From Biology to Diagnosis and Treatment: The Ariadne’s Thread in Cancer of Unknown Primary
Abstract
1. Introduction
2. Epidemiology of CUP
3. Risk Factors of CUP
4. Biology of CUP
4.1. Chromosomal Abnormalities
4.2. Oncogenes and Proteins
4.3. Angiogenesis
4.4. Evasion of Immune Destruction
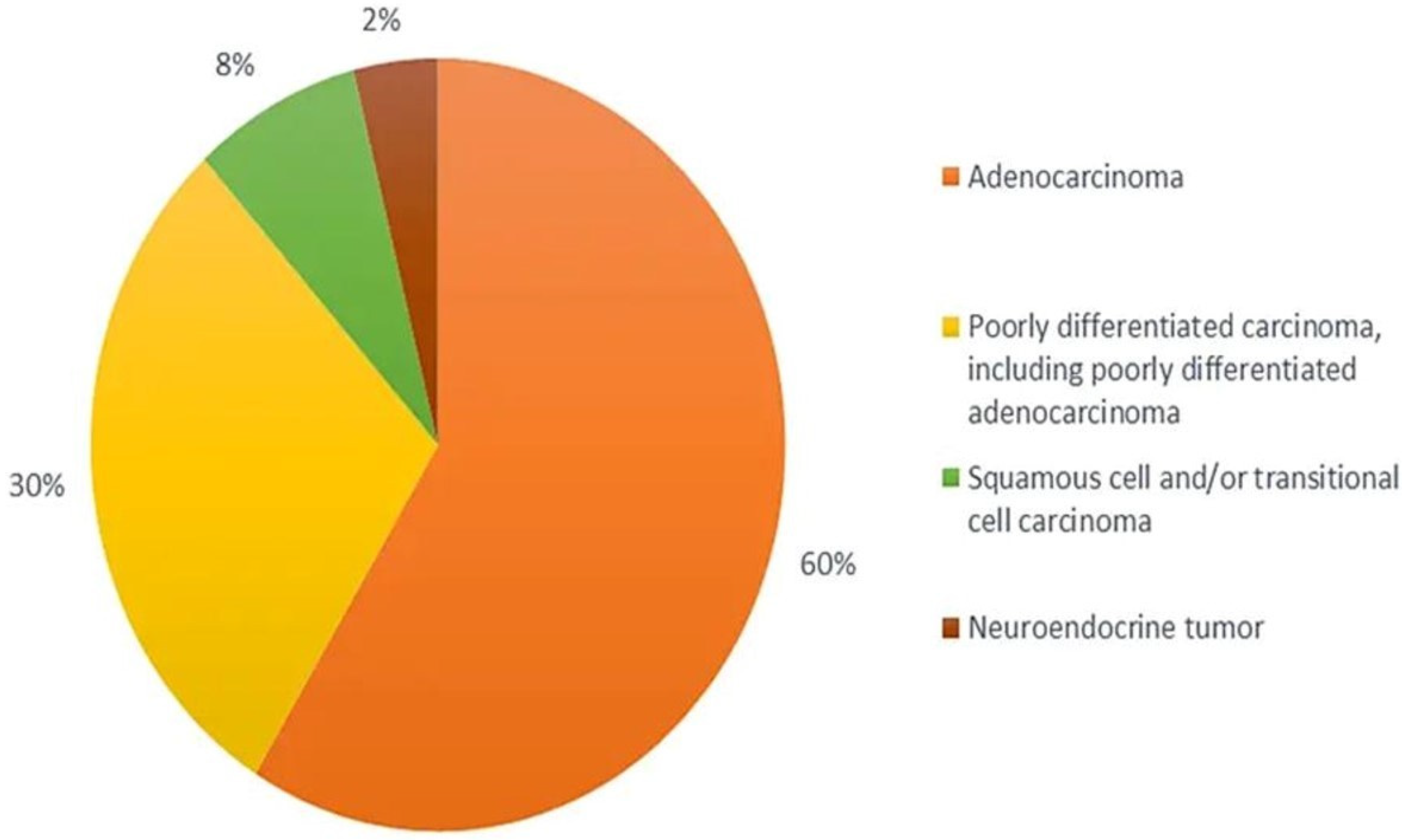
5. Classification of CUP
6. Diagnostic Workup
6.1. Pathology and Immunohistochemistry
6.2. Diagnostic Radiology
6.3. Endoscopy
6.4. Serum Tumour Markers
6.5. Liquid Biopsy
6.6. Molecular Profiling for the Tissue of Origin
7. Treatment of CUP
8. Conclusions and Future Directions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Fizazi, K.; Greco, F.A.; Pavlidis, N.; Daugaard, G.; Oien, K.; Pentheroudakis, G.; ESMO Guidelines Committee. Cancers of unknown primary site: ESMO Clinical Practice Guidelines for diagnosis, treatment and follow-up. Ann. Oncol. 2015, 26, v133–v138. [Google Scholar] [CrossRef] [PubMed]
- Rassy, E.; Pavlidis, N. The currently declining incidence of cancer of unknown primary. Cancer Epidemiol. 2019, 61, 139–141. [Google Scholar] [CrossRef] [PubMed]
- van der Strate, I.; Kazemzadeh, F.; Nagtegaal, I.D.; Robbrecht, D.; van de Wouw, A.; Padilla, C.S.; Duijts, S.; Esteller, M.; Greco, F.A.; Pavlidis, N.; et al. International consensus on the initial diagnostic workup of cancer of unknown primary. Crit. Rev. Oncol. Hematol. 2023, 181, 103868. [Google Scholar] [CrossRef] [PubMed]
- Pisacane, A.; Cascardi, E.; Berrino, E.; Polidori, A.; Sarotto, I.; Casorzo, L.; Panero, M.; Boccaccio, C.; Verginelli, F.; Benvenuti, S.; et al. Real-world histopathological approach to malignancy of undefined primary origin (MUO) to diagnose cancers of unknown primary (CUPs). Virchows Arch. 2022, 1–13. [Google Scholar] [CrossRef]
- National Comprehensive Cancer Network. NCCN Clinical Practice Guidelines in Oncology: Occult Primary (Cancer of Unknown Primary [CUP]). NCCN. Version 1.2022—2 September 2021. Available online: http://www.nccn.org/professionals/physician_gls/pdf/occult.pdf (accessed on 11 December 2022).
- Losa, F.; Soler, G.; Casado, A.; Estival, A.; Fernández, I.; Giménez, S.; Longo, F.; Pazo-Cid, R.; Salgado, J.; Seguí, M.Á. SEOM clinical guideline on unknown primary cancer (2017). Clin. Transl. Oncol. 2018, 20, 89–96. [Google Scholar] [CrossRef]
- National Institute for Health and Care Excellence (NICE). Surveillance Report (Exceptional Review) 2017—Metastatic Malignant Disease of Unknown Primary Origin in Adults (2010) NICE Guideline CG104; National Institute for Health and Care Excellence (NICE): London, UK, 2017. [Google Scholar]
- Cancer Research UK. 2022. Available online: https://www.cancerresearchuk.org/health-professional/cancer-statistics/statistics-by-cancer-type/cancer-of-unknown-primary#heading-Zero (accessed on 1 February 2023).
- Dyrvig, A.K.; Yderstræde, K.B.; Gerke, O.; Jensen, P.B.; Hess, S.; Høilund-Carlsen, P.F.; Green, A. Cancer of unknown primary: Registered procedures compared with national integrated cancer pathway for illuminating external validity. Medicine 2017, 96, e6693. [Google Scholar] [CrossRef]
