Anticancer and Targeting Activity of Phytopharmaceutical Structural Analogs of a Natural Peptide from Trichoderma longibrachiatum and Related Peptide-Decorated Gold Nanoparticles
Abstract
:1. Introduction
2. Results
2.1. Peptide Synthesis
2.2. Synthesis and Characterization of Peptide-Decorated Gold Nanoparticles
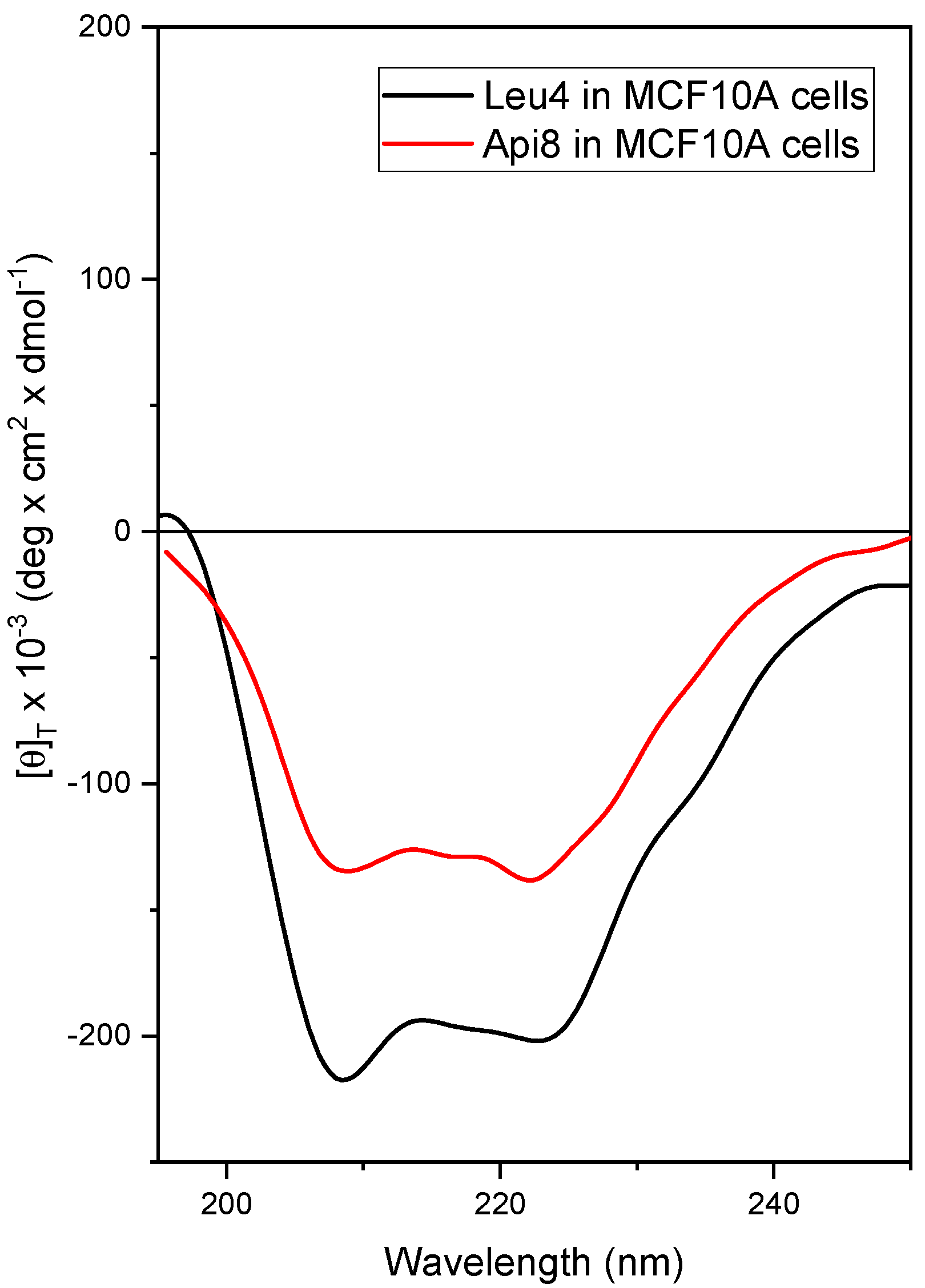
2.3. Conformational Study by Circular Dichroism (CD)
2.4. Membrane Leakage Study
2.5. In Vitro Cytotoxicity of Trichogin GA IV Analogs in Breast Cells
| Acronym | IC50 a (µM) and Target Cancer Cell Lines b | Hemolysis/ Healthy Cells | References | ||||
|---|---|---|---|---|---|---|---|
| HeLa | A549 | A431 | T67 | Ov-HL | |||
| TRIC | 4–8 | 4–6 | 4–6 | 2 | n.d. c | nonhemolytic | [29,30] |
| Api8-NH2 | - d | this work | |||||
| Api8-Lol e | >20 f | n.d. | n.d. | 13 | n.d. | nonhemolytic f/ nontoxic g | [29] |
| Leu4-NH2 | - | this work | |||||
| Leu4-Lol e | >20 | nt | nt | >20 | n.d. | nonhemolytic/ nontoxic | [29,68] |
| K2569-Lol | - | this work | |||||
| K259-NH2 | - | this work | |||||
| K259-Lol | 6 | 8 | 5 | n.d. | n.d. | nonhemolytic | [68] |
| K25-Lol | 7 | 10 | 5 | n.d. | 7–13 | nonhemolytic <16 µM | [68] |
| K56-Lol | 10 | 16 | 8 | 7 | n.d. | nonhemolytic/ nontoxic | [29,68] |
| K6-NH2 | n.d. | n.d. | n.d. | n.d. | 7–13 | n.d. | [28] |
| K6-Lol e | 2–8 | 8 | 5 | 4 | n.d. | nonhemolytic <16 µM | [28,29,30] |
| K2-NH2 | - | this work | |||||
| K2-Lol e | 7 | 8 | 6 | n.d. | n.d. | nonhemolytic | [30,68] |
| Api8-FITC | - | this work | |||||
| Leu4-FITC | - | this work | |||||
| Api8-SH | - | this work | |||||
| Leu4-SH | - | this work | |||||
2.6. In Vitro Uptake of Selected Peptaibols
2.7. Uptake of GNPs Decorated with Peptaibols in Breast Cancer Cells and Macrophages
3. Discussion
4. Materials and Methods
4.1. Peptide Synthesis
4.2. Circular Dichroism
4.3. Gold Nanoparticles Synthesis and Characterization
4.4. Leakage
4.5. Cell Lines
4.6. In Vitro Cytotoxicity of Peptaibols and Peptaibol-Decorated GNPs
4.7. Cellular Uptake of FITC-Conjugated Peptides
4.8. Cellular Uptake of GNPs Measured by TEM
4.9. Cellular Uptake of GNPs Measured by Atomic Absorption Spectroscopy
5. Conclusions and Future Perspectives
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Conflicts of Interest
References
- Sorolla, A.; Wang, E.; Golden, E.; Duffy, C.; Henriques, S.T.; Redfern, A.D.; Blancafort, P. Precision Medicine by Designer Interference Peptides: Applications in Oncology and Molecular Therapeutics. Oncogene 2020, 39, 1167–1184. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Al Musaimi, O.; Al Shaer, D.; de la Torre, B.G.; Albericio, F. 2017 FDA Peptide Harvest. Pharmaceuticals 2018, 11, 42. [Google Scholar] [CrossRef] [Green Version]
- Muttenthaler, M.; King, G.F.; Adams, D.J.; Alewood, P.F. Trends in peptide drug discovery. Nat. Rev. 2021, 20, 309–325. [Google Scholar] [CrossRef] [PubMed]
