The ENA1 Na+-ATPase Gene Is Regulated by the SPS Sensing Pathway and the Stp1/Stp2 Transcription Factors
Abstract
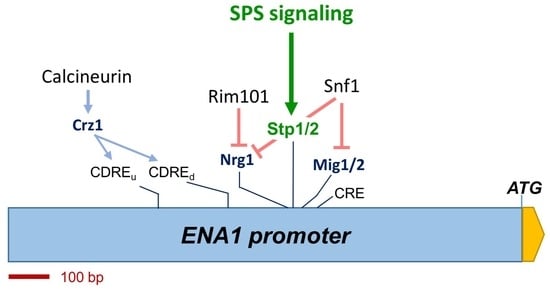
1. Introduction
2. Results
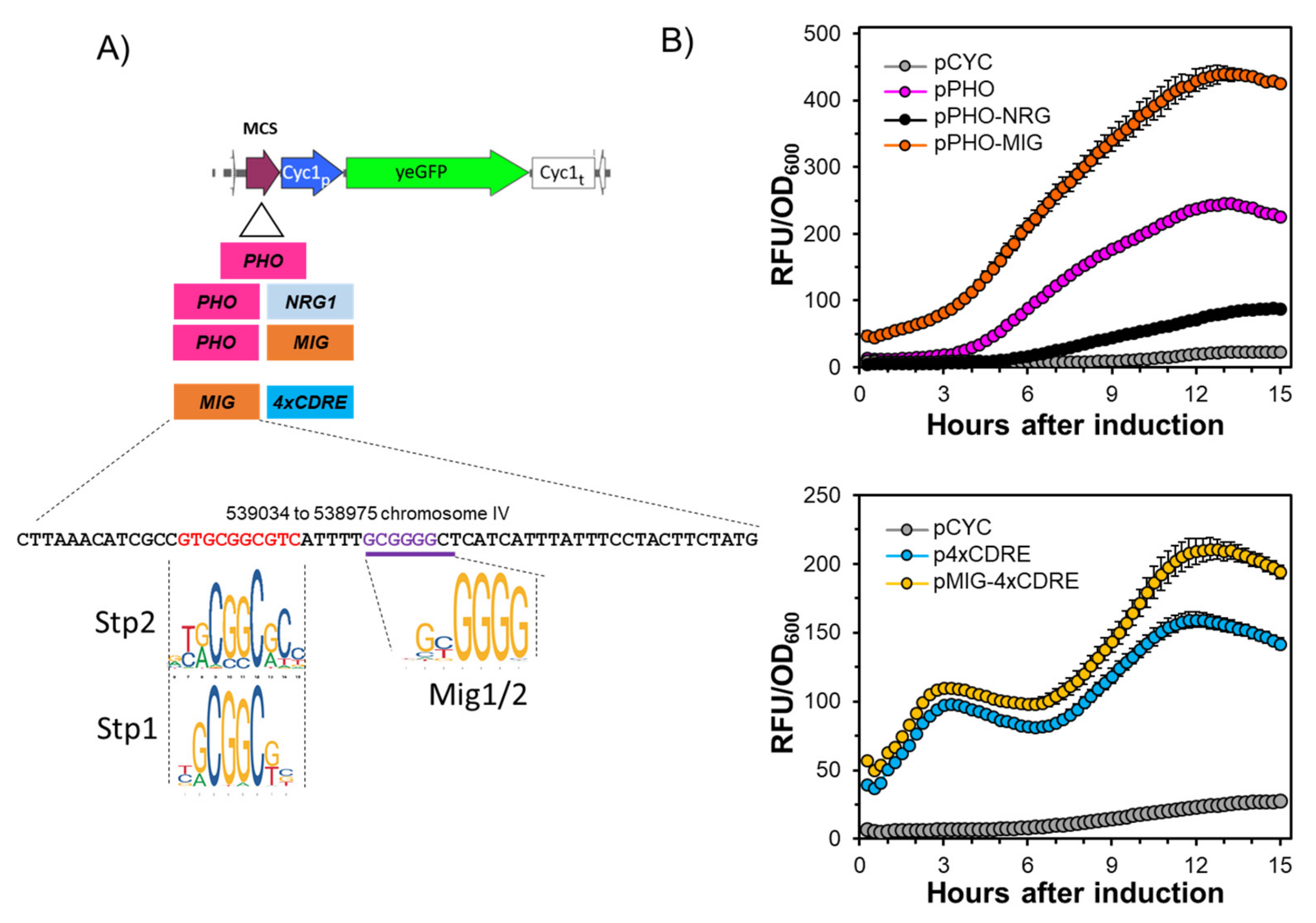
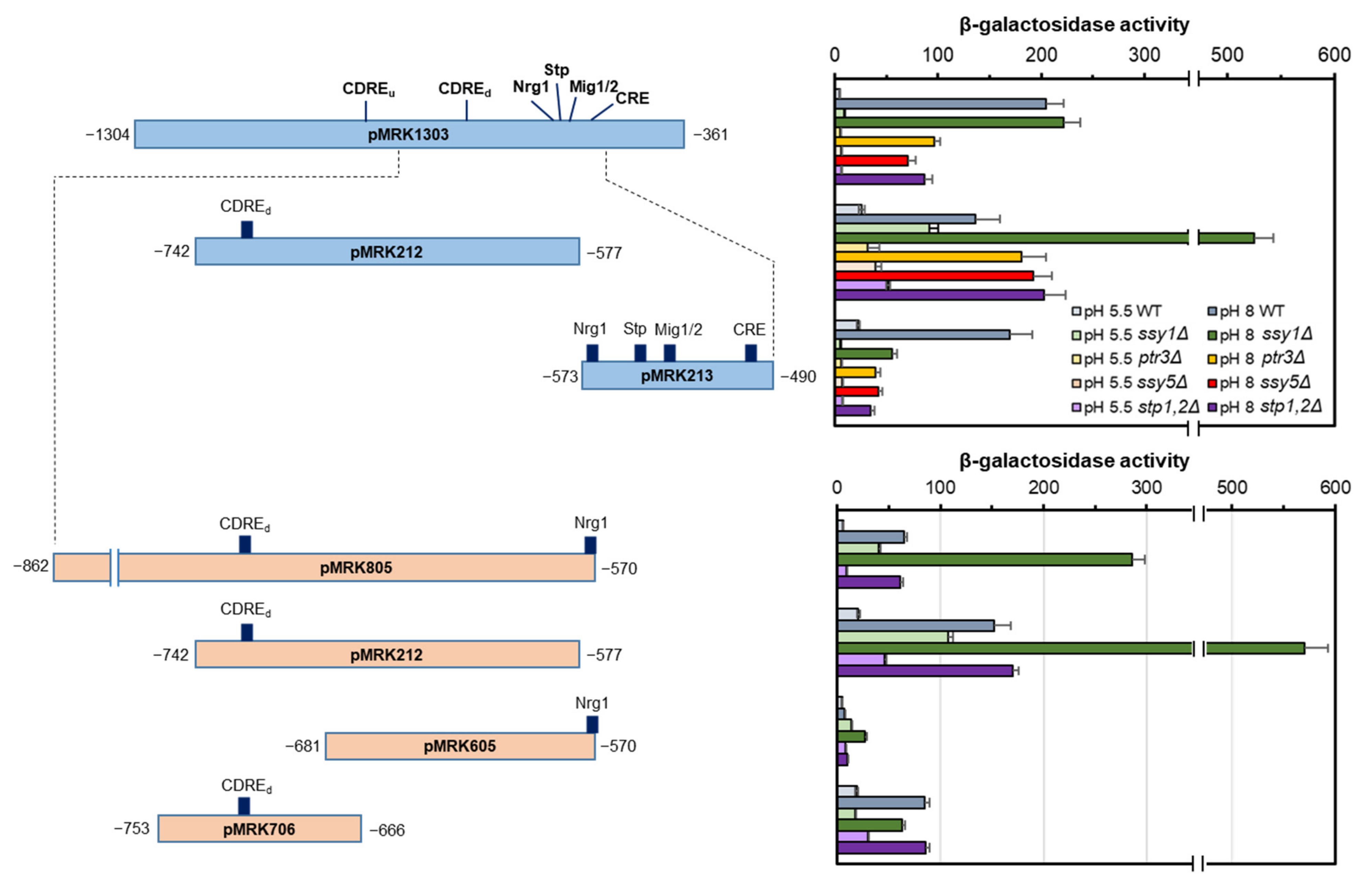
2.1. A Short Region of the ENA1 Promoter Containing the Mig1/2 Consensus Sequence Unexpectedly Increases the Activity of a Hybrid Promoter
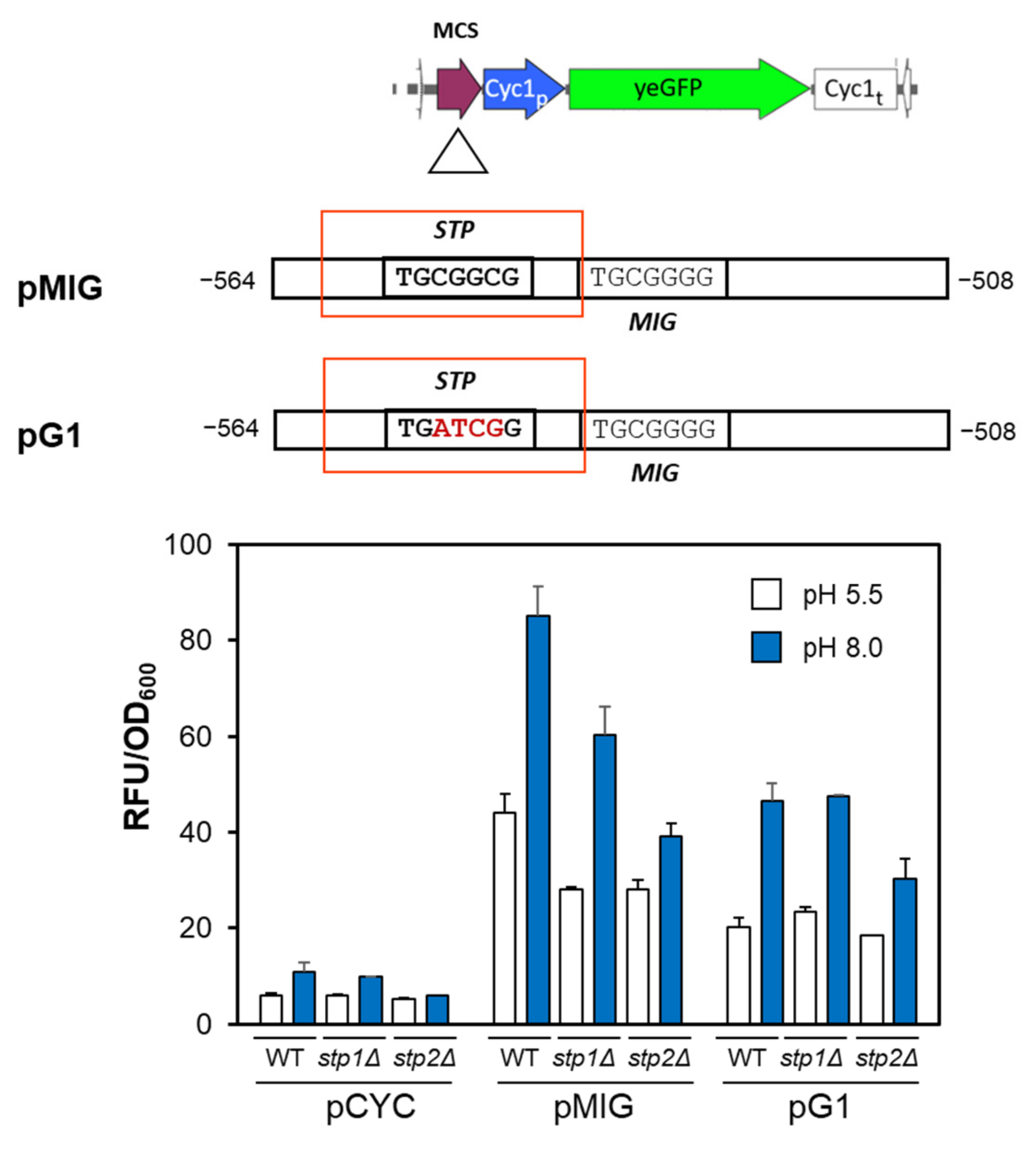
2.2. The Putative Stp1/2 Site Present in the ENA1 Promoter Is Relevant for Transcriptional Induction
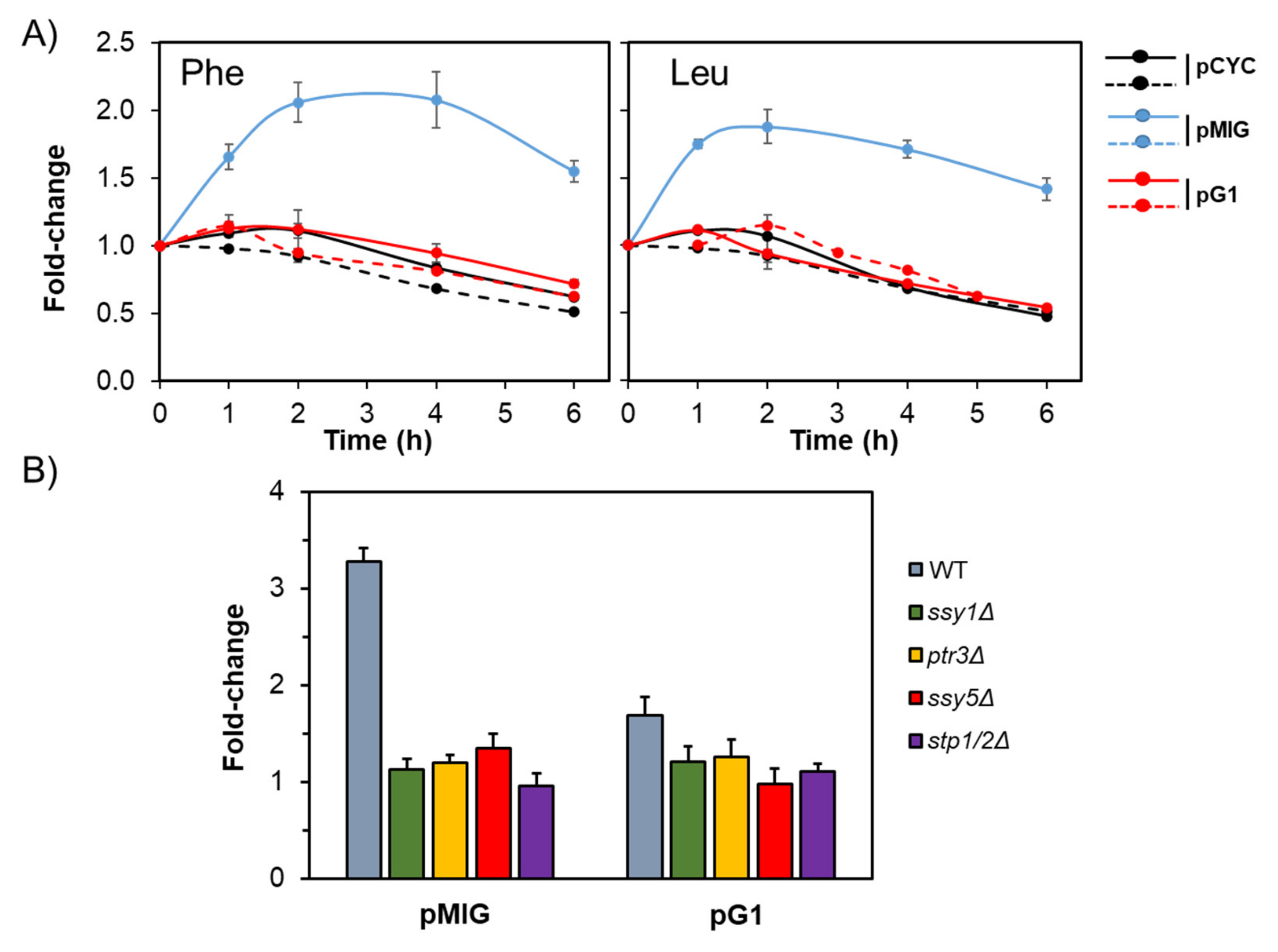
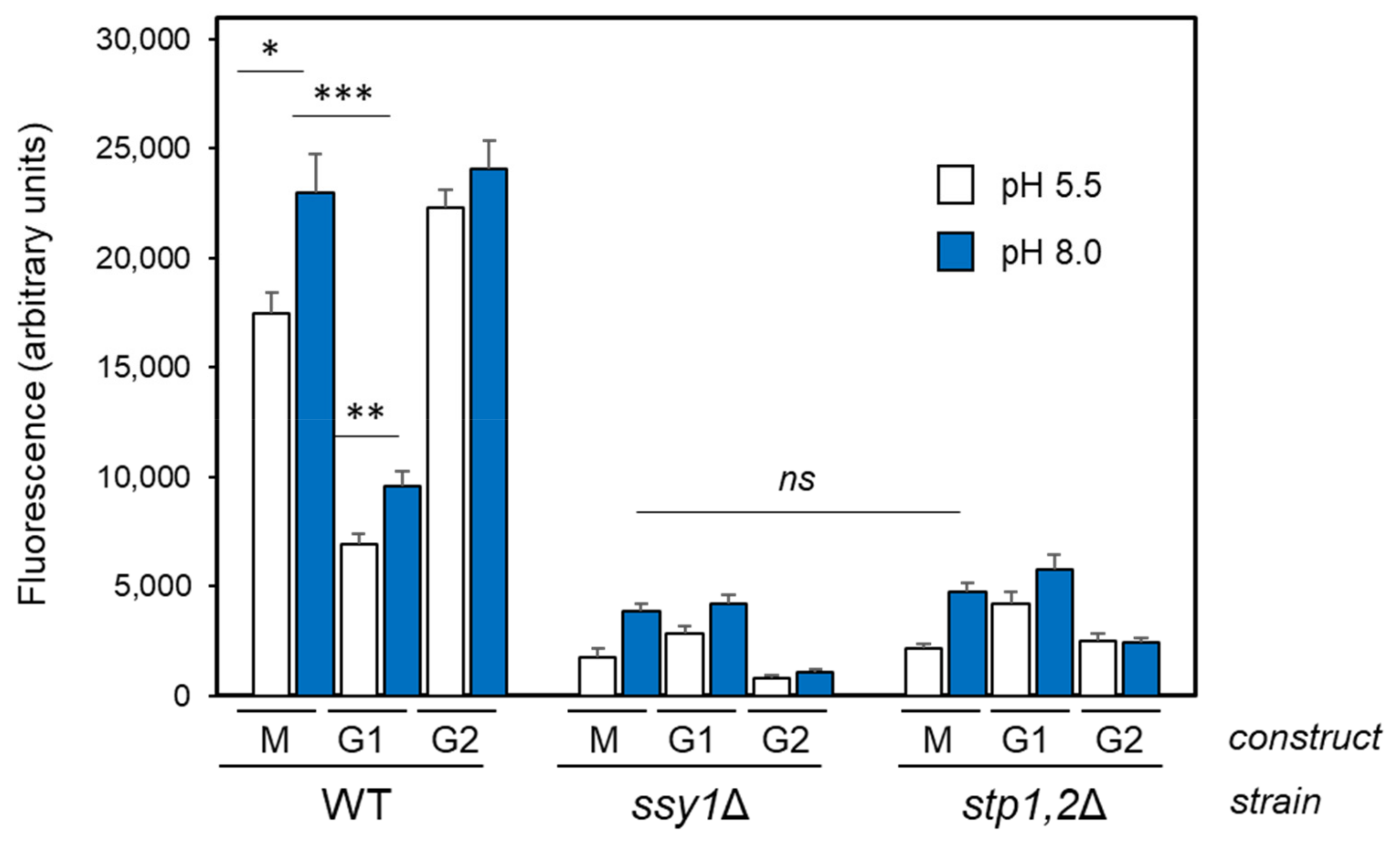
2.3. The Stp1/2 Site in the ENA1 Promoter Responds to the Activation of the SPS Pathway
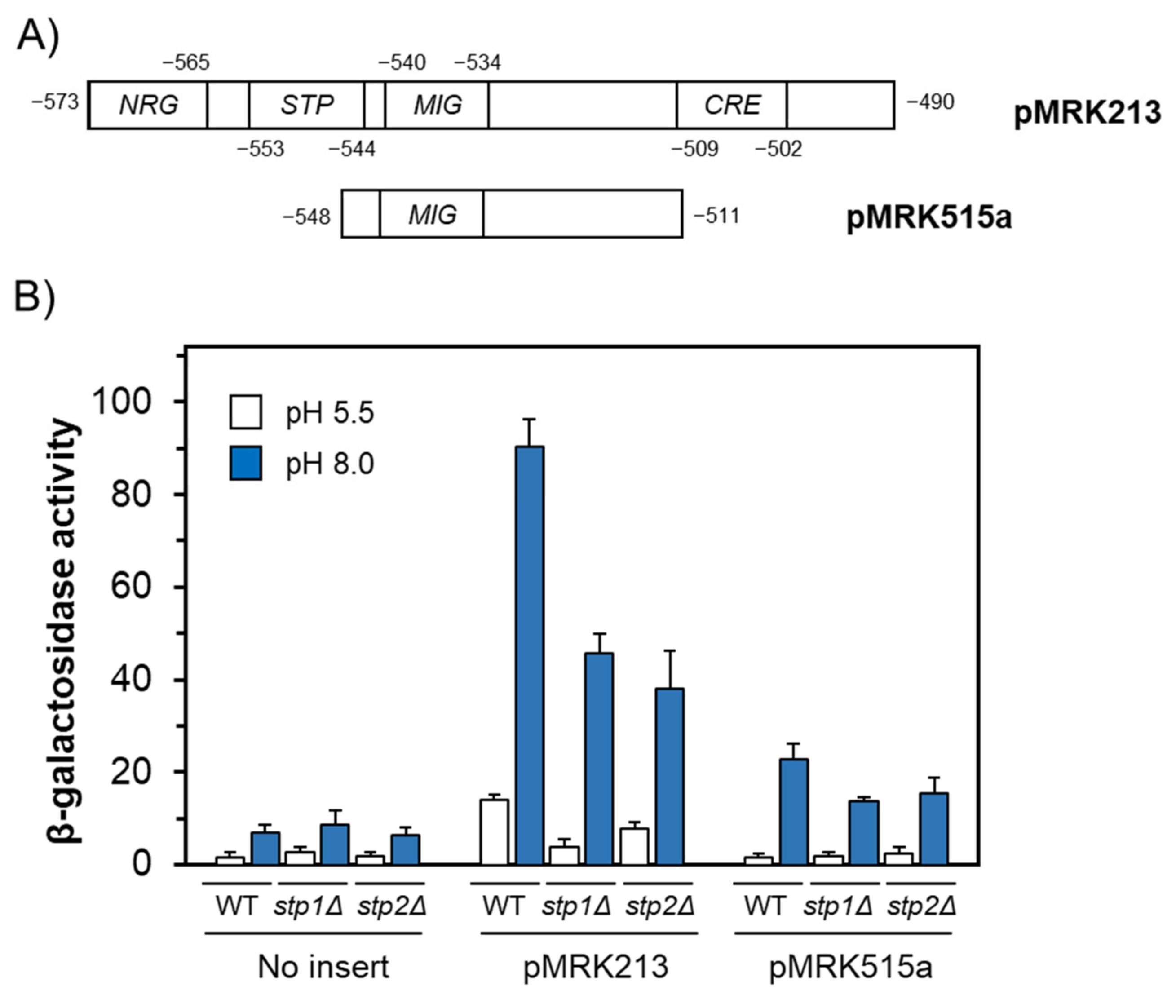
2.4. Non-Homogenous Impact of Mutations in the SPS Pathway on the Expression Driven from the Entire ENA1 Promoter
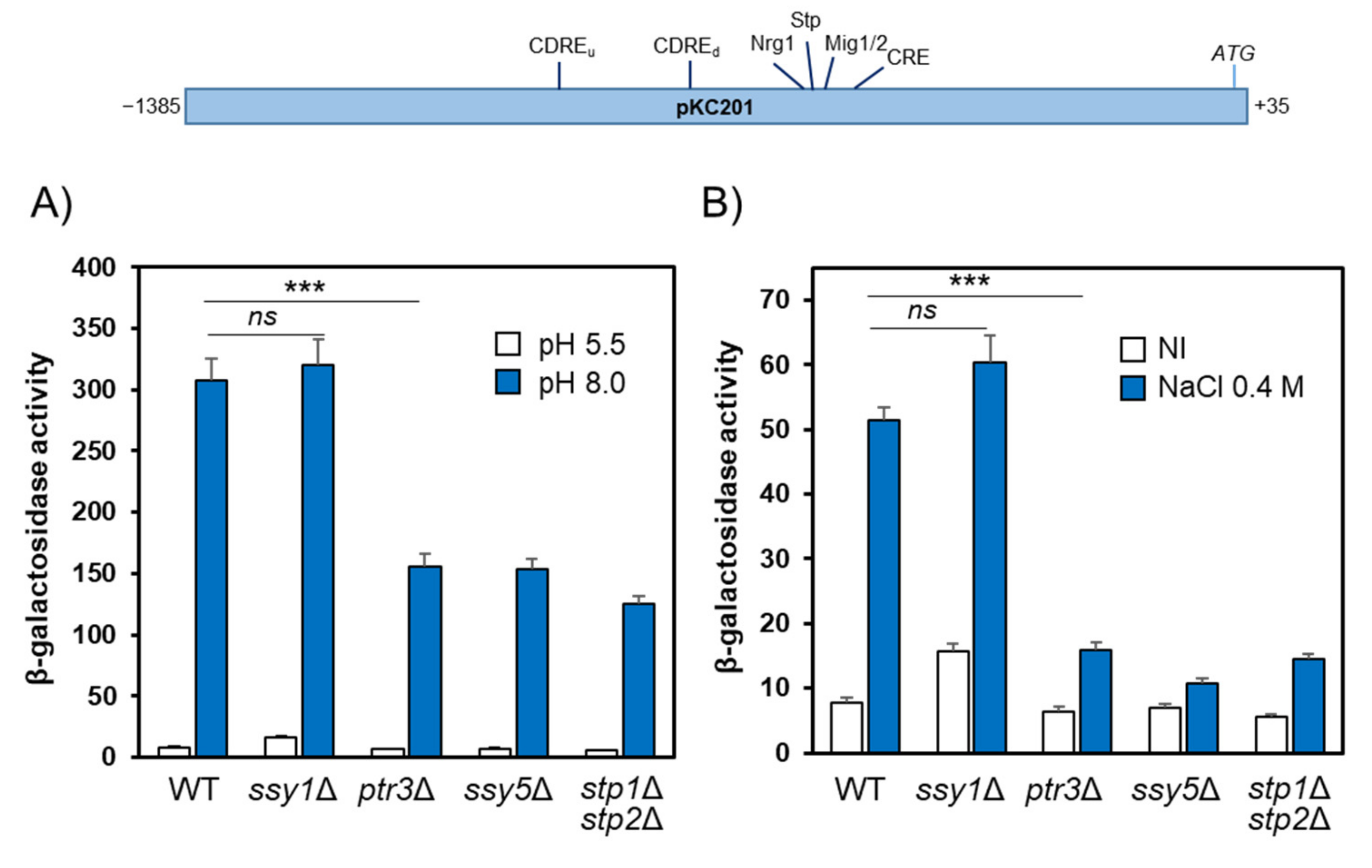
2.5. Functional Mapping of the Alkaline pH Response of the ENA1 Promoter and Its Relationship with the SPS Pathway
2.6. Participation of Stp1/2 in the Expression of Other Genes Responsive to Alkaline pH
3. Discussion
4. Materials and Methods
4.1. Yeast Strains and Culture Media
4.2. Recombinant DNA Techniques and Reporter Construction
4.3. Fluorimetric Determination of yeGFP Production
4.4. Flow Cytometry Analysis
4.5. β-Galactosidase Assays
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Ariño, J. Integrative responses to high pH stress in S. cerevisiae. OMICS 2010, 14, 517–523. [Google Scholar] [CrossRef] [PubMed]
- Serra-Cardona, A.; Canadell, D.; Ariño, J. Coordinate responses to alkaline pH stress in budding yeast. Microb. Cell 2015, 2, 182–196. [Google Scholar] [CrossRef] [PubMed]
- Garciadeblas, B.; Rubio, F.; Quintero, F.J.; Banuelos, M.A.; Haro, R.; Rodriguez-Navarro, A. Differential expression of two genes encoding isoforms of the ATPase involved in sodium efflux in Saccharomyces cerevisiae. Mol. Gen. Genet. 1993, 236, 363–368. [Google Scholar] [CrossRef] [PubMed]
- Mendoza, I.; Rubio, F.; Rodriguez-Navarro, A.; Pardo, J.M. The protein phosphatase calcineurin is essential for NaCl tolerance of Saccharomyces cerevisiae. J. Biol. Chem. 1994, 269, 8792–8796. [Google Scholar] [CrossRef]