- Mnatsakanyan, E.; Tung, W.C.; Caine, B.; Smith-Gagen, J. Cancer of unknown primary: Time trends in incidence, United States. Cancer Causes Control 2014, 25, 747–757. [Google Scholar] [CrossRef]
- Brewster, D.H.; Lang, J.; Bhatti, L.A.; Thomson, C.S.; Oien, K.A. Descriptive epidemiology of cancer of unknown primary site in Scotland, 1961–2010. Cancer Epidemiol. 2014, 38, 227–234. [Google Scholar] [CrossRef]
- Moran, S.; Martinez-Cardús, A.; Boussios, S.; Esteller, M. Precision medicine based on epigenomics: The paradigm of carcinoma of unknown primary. Nat. Rev. Clin. Oncol. 2017, 14, 682–694. [Google Scholar] [CrossRef]
- Boo, Y.K.; Park, D.; Lim, J.; Lim, H.S.; Won, Y.J. Descriptive epidemiology of cancer of unknown primary in South Korea, 1999–2017. Cancer Epidemiol. 2021, 74, 102000. [Google Scholar] [CrossRef]
- Randn, M.; Rutqvist, L.E.; Johansson, H. Cancer patients without a known primary: Incidence and survival trends in Sweden 1960–2007. Acta Oncol. 2009, 48, 915–920. [Google Scholar] [CrossRef]
- Agudo, A.; Bonet, C.; Travier, N.; González, C.A.; Vineis, P.; Bueno-de-Mesquita, H.B.; Trichopoulos, D.; Boffetta, P.; Clavel-Chapelon, F.; Boutron-Ruault, M.C.; et al. Impact of cigarette smoking on cancer risk in the European prospective investigation into cancer and nutrition study. J. Clin. Oncol. 2012, 30, 4550–4557. [Google Scholar] [CrossRef]
- Kaaks, R.; Sookthai, D.; Hemminki, K.; Krämer, A.; Boeing, H.; Wirfält, E.; Weiderpass, E.; Overvad, K.; Tjønneland, A.; Olsen, A.; et al. Risk factors for cancers of unknown primary site: Results from the prospective EPIC cohort. Int. J. Cancer 2014, 135, 2475–2481. [Google Scholar] [CrossRef]
- Rassy, E.; Kattan, J.; Pavlidis, N. Familial cancer of unknown primary. Int. J. Clin. Oncol. 2019, 24, 1328–1331. [Google Scholar] [CrossRef]
- Samadder, N.J.; Smith, K.R.; Hanson, H.; Pimentel, R.; Wong, J.; Boucher, K.; Akerley, W.; Gilcrease, G.; Ulrich, C.M.; Burt, R.W.; et al. Familial Risk in Patients With Carcinoma of Unknown Primary. JAMA Oncol. 2016, 2, 340–346. [Google Scholar] [CrossRef]
- Tijhuis, A.E.; Johnson, S.C.; McClelland, S.E. The emerging links between chromosomal instability (CIN), metastasis, inflammation and tumour immunity. Mol. Cytogenet. 2019, 12, 17. [Google Scholar] [CrossRef]
- Rassy, E.; Assi, T.; Pavlidis, N. Exploring the biological hallmarks of cancer of unknown primary: Where do we stand today? Br. J. Cancer 2020, 122, 1124–1132. [Google Scholar] [CrossRef] [PubMed]
- Kamposioras, K.; Pentheroudakis, G.; Pavlidis, N. Exploring the biology of cancer of unknown primary: Breakthroughs and drawbacks. Eur. J. Clin. Investig. 2013, 43, 491–500. [Google Scholar] [CrossRef]
- Hedley, D.W.; Leary, J.A.; Kirsten, F. Metastatic adenocarcinoma of unknown primary site: Abnormalities of cellular DNA content and survival. Eur. J. Cancer Clin. Oncol. 1985, 21, 185–189. [Google Scholar] [CrossRef]
- Chebly, A.; Yammine, T.; Boussios, S.; Pavlidis, N.; Rassy, E. Chromosomal instability in cancers of unknown primary. Eur. J. Cancer 2022, 172, 323–325. [Google Scholar] [CrossRef]
- Luo, J.; Solimini, N.L.; Elledge, S.J. Principles of cancer therapy: Oncogene and non-oncogene addiction. Cell 2009, 136, 823–837. [Google Scholar] [CrossRef]