- Kaarsholm, N.C. Peptide Drug Design for Diabetes and Related Metabolic Diseases. In Translational Research Methods in Diabetes, Obesity, and Nonalcoholic Fatty Liver Disease; Krentz, A., Weyer, C., Hompesch, M., Eds.; Springer: Cham, Switzerland, 2019. [Google Scholar] [CrossRef]
- Baggio, L.L.; Drucker, D.J. Glucagon-like peptide-1 receptor co-agonists for treating metabolic disease. Mol. Metab. 2021, 46, 101090. [Google Scholar] [CrossRef] [PubMed]
- Lau, J.L.; Dunn, M.K. Therapeutic Peptides: Historical Perspectives, Current Development Trends, and Future Directions. Bioorg. Med. Chem. 2018, 26, 2700–2707. [Google Scholar] [CrossRef] [PubMed]
- Brogden, K.A. Antimicrobial peptides: Pore formers or metabolic inhibitors in bacteria? Nat. Rev. Microbiol. 2005, 3, 238–250. [Google Scholar] [CrossRef] [PubMed]
- Räder, A.F.B.; Weinmüller, M.; Reichart, F.; Schumacher-Klinger, A.; Merzbach, S.; Gilon, C.; Hoffman, A.; Kessler, H. Orally Active Peptides: Is There a Magic Bullet? Angew. Chem. Int. Ed. 2018, 57, 14414. [Google Scholar] [CrossRef]
- Stoppacher, N.; Neumann, N.K.N.; Burgstaller, L.; Zeilinger, S.; Degenkolb, T.; Brückner, H.; Schuhmacher, R. The comprehensive peptaibiotics database. Chem. Biodivers. 2013, 10, 734–743. [Google Scholar] [CrossRef]
- Rebuffat, S.; Goulard, C.; Bodo, B.; Roquebert, M.-F. The peptaibol antibiotics from Trichoderma soil fungi: Structural diversity and membrane properties. Recent Res. Devel. Org. Bioorg. Chem. 1999, 3, 65–91. [Google Scholar]
- Toniolo, C.; Brückner, H. Peptaibiotics: Fungal Peptidescontaining α-Dialkyl α-Amino Acids; Wiley-VCD: Weinheim, Germany; Zürich, Switzerland, 2009. [Google Scholar]
- Lizio, M.G.; Campana, M.; De Poli, M.; Jefferies, D.F.; Cullen, W.; Andrushchenko, V.; Chmel, N.P.; Bouř, P.; Khalid, S.; Clayden, J.; et al. Insight into the Mechanism of Action and Peptide-Membrane Interactions of Aib-Rich Peptides: Multitechnique Experimental and Theoretical Analysis. ChemBioChem 2021, 22, 1656–1667. [Google Scholar] [CrossRef] [PubMed]
- Szekeres, A.; Leitgeb, B.; Kredics, L.; Antal, Z.; Hatvani, L.; Manczinger, L.; Vágvölgyi, C. Peptaibols and related peptaibiotics of Trichoderma. A review. Acta Microbiol. Immunol. Hung. 2005, 52, 137–168. [Google Scholar] [CrossRef] [PubMed]
- Toniolo, C.; Crisma, M.; Formaggio, F.; Peggion, C.; Epand, R.F.; Epand, R.M. Lipopeptaibols, a novel family of membrane active, antimicrobial peptides. Cell Mol. Life Sci. 2001, 58, 1179–1188. [Google Scholar] [CrossRef] [PubMed]
- Xiao, D.; Zhang, M.; Wu, P.; Li, T.; Li, W.; Zhang, L.; Yue, Q.; Chen, X.; Wei, X.; Xu, Y.; et al. Halovirs I–K, antibacterial and cytotoxic lipopeptaibols from the plant pathogenic fungus Paramyrothecium roridum NRRL 2183. J. Antibiot. 2022, 75, 247–257. [Google Scholar] [CrossRef]