- Haro, R.; Garciadeblas, B.; Rodríguez-Navarro, A. A novel P-type ATPase from yeast involved in sodium transport. FEBS Lett. 1991, 291, 189–191. [Google Scholar] [CrossRef]
- Wieland, J.; Nitsche, A.M.; Strayle, J.; Steiner, H.; Rudolph, H.K. The PMR2 gene cluster encodes functionally distinct isoforms of a putative Na+ pump in the yeast plasma membrane. EMBO J. 1995, 14, 3870–3882. [Google Scholar] [CrossRef]
- Posas, F.; Camps, M.; Ariño, J. The PPZ protein phosphatases are important determinants of salt tolerance in yeast cells. J. Biol. Chem. 1995, 270, 13036–13041. [Google Scholar] [CrossRef]
- Marquez, J.A.; Serrano, R. Multiple transduction pathways regulate the sodium-extrusion gene PMR2/ENA1 during salt stress in yeast. FEBS Lett. 1996, 382, 89–92. [Google Scholar] [CrossRef]
- Proft, M.; Struhl, K. Hog1 kinase converts the Sko1-Cyc8-Tup1 repressor complex into an activator that recruits SAGA and SWI/SNF in response to osmotic stress. Mol. Cell 2002, 9, 1307–1317. [Google Scholar] [CrossRef]
- De Nadal, E.; Zapater, M.; Alepuz, P.M.; Sumoy, L.; Mas, G.; Posas, F. The MAPK Hog1 recruits Rpd3 histone deacetylase to activate osmoresponsive genes. Nature 2004, 427, 370–374. [Google Scholar] [CrossRef]
- Stathopoulos, A.M.; Cyert, M.S. Calcineurin acts through the CRZ1/TCN1-encoded transcription factor to regulate gene expression in yeast. Genes Dev. 1997, 11, 3432–3444. [Google Scholar] [CrossRef] [PubMed]
- Matheos, D.P.; Kingsbury, T.J.; Ahsan, U.S.; Cunningham, K.W. Tcn1p/Crz1p, a calcineurin-dependent transcription factor that differentially regulates gene expression in Saccharomyces cerevisiae. Genes Dev. 1997, 11, 3445–3458. [Google Scholar] [CrossRef]
- Mendizabal, I.; Pascual-Ahuir, A.; Serrano, R.; de Larrinoa, I.F. Promoter sequences regulated by the calcineurin-activated transcription factor Crz1 in the yeast ENA1 gene. Mol. Genet.Genom. 2001, 265, 801–811. [Google Scholar] [CrossRef]
- Serrano, R.; Ruiz, A.; Bernal, D.; Chambers, J.R.; Arino, J. The transcriptional response to alkaline pH in Saccharomyces cerevisiae: Evidence for calcium-mediated signalling. Mol. Microbiol. 2002, 46, 1319–1333. [Google Scholar] [CrossRef] [PubMed]
