enrichment analysis web server 2016 update. Nucleic Acids Res. 2016, 44, 90–97. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xie, Z.; Bailey, A.; Kuleshov, M.V.; Clarke, D.J.B.; Evangelista, J.E.; Jenkins, S.L.; Lachmann, A.; Wojciechowicz, M.L.; Kropiwnicki, E.; Jagodnik, K.M.; et al. Gene set knowledge discovery with Enrichr. Curr. Protoc. 2021, 1, e90. [Google Scholar] [CrossRef] [PubMed]
- Fan, Y.; Siklenka, K.; Arora, S.K.; Ribeiro, P.; Kimmins, S.; Xia, J. miRNet-dissecting miRNA-target interactions and functional associations through network-based visual analysis. Nucleic Acids Res. 2016, 44, W135–W141. [Google Scholar] [CrossRef] [PubMed]
- Chang, L.; Zhou, G.; Soufan, O.; Xia, J. miRNet 2.0: Network-based visual analytics for miRNA functional analysis and systems biology. Nucleic Acids Res. 2020, 48, W244–W251. [Google Scholar] [CrossRef]
- Chang, L.; Xia, J. MicroRNA Regulatory Network Analysis Using miRNet 2.0. Methods Mol. Biol. 2023, 2594, 185–204. [Google Scholar] [CrossRef] [PubMed]
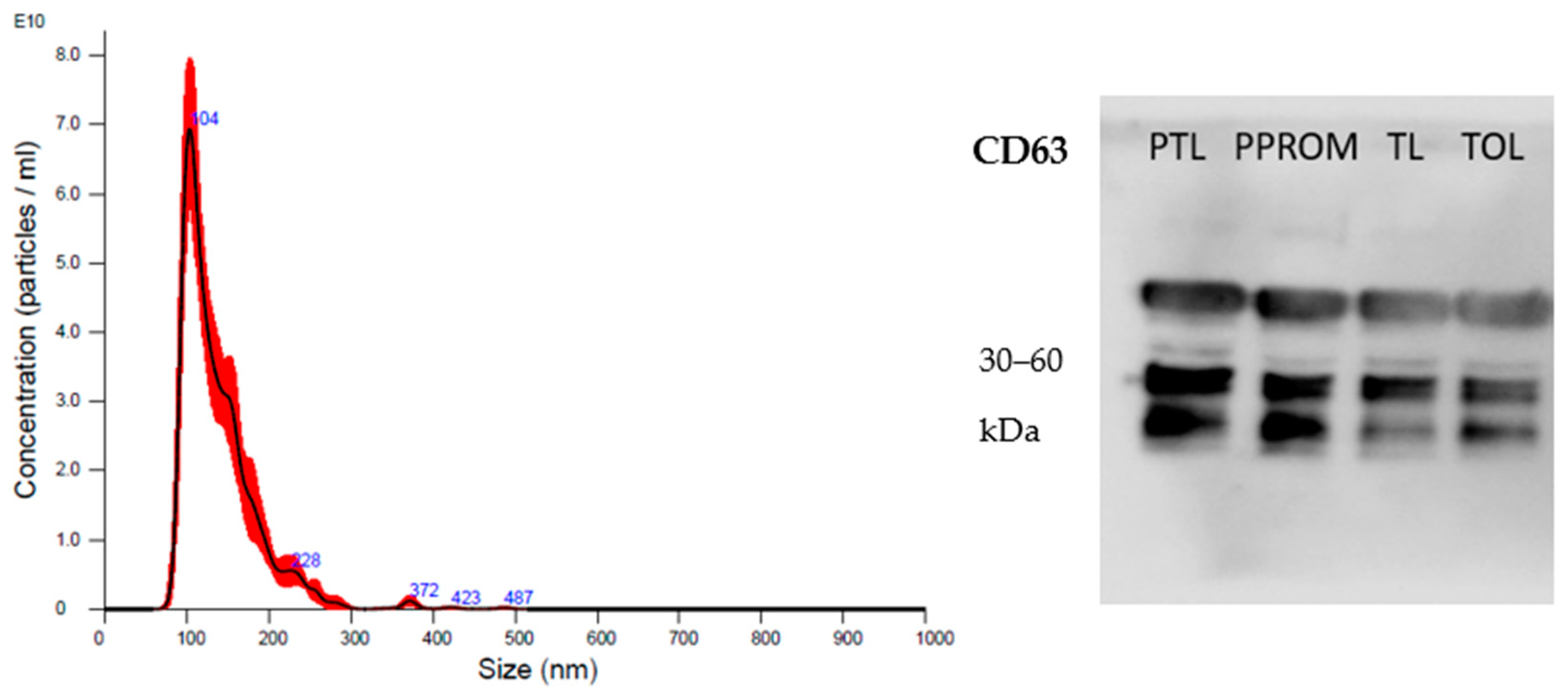
| Variables | PTL (n = 8) | PPROM (n = 7) | TL (n = 7) | T (n = 9) | p |
|---|---|---|---|---|---|
| Age (years) * | 25.6 ± 4.2 | 26.0 ± 7.4 | 23.8 ± 4.6 | 29.3 ± 4.9 | NS |
| GA at delivery (days) * | 247.0 ± 10.2 a | 239.6 ± 13.3 a | 279.9 ± 9.8 b | 272.1 ± 3.8 b | <0.0001 |
| BMI (kg/h2) * | 26.6 ± 3.8 | 29.6 ± 7.3 | 27.3 ± 3.1 | 28.6 ± 5.1 | NS |
| Delivery (%) | |||||
| Vaginal | 67 (4/6) | 57 (4/7) | 43 (3/7) | - | NS |
| Caesarean | 33 (2/6) | 43 (3/7) | 57 (4/7) | 100 (9/9) | |
| Marital status (%) | |||||
| Single | 13 (1/8) | 71 (5/7) | - | - | 0.001 |
| Civil union | 87 (7/8) a | 29 (2/7)b | 100 (7/7) a | 100 (9/9) a | |
| Self-reported ethnicity (%) | |||||
| White | 75 (6/8) a | 57 (4/7) b | 14 (1/7) b | 67 (6/9) b | 0.03 |
| Non-white | 25 (2/8) | 43 (3/7) | 86 (6/7) | 33 (3/9) | |
| Parturity (%) | |||||
| First pregnancy | 25% (2/8) | 57 (4/7) | 57 (4/7) | 44 (4/9) | NS |
| Multiple pregnancies | 75% (6/8) | 43 (3/7) | 43 (3/7) | 56 (5/9) | |
| Smoking (%) | |||||
| Smoking | - | 14 (1/7) | - | 11 (1/9) | NS |
| Not smoking | 100 (8/8) | 86 (6/7) | 100 (7/7) | 89 (8/9) | |
| Years of study (%) | |||||
| <8 years | - | 14 (1/7) | - | 11 (1/9) | NS |
| ≥8 years | 100 (8/8) | 86 (6/7) | 100 (7/7) | 89 (8/9) | |