- Auvin-Guette, C.; Rebuffat, S.; Prigent, Y.; Bodo, B. Trichogin A IV, an 11-residue lipopeptaibol from Trichoderma longibrachiatum. J. Am. Chem. Soc. 1992, 114, 2170–2174. [Google Scholar] [CrossRef]
- Peggion, C.; Formaggio, F.; Crisma, M.; Epand, R.F.; Epand, R.M.; Toniolo, C. Trichogin: A paradigm for lipopeptaibols. J. Pept. Sci. 2003, 9, 679–689. [Google Scholar] [CrossRef]
- Marik, T.; Tyagi, C.; Balázs, D.; Urbán, P.; Szepesi, Á.; Bakacsy, L.; Endr, G.; Rakk, D.; Szekeres, A.; Andersson, M.A.; et al. Structural diversity and bioactivities of peptaibol compounds from the Longibrachiatum clade of the filamentous fungal genus Trichoderma. Front. Microbiol. 2019, 10, 1437. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Marra, R.; Lombardi, N.; d’Errico, G.; Troisi, J.; Scala, G.; Vinale, F.; Woo, S.L.; Bonanomi, G.; Lorito, M. Application of Trichoderma Strains and Metabolites Enhances Soybean Productivity and Nutrient Content. J. Agric. Food Chem. 2019, 67, 1814–1822. [Google Scholar] [CrossRef]
- Tamandegani, P.R.; Marik, T.; Zafari, D.; Balázs, D.; Vágvölgyi, C.; Szekeres, A.; Kredics, L. Changes in peptaibol production of Trichoderma species during in vitro antagonistic interactions with fungal plant pathogens. Biomolecules 2020, 10, 730. [Google Scholar] [CrossRef] [PubMed]
- De Zotti, M.; Sella, L.; Bolzonello, A.; Gabbatore, L.; Peggion, C.; Bortolotto, A.; Elmaghraby, I.; Tundo, S.; Favaron, F. Targeted Amino Acid Substitutions in a Trichoderma Peptaibol Confer Activity against Fungal Plant Pathogens and Protect Host Tissues from Botrytis Cinerea Infection. Int. J. Mol. Sci. 2020, 21, 7521. [Google Scholar] [CrossRef]
- Sella, L.; Govind, R.; Caracciolo, R.; Quarantin, A.; Vu, V.V.; Tundo, S.; Nguyen, H.M.; Favaron, F.; Musetti, R.; De Zotti, M. Transcriptomic and Ultrastructural Analyses of Pyricularia oryzae Treated with Fungicidal Peptaibol Analogs of Trichoderma Trichogin. Front. Microbiol. 2021, 12, 753202. [Google Scholar] [CrossRef]
- Caracciolo, R.; Sella, L.; De Zotti, M.; Bolzonello, A.; Armellin, M.; Trainotti, L.; Favaron, F.; Tundo, S. Efficacy of Trichoderma longibrachiatum Trichogin GA IV Peptaibol analogs against the Black Rot Pathogen Xanthomonas campestris pv. campestris and other Phytopathogenic Bacteria. Microorganisms 2023, 11, 480. [Google Scholar] [CrossRef] [PubMed]
- Baccelli, I.; Luti, S.; Bernardi, R.; Favaron, F.; De Zotti, M.; Sella, L. Water-Soluble Trichogin GA IV-Derived Peptaibols Protect Tomato Plants from Botrytis cinerea Infection with Limited Impact on Plant Defenses. Front. Plant Sci. 2022, 13, 881961. [Google Scholar] [CrossRef] [PubMed]
- Bolzonello, A.; Morbiato, L.; Tundo, S.; Sella, L.; Baccelli, I.; Echeverrigaray, S.; Musetti, R.; De Zotti, M.; Favaron, F. Peptide analogs of a Trichoderma peptaibol effectively control downy mildew in the vineyard. Plant Dis. 2023; Advance online publication. [Google Scholar] [CrossRef] [PubMed]