- Ruiz, A.; Yenush, L.; Ariño, J. Regulation of ENA1 Na+-ATPase gene expression by the Ppz1 protein phosphatase is mediated by the calcineurin pathway. Eukaryot. Cell 2003, 2, 937–948. [Google Scholar] [CrossRef] [PubMed]
- Lamb, T.M.; Xu, W.; Diamond, A.; Mitchell, A.P. Alkaline response genes of Saccharomyces cerevisiae and their relationship to the RIM101 pathway. J. Biol. Chem. 2001, 276, 1850–1856. [Google Scholar] [CrossRef] [PubMed]
- Lamb, T.M.; Mitchell, A.P. The transcription factor Rim101p governs ion tolerance and cell differentiation by direct repression of the regulatory genes NRG1 and SMP1 in Saccharomyces cerevisiae. Mol. Cell. Biol. 2003, 23, 677–686. [Google Scholar] [CrossRef]
- Casamayor, A.; Serrano, R.; Platara, M.; Casado, C.; Ruiz, A.; Ariño, J. The role of the Snf1 kinase in the adaptive response of Saccharomyces cerevisiae to alkaline pH stress. Biochem. J. 2012, 444, 39–49. [Google Scholar] [CrossRef]
- Platara, M.; Ruiz, A.; Serrano, R.; Palomino, A.; Moreno, F.; Arino, J. The Transcriptional Response of the Yeast Na+-ATPase ENA1 Gene to Alkaline Stress Involves Three Main Signaling Pathways. J. Biol. Chem. 2006, 281, 36632–36642. [Google Scholar] [CrossRef]
- Serra-Cardona, A.; Petrezsélyová, S.; Canadell, D.; Ramos, J.; Ariño, J. Coregulated expression of the Na+/phosphate Pho89 transporter and Ena1 Na+-ATPase allows their functional coupling under high-pH stress. Mol. Cell. Biol. 2014, 34, 4420–4435. [Google Scholar] [CrossRef]
- Zhang, W.; Du, G.; Zhou, J.; Chen, J. Regulation of Sensing, Transportation, and Catabolism of Nitrogen Sources in Saccharomyces cerevisiae. Microbiol. Mol. Biol. Rev. 2018, 82, e00040-17. [Google Scholar] [CrossRef] [PubMed]
- Klasson, H.; Fink, G.R.; Ljungdahl, P.O. Ssy1p and Ptr3p Are Plasma Membrane Components of a Yeast System That Senses Extracellular Amino Acids. Mol. Cell. Biol. 1999, 19, 5405–5416. [Google Scholar] [CrossRef]
- Bernard, F.; André, B. Genetic analysis of the signalling pathway activated by external amino acids in Saccharomyces cerevisiae. Mol. Microbiol. 2001, 41, 489–502. [Google Scholar] [CrossRef]