- Negrini, S.; Gorgoulis, V.G.; Halazonetis, T.D. Genomic instability—An evolving hallmark of cancer. Nat. Rev. Mol. Cell Biol. 2010, 11, 220–228. [Google Scholar] [CrossRef] [PubMed]
- Gatalica, Z.; Millis, S.Z.; Vranic, S.; Bender, R.; Basu, G.D.; Voss, A.; Von Hoff, D.D. Comprehensive tumor profiling identifies numerous biomarkers of drug response in cancers of unknown primary site: Analysis of 1806 cases. Oncotarget 2014, 5, 12440–12447. [Google Scholar] [CrossRef] [PubMed]
- Massard, C.; Voigt, J.J.; Laplanche, A.; Culine, S.; Lortholary, A.; Bugat, R.; Theodore, C.; Priou, F.; Kaminsky, M.C.; Lesimple, T.; et al. Carcinoma of an unknown primary: Are EGF receptor, Her-2/neu, and c-Kit tyrosine kinases potential targets for therapy? Br. J. Cancer 2007, 97, 857–861. [Google Scholar] [CrossRef] [PubMed]
- Revythis, A.; Shah, S.; Kutka, M.; Moschetta, M.; Ozturk, M.A.; Pappas-Gogos, G.; Ioannidou, E.; Sheriff, M.; Rassy, E.; Boussios, S. Unraveling the Wide Spectrum of Melanoma Biomarkers. Diagnostics 2021, 11, 1341. [Google Scholar] [CrossRef]
- Verginelli, F.; Pisacane, A.; Gambardella, G.; D’Ambrosio, A.; Candiello, E.; Ferrio, M.; Panero, M.; Casorzo, L.; Benvenuti, S.; Cascardi, E.; et al. Cancer of unknown primary stem-like cells model multi-organ metastasis and unveil liability to MEK inhibition. Nat. Commun. 2021, 12, 2498. [Google Scholar] [CrossRef]
- Kato, S.; Krishnamurthy, N.; Banks, K.C.; De, P.; Williams, K.; Williams, C.; Leyland-Jones, B.; Lippman, S.M.; Lanman, R.B.; Kurzrock, R. Utility of Genomic Analysis In Circulating Tumor DNA from Patients with Carcinoma of Unknown Primary. Cancer Res. 2017, 77, 4238–4246. [Google Scholar] [CrossRef]
- Shah, S.; Rachmat, R.; Enyioma, S.; Ghose, A.; Revythis, A.; Boussios, S. BRCA Mutations in Prostate Cancer: Assessment, Implications and Treatment Considerations. Int. J. Mol. Sci. 2021, 22, 12628. [Google Scholar] [CrossRef]
- De Talhouet, S.; Peron, J.; Vuilleumier, A.; Friedlaender, A.; Viassolo, V.; Ayme, A.; Bodmer, A.; Treilleux, I.; Lang, N.; Tille, J.C.; et al. Clinical outcome of breast cancer in carriers of BRCA1 and BRCA2 mutations according to molecular subtypes. Sci. Rep. 2020, 10, 7073. [Google Scholar] [CrossRef]
- Shah, S.; Cheung, A.; Kutka, M.; Sheriff, M.; Boussios, S. Epithelial Ovarian Cancer: Providing Evidence of Predisposition Genes. Int. J. Environ. Res. Public. Health 2022, 19, 8113. [Google Scholar] [CrossRef]
- Boussios, S.; Rassy, E.; Shah, S.; Ioannidou, E.; Sheriff, M.; Pavlidis, N. Aberrations of DNA repair pathways in prostate cancer: A cornerstone of precision oncology. Expert Opin. Ther. Targets 2021, 25, 329–333. [Google Scholar] [CrossRef]
- Boussios, S.; Rassy, E.; Moschetta, M.; Ghose, A.; Adeleke, S.; Sanchez, E.; Sheriff, M.; Chargari, C.; Pavlidis, N. BRCA Mutations in Ovarian and Prostate Cancer: Bench to Bedside. Cancers 2022, 14, 3888. [Google Scholar] [CrossRef]
- Thompson, D.; Easton, D.F.; Breast Cancer Linkage Consortium. Cancer Incidence in BRCA1 mutation carriers. J. Natl. Cancer Inst. 2002, 94, 1358–1365. [Google Scholar] [CrossRef]
- Hunter, K.W. Host genetics and tumour metastasis. Br. J. Cancer 2004, 90, 752–755. [Google Scholar] [CrossRef][Green Version]