- De Zotti, M.; Biondi, B.; Peggion, C.; Formaggio, F.; Park, Y.; Hahm, K.-S.; Toniolo, C. Trichogin GA IV: A Versatile Template for the Synthesis of Novel Peptaibiotics. Org. Biomol. Chem. 2012, 10, 1285–1299. [Google Scholar] [CrossRef] [PubMed]
- De Zotti, M.; Biondi, B.; Park, Y.; Hahm, K.S.; Crisma, M.; Toniolo, C.; Formaggio, F. Antimicrobial lipopeptaibol trichogin GA IV: Role of the three Aib residues on conformation and bioactivity. Amino Acids 2012, 43, 1761–1777. [Google Scholar] [CrossRef] [PubMed]
- Casagrande, N.; Borghese, C.; Gabbatore, L.; Morbiato, L.; De Zotti, M.; Aldinucci, D. Analogs of a Natural Peptaibol Exert Anticancer Activity in Both Cisplatin- and Doxorubicin-Resistant Cells and in Multicellular Tumor Spheroids. Int. J. Mol. Sci. 2021, 22, 8362. [Google Scholar] [CrossRef]
- Dalzini, A.; Bergamini, C.; Biondi, B.; De Zotti, M.; Panighel, G.; Fato, R.; Peggion, C.; Bortolus, M.; Maniero, A.L. The Rational Search for Selective Anticancer Derivatives of the Peptide Trichogin GA IV: A Multi-Technique Biophysical Approach. Sci. Rep. 2016, 6, 24000. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tavano, R.; Malachin, G.; De Zotti, M.; Peggion, C.; Biondi, B.; Formaggio, F.; Papini, E. The peculiar N- and C-termini of trichogin GA IV are needed for membrane interaction and human cell death induction at doses lacking antibiotic activity. Biochim. Biophys. Acta Biomembr. 2015, 1848, 134–144. [Google Scholar] [CrossRef] [PubMed]
- Yin, L.; Duan, J.J.; Bian, X.W.; Yu, S.-C. Triple-negative breast cancer molecular subtyping and treatment progress. Breast Cancer Res. 2020, 22, 61. [Google Scholar] [CrossRef]
- Almansour, N.M. Triple-Negative Breast Cancer: A Brief Review About Epidemiology, Risk Factors, Signaling Pathways, Treatment and Role of Artificial Intelligence. Front. Mol. Biosci. 2022, 9, 836417. [Google Scholar] [CrossRef]
- Bianchini, G.; De Angelis, C.; Licata, L.; Gianni, L. Treatment landscape of triple-negative breast cancer—Expanded options, evolving needs. Nat. Rev. Clin. Oncol. 2022, 19, 91–113. [Google Scholar] [CrossRef] [PubMed]
- Wu, Q.; Siddharth, S.; Sharma, D. Triple Negative Breast Cancer: A Mountain Yet to Be Scaled Despite the Triumphs. Cancers 2021, 13, 3697. [Google Scholar] [CrossRef] [PubMed]
- Hou, K.; Ning, Z.; Chen, H.; Wu, Y. Nanomaterial Technology and Triple Negative Breast Cancer. Front. Oncol. 2022, 11, 828810. [Google Scholar] [CrossRef]
- Ganesan, K.; Wang, Y.; Gao, F.; Liu, Q.; Zhang, C.; Li, P.; Zhang, J.; Chen, J. Targeting Engineered Nanoparticles for Breast Cancer Therapy. Pharmaceutics 2021, 13, 1829. [Google Scholar] [CrossRef] [PubMed]
- Shan, X.; Gong, X.; Li, J.; Wen, J.; Li, Y.; Zhang, Z. Current approaches of nanomedicines in the market and various stage of clinical translation. Acta Pharm. Sin. B 2022, 12, 3028–3048. [Google Scholar] [CrossRef]