- Didion, T.; Regenberg, B.; Jørgensen, M.U.; Kielland-Brandt, M.C.; Andersen, H.A. The permease homologue Ssy1p controls the expression of amino acid and peptide transporter genes in Saccharomyces cerevisiae. Mol. Microbiol. 1998, 27, 643–650. [Google Scholar] [CrossRef]
- Iraqui, I.; Vissers, S.; Bernard, F.; de Craene, J.-O.; Boles, E.; Urrestarazu, A.; André, B. Amino acid signaling in Saccharomyces cerevisiae: A permease-like sensor of external amino acids and F-Box protein Grr1p are required for transcriptional induction of the AGP1 gene, which encodes a broad-specificity amino acid permease. Mol. Cell. Biol. 1999, 19, 989–1001. [Google Scholar] [CrossRef]
- Liu, Z.; Thornton, J.; Spírek, M.; Butow, R.A. Activation of the SPS amino acid-sensing pathway in Saccharomyces cerevisiae correlates with the phosphorylation state of a sensor component, Ptr3. Mol. Cell. Biol. 2008, 28, 551–563. [Google Scholar] [CrossRef] [PubMed]
- Andréasson, C.; Ljungdahl, P.O. Receptor-mediated endoproteolytic activation of two transcription factors in yeast. Genes Dev. 2002, 16, 3158–3172. [Google Scholar] [CrossRef]
- Abdel-Sater, F.; El Bakkoury, M.; Urrestarazu, A.; Vissers, S.; André, B. Amino acid signaling in yeast: Casein kinase I and the Ssy5 endoprotease are key determinants of endoproteolytic activation of the membrane-bound Stp1 transcription factor. Mol. Cell. Biol. 2004, 24, 9771–9785. [Google Scholar] [CrossRef] [PubMed]
- Andréasson, C.; Heessen, S.; Ljungdahl, P.O. Regulation of transcription factor latency by receptor-activated proteolysis. Genes Dev. 2006, 20, 1563–1568. [Google Scholar] [CrossRef]
- Poulsen, P.; Leggio, L.L.; Kielland-Brandt, M.C. Mapping of an internal protease cleavage site in the Ssy5p component of the amino acid sensor of Saccharomyces cerevisiae and functional characterization of the resulting pro- and protease domains by gain-of-function genetics. Eukaryot. Cell 2006, 5, 601–608. [Google Scholar] [CrossRef]
- Wielemans, K.; Jean, C.; Vissers, S.; André, B. Amino acid signaling in yeast: Post-genome duplication divergence of the Stp1 and Stp2 transcription factors. J. Biol. Chem. 2010, 285, 855–865. [Google Scholar] [CrossRef]
- Viladevall, L.; Serrano, R.; Ruiz, A.; Domenech, G.; Giraldo, J.; Barcelo, A.; Arino, J. Characterization of the calcium-mediated response to alkaline stress in Saccharomyces cerevisiae. J. Biol. Chem. 2004, 279, 43614–43624. [Google Scholar] [CrossRef]