- Ioannidou, E.; Moschetta, M.; Shah, S.; Parker, J.S.; Ozturk, M.A.; Pappas-Gogos, G.; Sheriff, M.; Rassy, E.; Boussios, S. Angiogenesis and Anti-Angiogenic Treatment in Prostate Cancer: Mechanisms of Action and Molecular Targets. Int. J. Mol. Sci. 2021, 22, 9926. [Google Scholar] [CrossRef]
- Rassy, E.; Pavlidis, N. Progress in refining the clinical management of cancer of unknown primary in the molecular era. Nat. Rev. Clin. Oncol. 2020, 17, 541–554. [Google Scholar] [CrossRef]
- Yadav, L.; Puri, N.; Rastogi, V.; Satpute, P.; Sharma, V. Tumour Angiogenesis and Angiogenic Inhibitors: A Review. J. Clin. Diagn. Res. 2015, 9, XE01–XE05. [Google Scholar] [CrossRef]
- van de Wouw, A.J.; Jansen, R.L.; Griffioen, A.W.; Hillen, H.F. Clinical and immunohistochemical analysis of patients with unknown primary tumour. A search for prognostic factors in UPT. Anticancer Res. 2004, 24, 297–301. [Google Scholar]
- Hillen, H.F.; Hak, L.E.; Joosten-Achjanie, S.R.; Arends, J.W. Microvessel density in unknown primary tumors. Int. J. Cancer 1997, 74, 81–85. [Google Scholar] [CrossRef]
- Agarwal, B.; Das, P.; Naresh, K.N.; Borges, A.M. Angiogenic ability of metastatic squamous carcinoma in the cervical lymph nodes from unknown primary tumours. J. Clin. Pathol. 2011, 64, 765–770. [Google Scholar] [CrossRef]
- Thomas, D.A.; Massagué, J. TGF-β directly targets cytotoxic T cell functions during tumor evasion of immune surveillance. Cancer Cell 2005, 8, 369–380. [Google Scholar] [CrossRef] [PubMed]
- Gatalica, Z.; Xiu, J.; Swensen, J.; Vranic, S. Comprehensive analysis of cancers of unknown primary for the biomarkers of response to immune checkpoint blockade therapy. Eur. J. Cancer 2018, 94, 179–186. [Google Scholar] [CrossRef] [PubMed]
- Adeleke, S.; Haslam, A.; Choy, A.; Diaz-Cano, S.; Galante, J.R.; Mikropoulos, C.; Boussios, S. Microsatellite instability testing in colorectal patients with Lynch syndrome: Lessons learned from a case report and how to avoid such pitfalls. Per. Med. 2022, 19, 277–286. [Google Scholar] [CrossRef] [PubMed]
- Rassy, E.; Parent, P.; Lefort, F.; Boussios, S.; Baciarello, G.; Pavlidis, N. New rising entities in cancer of unknown primary: Is there a real therapeutic benefit? Crit. Rev. Oncol. Hematol. 2020, 147, 102882. [Google Scholar] [CrossRef]
- Pavlidis, N.; Pentheroudakis, G. Cancer of unknown primary site. Lancet 2012, 379, 1428–1435. [Google Scholar] [CrossRef]
- Nguyen, L.; Van Hoeck, A.; Cuppen, E. Machine learning-based tissue of origin classification for cancer of unknown primary diagnostics using genome-wide mutation features. Nat. Commun. 2022, 13, 4013. [Google Scholar] [CrossRef]
- Boussios, S.; Rassy, E.; Samartzis, E.; Moschetta, M.; Sheriff, M.; Pérez-Fidalgo, J.A.; Pavlidis, N. Melanoma of unknown primary: New perspectives for an old story. Crit. Rev. Oncol. Hematol. 2021, 158, 103208. [Google Scholar] [CrossRef]
- Rassy, E.; Boussios, S.; Chebly, A.; Farra, C.; Kattan, J.; Pavlidis, N. Comparative genomic characterization of melanoma of known and unknown primary. Clin. Transl. Oncol. 2021, 23, 2302–2308. [Google Scholar] [CrossRef]