- Mitchell, M.J.; Billingsley, M.M.; Haley, R.M.; Wechsler, M.E.; Peppas, N.A.; Langer, R. Engineering precision nanoparticles for drug delivery. Nat. Rev. Drug Discov. 2021, 20, 101–124. [Google Scholar] [CrossRef]
- Crivellaro, S.; Guadagnini, A.; Arboleda, D.M.; Schinca, D.; Amendola, V. A system for the synthesis of nanoparticles by laser ablation in liquid that is remotely controlled with PC or smartphone. Rev. Sci. Instrum. 2019, 90, 033902. [Google Scholar] [CrossRef] [PubMed]
- Petersen, S.; Barcilowski, S. Conjugation Efficiency of Laser-Based Bioconjugation of Gold Nanoparticles with Nucleic Acids. J. Phys. Chem. C 2009, 113, 19830–19835. [Google Scholar] [CrossRef]
- Mutisya, S.; Franzel, L.; Barnstein, B.O.; Faber, T.W.; Ryan, J.J.; Bertino, M.F. Comparison of in situ and ex situ bioconjugation of Au nanoparticles generated by laser ablation. Appl. Surf. Sci. 2013, 264, 27–30. [Google Scholar] [CrossRef]
- Amendola, V.; Meneghetti, M. Controlled size manipulation of free gold nanoparticles by laser irradiation and their facile bioconjugation. J. Mater. Chem. 2007, 17, 4705–4710. [Google Scholar] [CrossRef]
- Bravin, C.; Amendola, V. Wide range detection of C-Reactive protein with a homogeneous immunofluorimetric assay based on cooperative fluorescence quenching assisted by gold nanoparticles. Biosens. Bioelectron. 2020, 169, 112591. [Google Scholar] [CrossRef] [PubMed]
- Zhang, D.; Gökce, B.; Barcikowski, S. Laser Synthesis and Processing of Colloids: Fundamentals and Applications. Chem. Rev. 2017, 117, 3990–4103. [Google Scholar] [CrossRef]
- Subirós-Funosas, R.; Prohens, R.; Barbas, R.; El-Faham, A.; Albericio, F. Oxyma: An Efficient Additive for Peptide Synthesis to Replace the Benzotriazole-Based HOBt and HOAt with a Lower Risk of Explosion. Chem. Eur. J. 2009, 15, 9394–9403. [Google Scholar] [CrossRef] [PubMed]
- Musaimi, O.A.; de la Torre, B.G.; Albericio, F. Greening Fmoc/TBu Solid-Phase Peptide Synthesis. Green Chem. 2020, 22, 996–1018. [Google Scholar] [CrossRef]
- Pawlas, J.; Rasmussen, J.H. ReGreen SPPS: Enabling circular chemistry in environmentally sensible solid-phase peptide synthesis. Green Chem. 2019, 21, 5990–5998. [Google Scholar] [CrossRef]
- Mulder, K.C.; Lima, L.A.; Miranda, V.J.; Dias, S.C.; Franco, O.L. Current scenario of peptide-based drugs: The key roles of cationic antitumor and antiviral peptides. Front. Microbiol. 2013, 4, 321. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, Z.; Ruan, J.; Zhuang, X. Effective Capture of Circulating Tumor Cells from an S180-Bearing Mouse Model Using Electrically Charged Magnetic Nanoparticles. J. Nanobiotechnol. 2019, 17, 59. [Google Scholar] [CrossRef]
- Szlasa, W.; Zendran, I.; Zalesińska, A.; Tarek, M.; Kulbacka, J. Lipid Composition of the Cancer Cell Membrane. J. Bioenerg. Biomembr. 2020, 52, 321–342. [Google Scholar] [CrossRef] [PubMed]