- Casado, C.; González, A.; Platara, M.; Ruiz, A.; Ariño, J. The role of the protein kinase A pathway in the response to alkaline pH stress in yeast. Biochem. J. 2011, 438, 523–533. [Google Scholar] [CrossRef]
- Serrano, R.; Martín, H.; Casamayor, A.; Ariño, J. Signaling alkaline pH stress in the yeast Saccharomyces cerevisiae through the Wsc1 cell surface sensor and the Slt2 MAPK pathway. J. Biol. Chem. 2006, 281, 39785–39795. [Google Scholar] [CrossRef]
- Canadell, D.; García-Martínez, J.; Alepuz, P.; Pérez-Ortín, J.E.; Ariño, J. Impact of high pH stress on yeast gene expression: A comprehensive analysis of mRNA turnover during stress responses. Biochim. Biophys. Acta-Gene Regul. Mech. 2015, 1849, 653–664. [Google Scholar] [CrossRef]
- Causton, H.C.; Ren, B.; Koh, S.S.; Harbison, C.T.; Kanin, E.; Jennings, E.G.; Lee, T.I.; True, H.L.; Lander, E.S.; Young, R.A. Remodeling of yeast genome expression in response to environmental changes. Mol. Biol. Cell 2001, 12, 323–337. [Google Scholar] [CrossRef] [PubMed]
- Ljungdahl, P.O. Amino-acid-induced signalling via the SPS-sensing pathway in yeast. Biochem. Soc. Trans. 2009, 37, 242–247. [Google Scholar] [CrossRef]
- Ruiz, A.; Ariño, J. Function and regulation of the Saccharomyces cerevisiae ENA sodium ATPase system. Eukaryot. Cell 2007, 6, 2175–2183. [Google Scholar] [CrossRef] [PubMed]
- Yenush, L. Potassium and Sodium Transport in Yeast. Adv. Exp. Med. Biol. 2016, 892, 187–228. [Google Scholar] [PubMed]
- Alepuz, P.M.; Cunningham, K.W.; Estruch, F. Glucose repression affects ion homeostasis in yeast through the regulation of the stress-activated ENA1 gene. Mol. Microbiol. 1997, 26, 91–98. [Google Scholar] [CrossRef]
- Proft, M.; Serrano, R. Repressors and upstream repressing sequences of the stress-regulated ENA1 gene in Saccharomyces cerevisiae: bZIP protein Sko1p confers HOG-dependent osmotic regulation. Mol. Cell Biol. 1999, 19, 537–546. [Google Scholar] [CrossRef]
- Roque, A.; Petrezsélyová, S.; Serra-Cardona, A.; Ariño, J. Genome-wide recruitment profiling of transcription factor Crz1 in response to high pH stress. BMC Genom. 2016, 17, 662. [Google Scholar] [CrossRef] [PubMed]