- Rassy, E.; Abou-Jaoude, R.; Boussios, S.; Assi, T.; Kattan, J.; Khaled, H.; Pavlidis, N. Sarcoma of unknown primary: Myth or reality? J. Egypt. Natl. Canc. Inst. 2022, 34, 27. [Google Scholar] [CrossRef]
- Rassy, E.; Pavlidis, N. The diagnostic challenges of patients with carcinoma of unknown primary. Expert. Rev. Anticancer Ther. 2020, 20, 775–783. [Google Scholar] [CrossRef]
- Ross, J.S.; Sokol, E.S.; Moch, H.; Mileshkin, L.; Baciarello, G.; Losa, F.; Beringer, A.; Thomas, M.; Elvin, J.A.; Ngo, N.; et al. Comprehensive Genomic Profiling of Carcinoma of Unknown Primary Origin: Retrospective Molecular Classification Considering the CUPISCO Study Design. Oncologist 2021, 26, e394–e402. [Google Scholar] [CrossRef]
- Varadhachary, G.R.; Spector, Y.; Abbruzzese, J.L.; Rosenwald, S.; Wang, H.; Aharonov, R.; Carlson, H.R.; Cohen, D.; Karanth, S.; Macinskas, J.; et al. Prospective gene signature study using microRNA to identify the tissue of origin in patients with carcinoma of unknown primary. Clin. Cancer Res. 2011, 17, 4063–4070. [Google Scholar] [CrossRef]
- Pavlidis, N.; Fizazi, K. Carcinoma of unknown primary (CUP). Crit. Rev. Oncol. Hematol. 2009, 69, 271–278. [Google Scholar] [CrossRef]
- Kwee, T.C.; Kwee, R.M. Combined FDG-PET/CT for the detection of unknown primary tumors: Systematic review and meta-analysis. Eur. Radiol. 2009, 19, 731–744. [Google Scholar] [CrossRef]
- Prasad, V.; Ambrosini, V.; Hommann, M.; Hoersch, D.; Fanti, S.; Baum, R.P. Detection of unknown primary neuroendocrine tumours (CUP-NET) using (68)Ga-DOTA-NOC receptor PET/CT. Eur. J. Nucl. Med. Mol. Imaging 2010, 37, 67–77. [Google Scholar] [CrossRef]
- Takamizawa, S.; Shimoi, T.; Yoshida, M.; Tokura, M.; Yazaki, S.; Mizoguchi, C.; Saito, A.; Kita, S.; Yamamoto, K.; Kojima, Y.; et al. Diagnostic value of tumor markers in identifying favorable or unfavorable subsets in patients with cancer of unknown primary: A retrospective study. BMC Cancer 2022, 22, 412. [Google Scholar] [CrossRef]
- Oltmann, S.C.; Leverson, G.; Lin, S.H.; Schneider, D.F.; Chen, H.; Sippel, R.S. Markedly elevated thyroglobulin levels in the preoperative thyroidectomy patient correlates with metastatic burden. J. Surg. Res. 2014, 187, 1–5. [Google Scholar] [CrossRef][Green Version]
- Esteller, M.; Sanchez-Cespedes, M.; Rosell, R.; Sidransky, D.; Baylin, S.B.; Herman, J.G. Detection of aberrant promoter hypermethylation of tumor suppressor genes in serum DNA from non-small cell lung cancer patients. Cancer Res. 1999, 59, 67–70. [Google Scholar]
- Kristensen, L.S.; Hansen, J.W.; Kristensen, S.S.; Tholstrup, D.; Harsløf, L.B.; Pedersen, O.B.; De Nully Brown, P.; Grønbæk, K. Aberrant methylation of cell-free circulating DNA in plasma predicts poor outcome in diffuse large B cell lymphoma. Clin. Epigenetics 2016, 8, 95. [Google Scholar] [CrossRef]
- Vaca-Paniagua, F.; Oliver, J.; Nogueira da Costa, A.; Merle, P.; McKay, J.; Herceg, Z.; Holmila, R. Targeted deep DNA methylation analysis of circulating cell-free DNA in plasma using massively parallel semiconductor sequencing. Epigenomics 2015, 7, 353–362. [Google Scholar] [CrossRef]
- Lubotzky, A.; Zemmour, H.; Neiman, D.; Gotkine, M.; Loyfer, N.; Piyanzin, S.; Ochana, B.L.; Lehmann-Werman, R.; Cohen, D.; Moss, J.; et al. Liquid biopsy reveals collateral tissue damage in cancer. JCI Insight 2022, 7, e153559. [Google Scholar] [CrossRef]