- Ferrer-Gago, F.J.; Koh, L.; Lane, D. Functionalized Resins for the Synthesis of Peptide Alcohols. Chem. Eur. J. 2020, 26, 379. [Google Scholar] [CrossRef]
- Ferrer-Gago, F.J.; Koh, L. Methods and Approaches for the Solid-Phase Synthesis of Peptide Alcohols. ChemPlusChem 2020, 85, 641. [Google Scholar] [CrossRef]
- Akrami, M.; Balalaie, S.; Hosseinkhani, S.; Alipour, M.; Salehi, F.; Bahador, A.; Haririan, I. Tuning the anticancer activity of a novel pro-apoptotic peptide using gold nanoparticle platforms. Sci. Rep. 2016, 6, 31030. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Amendola, V.; Meneghetti, M. Size Evaluation of Gold Nanoparticles by UV−vis Spectroscopy. J. Phys. Chem. C 2009, 113, 4277–4285. [Google Scholar] [CrossRef]
- Bravin, C.; Amendola, V. Plasmonic Absorption in Antigen-Induced Aggregated Gold Nanoparticles: Toward a Figure of Merit for Optical Nanosensors. ACS Appl. Nano Mater. 2022, 5, 578–586. [Google Scholar] [CrossRef]
- Fasman, G.D. (Ed.) Circular Dichroism and the Conformational Analysis of Biomolecules; Plenum Press: New York, NY, USA, 1996. [Google Scholar]
- Whitmore, L.; Wallace, B.A. Handbook of Biologically Active Peptides; Kastin, A.J., Ed.; Academic Press: Cambridge, MA, USA, 2006. [Google Scholar] [CrossRef]
- Gatto, E.; Porchetta, A.; Stella, L.; Guryanov, I.; Formaggio, F.; Toniolo, C.; Kaptein, B.; Broxterman, Q.; Venanzi, M. Conformational effects on the electron-transfer efficiency in peptide foldamers based on alpha,alpha-disubstituted glycyl residues. Chem. Biodivers. 2008, 5, 1263–1278. [Google Scholar] [CrossRef] [PubMed]
- Cho, J.-I.; Tanaka, M.; Sato, S.; Kinbara, K.; Aida, T. Oligo(4-aminopiperidine-4-carboxylic acid): An unusual basic oligopeptide with an acid-induced helical conformation. J. Am. Chem. Soc. 2010, 132, 13176–13178. [Google Scholar] [CrossRef]
- Toniolo, C.; Peggion, C.; Crisma, M.; Formaggio, F.; Shui, X.; Eggleston, D.S. Structure determination of racemic trichogin A IV using centrosymmetric crystals. Nat. Struct. Biol. 1994, 1, 908–914. [Google Scholar] [CrossRef]
- Toniolo, C.; Crisma, M.; Formaggio, F.; Peggion, C.; Monaco, V.; Goulard, C.; Rebuffat, S.; Bodo, B. Effect of Nα-acyl chain length on the membrane modifying properties of synthetic analogs of the lipopeptaibol trichogin GA IV. J. Am. Chem. Soc. 1996, 118, 4952–4958. [Google Scholar] [CrossRef]
- Toniolo, C.; Polese, A.; Formaggio, F.; Crisma, M.; Kamphuis, J. Circular Dichroism Spectrum of a Peptide 310-Helix. J. Am. Chem. Soc. 1996, 118, 2744–2745. [Google Scholar] [CrossRef]
- Avitabile, C.; D’Andrea, L.D.; Romanelli, A. Circular Dichroism studies on the interactions of antimicrobial peptides with bacterial cells. Sci. Rep. 2014, 4, 4293. [Google Scholar] [CrossRef] [Green Version]