- Petrezsélyová, S.; López-Malo, M.; Canadell, D.; Roque, A.; Serra-Cardona, A.; Marqués, M.; Vilaprinyó, E.; Alves, R.; Yenush, L.; Ariño, J. Regulation of the Na+/K+-ATPase Ena1 Expression by Calcineurin/Crz1 under High pH Stress: A Quantitative Study. PLoS ONE 2016, 11, e0158424. [Google Scholar] [CrossRef] [PubMed]
- Forsberg, H.; Gilstring, C.F.; Zargari, A.; Martínez, P.; Ljungdahl, P.O. The role of the yeast plasma membrane SPS nutrient sensor in the metabolic response to extracellular amino acids. Mol. Microbiol. 2001, 42, 215–228. [Google Scholar] [CrossRef] [PubMed]
- Crespo, J.L.; Daicho, K.; Ushimaru, T.; Hall, M.N. The GATA transcription factors GLN3 and GAT1 link TOR to salt stress in Saccharomyces cerevisiae. J. Biol. Chem. 2001, 276, 34441–34444. [Google Scholar] [CrossRef]
- Rodríguez, C.; Galindo, L.R.; Siverio, J.M. Nitrogen-dependent calcineurin activation in the yeast Hansenula polymorpha. Fungal Genet. Biol. 2013, 53, 34–41. [Google Scholar] [CrossRef]
- Ruiz, A.; Serrano, R.; Arino, J. Direct regulation of genes involved in glucose utilization by the calcium/calcineurin pathway. J. Biol. Chem. 2008, 283, 13923–13933. [Google Scholar] [CrossRef] [PubMed]
- Scott, T.D.; Xu, P.; McClean, M.N. Strain-dependent differences in coordination of yeast signalling networks. FEBS J. 2022. [Google Scholar] [CrossRef] [PubMed]
- Dowell, R.D.; Ryan, O.; Jansen, A.; Cheung, D.; Agarwala, S.; Danford, T.; Bernstein, D.A.; Alexander Rolfe, P.; Heisler, L.E.; Chin, B.; et al. Genotype to phenotype: A complex problem. Science 2010, 328, 469. [Google Scholar] [CrossRef]
- Galardini, M.; Busby, B.P.; Vieitez, C.; Dunham, A.S.; Typas, A.; Beltrao, P. The impact of the genetic background on gene deletion phenotypes in Saccharomyces cerevisiae. Mol. Syst. Biol. 2019, 15, e8831. [Google Scholar] [CrossRef]
- Boer, V.M.; Tai, S.L.; Vuralhan, Z.; Arifin, Y.; Walsh, M.C.; Piper, M.D.W.; De Winde, J.H.; Pronk, J.T.; Daran, J.M. Transcriptional responses of Saccharomyces cerevisiae to preferred and nonpreferred nitrogen sources in glucose-limited chemostat cultures. FEMS Yeast Res. 2007, 7, 604–620. [Google Scholar] [CrossRef] [PubMed]