- Handorf, C.R.; Kulkarni, A.; Grenert, J.P.; Weiss, L.M.; Rogers, W.M.; Kim, O.S.; Monzon, F.A.; Halks-Miller, M.; Anderson, G.G.; Walker, M.G.; et al. A multicenter study directly comparing the diagnostic accuracy of gene expression profiling and immunohistochemistry for primary site identification in metastatic tumors. Am. J. Surg. Pathol. 2013, 37, 1067–1075. [Google Scholar] [CrossRef] [PubMed]
- Conway, A.M.; Mitchell, C.; Kilgour, E.; Brady, G.; Dive, C.; Cook, N. Molecular characterisation and liquid biomarkers in Carcinoma of Unknown Primary (CUP): Taking the ‘U’ out of ‘CUP’. Br. J. Cancer 2019, 120, 141–153. [Google Scholar] [CrossRef] [PubMed]
- Rassy, E.; Labaki, C.; Chebel, R.; Boussios, S.; Smith-Gagen, J.; Greco, F.A.; Pavlidis, N. Systematic review of the CUP trials characteristics and perspectives for next-generation studies. Cancer Treat. Rev. 2022, 107, 102407. [Google Scholar] [CrossRef] [PubMed]
- Le, A.P.; Huang, Y.; Pingle, S.C.; Kesari, S.; Wang, H.; Yong, R.L.; Zou, H.; Friedel, R.H. Plexin-B2 promotes invasive growth of malignant glioma. Oncotarget 2015, 6, 7293–7304. [Google Scholar] [CrossRef] [PubMed]
- Yu, W.; Goncalves, K.A.; Li, S.; Kishikawa, H.; Sun, G.; Yang, H.; Vanli, N.; Wu, Y.; Jiang, Y.; Hu, M.G.; et al. Plexin-B2 Mediates Physiologic and Pathologic Functions of Angiogenin. Cell 2017, 171, 849–864. [Google Scholar] [CrossRef]
- Gurrapu, S.; Pupo, E.; Franzolin, G.; Lanzetti, L.; Tamagnone, L. Sema4C/PlexinB2 signaling controls breast cancer cell growth, hormonal dependence and tumorigenic potential. Cell Death Differ. 2018, 25, 1259–1275. [Google Scholar] [CrossRef]
- Huang, Y.; Tejero, R.; Lee, V.K.; Brusco, C.; Hannah, T.; Bertucci, T.B.; Junqueira Alves, C.; Katsyv, I.; Kluge, M.; Foty, R.; et al. Plexin-B2 facilitates glioblastoma infiltration by modulating cell biomechanics. Commun. Biol. 2021, 4, 145. [Google Scholar] [CrossRef]
- Ma, Q.; Zhang, K.; Yao, H.P.; Zhou, Y.Q.; Padhye, S.; Wang, M.H. Inhibition of MSP-RON signaling pathway in cancer cells by a novel soluble form of RON comprising the entire sema sequence. Int. J. Oncol. 2010, 36, 1551–1561. [Google Scholar]
- Navis, A.C.; van Lith, S.A.; van Duijnhoven, S.M.; de Pooter, M.; Yetkin-Arik, B.; Wesseling, P.; Hendriks, W.J.; Venselaar, H.; Timmer, M.; van Cleef, P.; et al. Identification of a novel MET mutation in high-grade glioma resulting in an auto-active intracellular protein. Acta Neuropathol. 2015, 130, 131–144. [Google Scholar] [CrossRef]
- Cagnoni, G.; Tamagnone, L. Semaphorin receptors meet receptor tyrosine kinases on the way of tumor progression. Oncogene 2014, 33, 4795–4802. [Google Scholar] [CrossRef]
- Brundu, S.; Napolitano, V.; Franzolin, G.; Lo Cascio, E.; Mastrantonio, R.; Sardo, G.; Cascardi, E.; Verginelli, F.; Sarnataro, S.; Gambardella, G.; et al. Mutated axon guidance gene PLXNB2 sustains growth and invasiveness of stem cells isolated from cancers of unknown primary. EMBO Mol. Med. 2023, 15, e16104. [Google Scholar] [CrossRef]