- De Zotti, M.; Biondi, B.; Formaggio, F.; Toniolo, C.; Stella, L.; Park, Y.; Hahm, K.-S. Trichogin GA IV: An antibacterial and protease-resistant peptide. J. Pept. Sci. 2009, 15, 615–619. [Google Scholar] [CrossRef]
- Manning, M.C.; Woody, R.W. Theoretical CD studies of polypeptide helices: Examination of important electronic and geometric factors. Biopolymers 1991, 31, 569–586. [Google Scholar] [CrossRef] [PubMed]
- el Hajji, M.; Rebuffat, S.; Le Doan, T.; Klein, G.; Satre, M.; Bodo, B. Interaction of trichorzianines A and B with model membranes and with the amoeba. Dictyostelium. Biochim. Biophys. Acta 1989, 978, 97–104. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Lykotrafitis, G. Erythrocyte membrane model with explicit description of the lipid bilayer and the spectrin network. Biophys. J. 2014, 107, 642–653. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tavano, R.; Malachin, G.; De Zotti, M.; Peggion, C.; Biondi, B.; Formaggio, F.; Papini, E. Comparison of Bactericidal and Cytotoxic Activities of Trichogin Analogs. Data Brief 2016, 6, 359–367. [Google Scholar] [CrossRef] [PubMed]
- Nie, S. Understanding and overcoming major barriers in cancer nanomedicine. Nanomedicine 2010, 5, 523–528. [Google Scholar] [CrossRef] [Green Version]
- Tornesello, A.L.; Borrelli, A.; Buonaguro, L.; Buonaguro, F.M.; Tornesello, M.L. Antimicrobial Peptides as Anticancer Agents: Functional Properties and Biological Activities. Molecules 2020, 25, 2850. [Google Scholar] [CrossRef]
- Deslouches, B.; Di, Y.P. Antimicrobial peptides with selective antitumor mechanisms: Prospect for anticancer applications. Oncotarget 2017, 8, 46635–46651. [Google Scholar] [CrossRef] [Green Version]
- Yamaguchi, H.; Kodama, H.; Osada, S.; Kato, F.; Jelokhani-Niaraki, M.; Kondo, M. Effect of α,α-dialkyl amino acids on the protease resistance of peptides. Biosci. Biotechnol. Biochem. 2003, 67, 2269–2272. [Google Scholar] [CrossRef] [Green Version]
- Sadowski, J.D.; Murray, J.K.; Tomita, Y.; Gellman, S.H. Exploration of backbone space in foldamers containing α- and β-amino acid residues: Developing protease-resistant oligomers that bind tightly to the BH3-recognition cleft of Bcl-xL. ChemBioChem 2007, 8, 903–916. [Google Scholar] [CrossRef]
- Mohtar, N.; Parumasivam, T.; Gazzali, A.M.; Tan, C.S.; Tan, M.L.; Othman, R.; Fazalul Rahiman, S.S.; Wahab, H.A. Advanced Nanoparticle-Based Drug Delivery Systems and Their Cellular Evaluation for Non-Small Cell Lung Cancer Treatment. Cancers 2021, 13, 3539. [Google Scholar] [CrossRef]
- Cai, D.; Gao, W.; Li, Z.; Zhang, Y.; Xiao, L.; Xiao, Y. Current Development of Nano-Drug Delivery to Target Macrophages. Biomedicines 2022, 10, 1203. [Google Scholar] [CrossRef] [PubMed]
- Bastús, N.G.; Sánchez-Tilló, E.; Pujals, S.; Farrera, C.; López, C.; Giralt, E.; Celada, A.; Lloberas, J.; Puntes, V. Homogeneous conjugation of peptides onto gold nanoparticles enhances macrophage response. ACS Nano 2009, 3, 1335–1344. [Google Scholar] [CrossRef] [PubMed]