- Adams, A.; Gottschling, D.E.; Kaiser, C.A.; Stearns, T. Methods in Yeast Genetics: A Cold Spring Harbor Laboratory Course Manual; Cold Spring Harbor Laboratory Press: Suffolk County, NY, USA, 1998; ISBN 0879695080. [Google Scholar]
- Santana-Garcia, W.; Castro-Mondragon, J.A.; Padilla-Gálvez, M.; Nguyen, N.T.T.; Elizondo-Salas, A.; Ksouri, N.; Gerbes, F.; Thieffry, D.; Vincens, P.; Contreras-Moreira, B.; et al. RSAT 2022: Regulatory sequence analysis tools. Nucleic Acids Res. 2022, 50, W670–W676. [Google Scholar] [CrossRef] [PubMed]
- Brachmann, C.B.; Davies, A.; Cost, G.J.; Caputo, E.; Li, J.; Hieter, P.; Boeke, J.D. Designer deletion strains derived from Saccharomyces cerevisiae S288C: A useful set of strains and plasmids for PCR-mediated gene disruption and other applications. Yeast 1998, 14, 115–132. [Google Scholar] [CrossRef]
- Winzeler, E.A.; Shoemaker, D.D.; Astromoff, A.; Liang, H.; Anderson, K.; Andre, B.; Bangham, R.; Benito, R.; Boeke, J.D.; Bussey, H.; et al. Functional characterization of the S. cerevisiae genome by gene deletion and parallel analysis. Science 1999, 285, 901–906. [Google Scholar] [CrossRef] [PubMed]
- Sambrook, J.; Russell, D.W. Molecular Cloning: A Laboratory Manual, 3rd ed.; Cold Spring Harbor Laboratory Press: Cold Spring Harbor, NY, USA, 2001. [Google Scholar]
- Ito, H.; Fukuda, Y.; Murata, K.; Kimura, A. Transformation of intact yeast cells treated with alkali cations. J. Bacteriol. 1983, 153, 163–168. [Google Scholar] [CrossRef]
- Christianson, T.W.; Sikorski, R.S.; Dante, M.; Shero, J.H.; Hieter, P. Multifunctional yeast high-copy-number shuttle vectors. Gene 1992, 110, 119–122. [Google Scholar] [CrossRef]
- Ogawa, N.; Saitoh, H.; Miura, K.; Magbanua, J.P.; Bun-ya, M.; Harashima, S.; Oshima, Y. Structure and distribution of specific cis-elements for transcriptional regulation of PHO84 in Saccharomyces cerevisiae. Mol. Gen.Genet. 1995, 249, 406–416. [Google Scholar] [CrossRef]
- Cunningham, K.W.; Fink, G.R. Calcineurin inhibits VCX1-dependent H+/Ca2+ exchange and induces Ca2+ ATPases in Saccharomyces cerevisiae. Mol. Cell Biol. 1996, 16, 2226–2237. [Google Scholar] [CrossRef]