| Previous history of PTL/PPROM (%) | |||||
| Presence | 38 (3/8) | - | - | - | |
| Absence | 62 (5/8) b | 100 (7/7)a | 100 (7/7) a | 100 (9/9) a | 0.02 |
| Prior abortion (%) | |||||
| Presence | 38 (3/8) | - | - | 22 (2/9) | NS |
| Absence | 62 (5/8) | 100 (7/7) | 100 (7/7) | 88 (7/9) |
| Variables | PTL (n = 8) | PPROM (n = 7) | TL (n = 7) | T (n = 9) | p |
|---|---|---|---|---|---|
| Weight (g) * | 2409 ± 677.0 a | 2208 ± 401.6 a | 3256 ± 459.1 b | 3358 ± 470.2 b | 0.004 |
| Apgar 10 * | 9.3 ± 0.8 | 9.0 ± 0.9 | 9.7 ± 0.5 | 9.8 ± 0.4 | NS |
| Sex (%) | |||||
| Female | 38 (3/8) | 14 (1/7) | 43 (3/7) | 56 (5/9) | NS |
| Male | 62 (5/8) | 86 (6/7) | 57 (4/7) | 44 (4/9) |
| Variables | Protein (µg/mL) | Particles/mL * | Mode | p |
|---|---|---|---|---|
| PTL | 2964.1 ± 1595 | 9.85 × 1011 | 105.2 ± 13.5 | NS |
| PPROM | 2528.0 ± 1092 | 5.81 × 1011 | 107.6 ± 11.5 | |
| TL | 2576.5 ± 1940 | 5.60 × 1011 | 103.8 ± 12.0 | |
| T | 2338.9 ± 926 | 5.05 × 1011 | 101.8 ± 7.9 |
| miRNAs | PTL (n = 8) | PPROM (n = 7) | TL (n = 7) | T (n = 9) | PTL vs. TL | PPROM vs. T | PTL vs. PPROM | TL vs. T |
|---|---|---|---|---|---|---|---|---|
| let-7i-5p | 831.2 ± 84.3 | 527.7 ± 121.1 | 814.7 ± 86.8 | 742.1 ± 154.9 | NS | NS | RR = 1.54 (1.16–2.05) | NS |
| miR-1253 | 224.0 ± 28.2 | 153.9 ± 8.4 | 214.6 ± 32.0 | 211.3 ± 21.9 | NS | RR = 0.73 (0.65–0.82) | RR = 1.47 (1.30–1.65) | NS |
| miR-1283 | 134.1 ± 20.4 | 83.4 ± 16.6 | 122.0 ± 16.8 | 125.4 ± 11.4 | NS | RR = 0.68 (0.58–0.78) | RR = 1.60 (1.37–1.87) | NS |
| miR-302-3p | 259.5 ± 40.8 | 201.0 ± 28.0 | 236.0 ± 36.9 | 196.7 ± 16.4 | NS | NS | RR = 1.29 (1.02–1.64) | RR = 1.21 (1.08–1.35) |
| miR-3144-3p | 103.6 ± 16.7 | 80.0 ± 7.0 | 96.8 ± 12.0 | 85.8 ± 9.4 | NS | NS | RR = 1.31 (1.11–1.55) | NS |
| miR-362-3p | 162.1 ± 24.2 | 140.2 ± 16.6 | 154.0 ± 9.5 | 145.2 ± 15.2 | NS | NS | RR = 1.16 (1.02–1.32) | NS |
| miR-378e | 301.1 ± 31.9 | 200.9 ± 19.6 | 284.7 ± 34.1 | 279.4 ± 38.0 | NS | RR = 0.71 (0.65–0.79) | RR = 1.50 (1.35–1.66) | NS |
| miR-451a | 35.6 ± 7.8 | 188.2 ± 53.6 | 40.0 ± 12.1 | 122.6 ± 72.6 | NS | NS | RR = 0.19 (0.12–0.29) | RR = 0.30 (0.15–0.59) |
| miR-520f | 28.4 ± 3.7 | 83.8 ± 62.8 | 32.0 ± 2.6 | 96.9 ± 87.9 | NS | NS | RR = 0.28 (0.13–0.60) | RR = 0.26 (0.12–0.58) |