- McHale, A.P.; Callan, J.F.; Nomikou, N.; Fowley, C.; Callan, B. Sonodynamic Therapy: Concept, Mechanism and Application to Cancer Treatment. Adv. Exp. Med. Biol. 2016, 880, 429–450. [Google Scholar] [CrossRef] [PubMed]





| Acronym | Sequence a |
|---|---|
| TRIC b | Oct-Aib1-Gly-Leu-Aib4-Gly-Gly-Leu-Aib8-Gly-Ile-Lol11 |
| Api8-NH2 | Oct-Aib1-Gly-Leu-Aib4-Gly-Gly-Leu-Api8-Gly-Ile-Leu11-NH2 |
| Leu4-NH2 | Oct-Aib1-Gly-Leu-Leu4-Gly-Gly-Leu-Aib8-Gly-Ile-Leu11-NH2 |
| K2569-Lol | Oct-Aib1-Lys-Leu-Aib4-Lys-Lys-Leu-Aib8-Lys-Ile-Lol11 |
| K259-NH2 | Oct-Aib1-Lys-Leu-Aib4-Lys-Gly-Leu-Aib8-Lys-Ile-Leu11-NH2 |
| K259-Lol | Oct-Aib1-Lys-Leu-Aib4-Lys-Gly-Leu-Aib8-Lys-Ile-Lol11 |
| K25-Lol | Oct-Aib1-Lys-Leu-Aib4-Lys-Gly-Leu-Aib8-Gly-Ile-Lol11 |
| K56-Lol | Oct-Aib1-Gly-Leu-Aib4-Lys-Lys-Leu-Aib8-Gly-Ile-Lol11 |
| K6-NH2 | Oct-Aib1-Gly-Leu-Aib4-Gly-Lys-Leu-Aib8-Gly-Ile-Leu11-NH2 |
| K2-NH2 | Oct-Aib1-Lys-Leu-Aib4-Gly-Gly-Leu-Aib8-Gly-Ile-Leu11-NH2 |
| Api8-FITC | Oct-Aib1-Gly-Leu-Aib4-Gly-Gly-Leu-Api8-Gly-Ile-Leu11-NH-(CH2)2-NH-FITC |
| Leu4-FITC | Oct-Aib1-Gly-Leu-Leu4-Gly-Gly-Leu-Aib8-Gly-Ile-Leu11-NH-(CH2)2-NH-FITC |
| Api8-SH | Oct-Aib1-Gly-Leu-Aib4-Gly-Gly-Leu-Api8-Gly-Ile-Leu11-NH-(CH2)2-NH-Lipoyl |
| Leu4-SH | Oct-Aib1-Gly-Leu-Leu4-Gly-Gly-Leu-Aib8-Gly-Ile-Leu11-NH-(CH2)2-NH-Lipoyl |
| Acronym | MDA-MB-231 | MCF-10A |
|---|---|---|
| Api8-NH2 | >20 | >20 |
| Leu4-NH2 | >20 | >20 |
| Leu4-Lol | >20 | >20 |
| K2569-Lol | 12 | 14 |
| K259-NH2 | 13 | 17 |
| K259-Lol | 10 | 15 |
| K25-Lol | 9 | 16 |
| K56 | 12 | 18 |
| K6-NH2 | 8 | 15 |
| K2-NH2 | 9 | 17 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Moret, F.; Menilli, L.; Milani, C.; Di Cintio, G.; Dalla Torre, C.; Amendola, V.; De Zotti, M. Anticancer and Targeting Activity of Phytopharmaceutical Structural Analogs of a Natural Peptide from Trichoderma longibrachiatum and Related Peptide-Decorated Gold Nanoparticles. Int. J. Mol. Sci. 2023, 24, 5537. https://doi.org/10.3390/ijms24065537
Moret F, Menilli L, Milani C, Di Cintio G, Dalla Torre C, Amendola V, De Zotti M. Anticancer and Targeting Activity of Phytopharmaceutical Structural Analogs of a Natural Peptide from Trichoderma longibrachiatum and Related Peptide-Decorated Gold Nanoparticles. International Journal of Molecular Sciences. 2023; 24(6):5537. https://doi.org/10.3390/ijms24065537
Chicago/Turabian StyleMoret, Francesca, Luca Menilli, Celeste Milani, Giorgia Di Cintio, Chiara Dalla Torre, Vincenzo Amendola, and Marta De Zotti. 2023. "Anticancer and Targeting Activity of Phytopharmaceutical Structural Analogs of a Natural Peptide from Trichoderma longibrachiatum and Related Peptide-Decorated Gold Nanoparticles" International Journal of Molecular Sciences 24, no. 6: 5537. https://doi.org/10.3390/ijms24065537