- Pérez-Sampietro, M.; Serra-Cardona, A.; Canadell, D.; Casas, C.; Ariño, J.; Herrero, E. The yeast Aft2 transcription factor determines selenite toxicity by controlling the low affinity phosphate transport system. Sci. Rep. 2016, 6, 32836. [Google Scholar] [CrossRef]
- Ozcan, S.; Johnston, M. Three different regulatory mechanisms enable yeast hexose transporter (HXT) genes to be induced by different levels of glucose. Mol. Cell Biol. 1995, 15, 1564–1572. [Google Scholar] [CrossRef]
- Idrissi, F.Z.; Fernandez-Larrea, J.B.; Pina, B. Structural and functional heterogeneity of Rap1p complexes with telomeric and UASrpg-like DNA sequences. J. Mol. Biol. 1998, 284, 925–935. [Google Scholar] [CrossRef] [PubMed]

| Strain | Genotype | Reference |
|---|---|---|
| BY4741 | MATa his3Δ1 leu2Δ0 met15Δ0 ura3Δ0 | [54] |
| BYstp1Δ | BY4741 stp1::kanMX4 | [55] |
| BYstp2Δ | BY4741 stp2::kanMX4 | [55] |
| CEN.PK113-5D | MATa ura3-52 TRP1 LEU2 HIS3 MAL2-8c SUC2 | Euroscarf |
| 23344c | MATα ura3-52 | [25] |
| 32501c | MATa ssy1::kanMX2 ura3 | [25] |
| FB98 | MATα ptr3::kanMX2 ura3 | [23] |
| FB90 | MATα ssy5::kanMX2 ura3 | [23] |
| KW023 | MATα stp1Δ stp2Δ ura3 | [31] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zekhnini, A.; Albacar, M.; Casamayor, A.; Ariño, J. The ENA1 Na+-ATPase Gene Is Regulated by the SPS Sensing Pathway and the Stp1/Stp2 Transcription Factors. Int. J. Mol. Sci. 2023, 24, 5548. https://doi.org/10.3390/ijms24065548
Zekhnini A, Albacar M, Casamayor A, Ariño J. The ENA1 Na+-ATPase Gene Is Regulated by the SPS Sensing Pathway and the Stp1/Stp2 Transcription Factors. International Journal of Molecular Sciences. 2023; 24(6):5548. https://doi.org/10.3390/ijms24065548
Chicago/Turabian StyleZekhnini, Abdelghani, Marcel Albacar, Antonio Casamayor, and Joaquín Ariño. 2023. "The ENA1 Na+-ATPase Gene Is Regulated by the SPS Sensing Pathway and the Stp1/Stp2 Transcription Factors" International Journal of Molecular Sciences 24, no. 6: 5548. https://doi.org/10.3390/ijms24065548
APA StyleZekhnini, A., Albacar, M., Casamayor, A., & Ariño, J. (2023). The ENA1 Na+-ATPase Gene Is Regulated by the SPS Sensing Pathway and the Stp1/Stp2 Transcription Factors. International Journal of Molecular Sciences, 24(6), 5548. https://doi.org/10.3390/ijms24065548