- Pavlidis, N.; Rassy, E.; Vermorken, J.B.; Assi, T.; Kattan, J.; Boussios, S.; Smith-Gagen, J. The outcome of patients with serous papillary peritoneal cancer, fallopian tube cancer, and epithelial ovarian cancer by treatment eras: 27 years data from the SEER registry. Cancer Epidemiol. 2021, 75, 102045. [Google Scholar] [CrossRef]
- Rassy, E.; Assi, T.; Boussios, S.; Kattan, J.; Smith-Gagen, J.; Pavlidis, N. Narrative review on serous primary peritoneal carcinoma of unknown primary site: Four questions to be answered. Ann. Transl. Med. 2020, 8, 1709. [Google Scholar] [CrossRef]
- Pouyiourou, M.; Wohlfromm, T.; Kraft, B.; Hielscher, T.; Stichel, D.; von Deimling, A.; Delorme, S.; Endris, V.; Neumann, O.; Stenzinger, A.; et al. Local ablative treatment with surgery and/or radiotherapy in single-site and oligometastatic carcinoma of unknown primary. Eur. J. Cancer 2021, 157, 179–189. [Google Scholar] [CrossRef]
- Deneve, J.L.; Messina, J.L.; Marzban, S.S.; Gonzalez, R.J.; Walls, B.M.; Fisher, K.J.; Chen, Y.A.; Cruse, C.W.; Sondak, V.K.; Zager, J.S. Merkel cell carcinoma of unknown primary origin. Ann. Surg. Oncol. 2012, 19, 2360–2366. [Google Scholar] [CrossRef]
- Hainsworth, J.D.; Daugaard, G.; Lesimple, T.; Hübner, G.; Greco, F.A.; Stahl, M.J.; Büschenfelde, C.M.; Allouache, D.; Penel, N.; Knoblauch, P.; et al. Paclitaxel/carboplatin with or without belinostat as empiric first-line treatment for patients with carcinoma of unknown primary site: A randomized, phase 2 trial. Cancer 2015, 121, 1654–1661. [Google Scholar] [CrossRef]
- El Rassy, E.; Pavlidis, N. The current evidence for a biomarker-based approach in cancer of unknown primary. Cancer Treat. Rev. 2018, 67, 21–28. [Google Scholar] [CrossRef]
- Rassy, E.; Bakouny, Z.; Choueiri, T.K.; Van Allen, E.M.; Fizazi, K.; Greco, F.A.; Pavlidis, N. The role of site-specific therapy for cancers of unknown of primary: A meta-analysis. Eur. J. Cancer 2020, 127, 118–122. [Google Scholar] [CrossRef]
- Ding, Y.; Jiang, J.; Xu, J.; Chen, Y.; Zheng, Y.; Jiang, W.; Mao, C.; Jiang, H.; Bao, X.; Shen, Y.; et al. Site-specific therapy in cancers of unknown primary site: A systematic review and meta-analysis. ESMO Open 2022, 7, 100407. [Google Scholar] [CrossRef]
- Naing, A.; Meric-Bernstam, F.; Stephen, B.; Karp, D.D.; Hajjar, J.; Rodon Ahnert, J.; Piha-Paul, S.A.; Colen, R.R.; Jimenez, C.; Raghav, K.P.; et al. Phase 2 study of pembrolizumab in patients with advanced rare cancers. J. Immunother. Cancer 2020, 8, e000347. [Google Scholar] [CrossRef] [PubMed]
- Sicklick, J.K.; Kato, S.; Okamura, R.; Patel, H.; Nikanjam, M.; Fanta, P.T.; Hahn, M.E.; De, P.; Williams, C.; Guido, J.; et al. Molecular profiling of advanced malignancies guides first-line N-of-1 treatments in the I-PREDICT treatment-naïve study. Genome Med. 2021, 13, 155. [Google Scholar] [CrossRef]
- Rassy, E.; Boussios, S.; Pavlidis, N. Genomic correlates of response and resistance to immune checkpoint inhibitors in carcinomas of unknown primary. Eur. J. Clin. Investig. 2021, 51, e13583. [Google Scholar] [CrossRef] [PubMed]
- Haratani, K.; Hayashi, H.; Takahama, T.; Nakamura, Y.; Tomida, S.; Yoshida, T.; Chiba, Y.; Sawada, T.; Sakai, K.; Fujita, Y.; et al. Clinical and immune profiling for cancer of unknown primary site. J. Immunother. Cancer 2019, 7, 251. [Google Scholar] [CrossRef] [PubMed]