| miR-579-3p | 2219.4 ± 438.1 | 1188.3 ± 206.8 | 2002.9 ± 451.3 | 2266.4 ± 479.0 | NS | RR = 0.52 (0.38–0.71) | RR = 1.88 (1.38–2.54) | NS |
| miR-612 | 187.7 ± 22.4 | 168.7 ± 18.6 | 155.7 ± 24.4 | 135.3 ± 16.7 | RR = 1.20 (1.06–1.36) | RR = 1.25 (1.09–1.42) | NS | RR = 1.16 (1.01–1.32) |
| Pathways | p | Genes |
|---|---|---|
| Endocytosis | 1.48 × 1012 | EHD2; TSG101; SMURF1; CAV1; CAPZ; GRK6; PSD3; CHMP7; RAB; FIP4; ARF6 |
| TGF-beta signaling pathway | 0.01218 | TGIF2; SMURF1; SMAD6; FMOD |
| Fc gamma R-mediated phagocytosis | 0.01354 | PAK1; PAK4; MARCKSL1; GAB2; ARF6 |
| Pathways | p | Genes |
|---|---|---|
| Cellular senescence | 3.62 × 1011 | NFATC3; PTEN; FOXO3; SIRT1; FOXO1; ZFP36L2; TGFBR1; HIPK3; PPP1CB; PPP2R1B; CDK6; CDK13; CCND1; RBBP4; RAD1 |
| Signaling pathways regulating pluripotency of stem cells | 5.98 × 1010 | FZD3; ZFHX3; FZD2; WNT10A; PCGF3; LIF; LIFR; PAX6; IGF1R; REST; KAT6A; IL6ST; SKIL |
| Focal adhesion | 1.59 × 1011 | SHC3; PRKCB; RASGRF1; PTEN; PARVA; IGF2R; PPP1CB; CDC42; PPP1CC; MAPK9; RAP1A; CCND1; PIP5K1A; PAK3; PPP1R12B |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ramos, B.R.A.; Tronco, J.A.; Carvalho, M.; Felix, T.F.; Reis, P.P.; Silveira, J.C.; Silva, M.G. Circulating Extracellular Vesicles microRNAs Are Altered in Women Undergoing Preterm Birth. Int. J. Mol. Sci. 2023, 24, 5527. https://doi.org/10.3390/ijms24065527
Ramos BRA, Tronco JA, Carvalho M, Felix TF, Reis PP, Silveira JC, Silva MG. Circulating Extracellular Vesicles microRNAs Are Altered in Women Undergoing Preterm Birth. International Journal of Molecular Sciences. 2023; 24(6):5527. https://doi.org/10.3390/ijms24065527
Chicago/Turabian StyleRamos, Bruna Ribeiro Andrade, Júlia Abbade Tronco, Márcio Carvalho, Tainara Francini Felix, Patrícia Pintor Reis, Juliano Coelho Silveira, and Márcia Guimarães Silva. 2023. "Circulating Extracellular Vesicles microRNAs Are Altered in Women Undergoing Preterm Birth" International Journal of Molecular Sciences 24, no. 6: 5527. https://doi.org/10.3390/ijms24065527
APA StyleRamos, B. R. A., Tronco, J. A., Carvalho, M., Felix, T. F., Reis, P. P., Silveira, J. C., & Silva, M. G. (2023). Circulating Extracellular Vesicles microRNAs Are Altered in Women Undergoing Preterm Birth. International Journal of Molecular Sciences, 24(6), 5527. https://doi.org/10.3390/ijms24065527