- Tanizaki, J.; Yonemori, K.; Akiyoshi, K.; Minami, H.; Ueda, H.; Takiguchi, Y.; Miura, Y.; Segawa, Y.; Takahashi, S.; Iwamoto, Y.; et al. Open-label phase II study of the efficacy of nivolumab for cancer of unknown primary. Ann. Oncol. 2022, 33, 216–226. [Google Scholar] [CrossRef]

| Favourable Subsets | Unfavourable Subsets | |
|---|---|---|
| 1 | Poorly differentiated carcinoma with midline distribution (extragonadal germ cell syndrome) | Adenocarcinoma metastatic to the liver or other organs |
| 2 | Women with papillary adenocarcinoma of the peritoneal cavity | Non-papillary malignant ascites (adenocarcinoma) |
| 3 | Women with adenocarcinoma involving only axillary lymph nodes | Multiple cerebral metastases (adeno or squamous carcinoma) |
| 4 | Squamous cell carcinoma involving cervical lymph nodes | Multiple lung/pleural metastases (adenocarcinoma) |
| 5 | Isolated inguinal adenopathy (squamous carcinoma) | Multiple metastatic bone disease (adenocarcinoma) |
| 6 | Poorly differentiated neuroendocrine carcinomas | Squamous abdominopelvic CUP |
| 7 | Men with blastic bone metastases and elevated PSA (adenocarcinoma) | |
| 8 | Patients with a single, small, potentially resectable tumour | |
| 9 | CUP patients with a single small metastasis | |
| 10 | Merkel cell adenopathy of unknown origin |
| Tumour Type | Immunoperoxidase Marker |
|---|---|
| Carcinoma | Cytokeratins, EMA |
| Lymphoma | CLA, EMA |
| Sarcoma | Vimentin, desmin, factor VIII antigen |
| Melanoma | S-100, HMB-45, NSE, vimentin |
| Neuroendocrine | Chromogranin, EMA, NSE, cytokeratins, synaptophysin |
| Germ cell | hCG, AFP, EMA, cytokeratins |
| Prostate cancer | PSA, EMA, cytokeratins (CK7−/CK20−) |
| Breast cancer | ER, PR, EMA, cytokeratins (CK7+/CK20−) |
| Thyroid cancer | Thyroglobulin, cytokeratins (CK7+/CK20−), calcitonin, EMA |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mathew, B.G.; Aliyuda, F.; Taiwo, D.; Adekeye, K.; Agada, G.; Sanchez, E.; Ghose, A.; Rassy, E.; Boussios, S. From Biology to Diagnosis and Treatment: The Ariadne’s Thread in Cancer of Unknown Primary. Int. J. Mol. Sci. 2023, 24, 5588. https://doi.org/10.3390/ijms24065588
Mathew BG, Aliyuda F, Taiwo D, Adekeye K, Agada G, Sanchez E, Ghose A, Rassy E, Boussios S. From Biology to Diagnosis and Treatment: The Ariadne’s Thread in Cancer of Unknown Primary. International Journal of Molecular Sciences. 2023; 24(6):5588. https://doi.org/10.3390/ijms24065588
Chicago/Turabian StyleMathew, Beatrice Gadiel, Fine Aliyuda, Denis Taiwo, Kehinde Adekeye, Godwin Agada, Elisabet Sanchez, Aruni Ghose, Elie Rassy, and Stergios Boussios. 2023. "From Biology to Diagnosis and Treatment: The Ariadne’s Thread in Cancer of Unknown Primary" International Journal of Molecular Sciences 24, no. 6: 5588. https://doi.org/10.3390/ijms24065588
APA StyleMathew, B. G., Aliyuda, F., Taiwo, D., Adekeye, K., Agada, G., Sanchez, E., Ghose, A., Rassy, E., & Boussios, S. (2023). From Biology to Diagnosis and Treatment: The Ariadne’s Thread in Cancer of Unknown Primary. International Journal of Molecular Sciences, 24(6), 5588. https://doi.org/10.3390/ijms24065588







