Functional and Phenotypic Characterisations of Common Syngeneic Tumour Cell Lines as Estrogen Receptor-Positive Breast Cancer Models
Abstract
:1. Introduction
2. Results
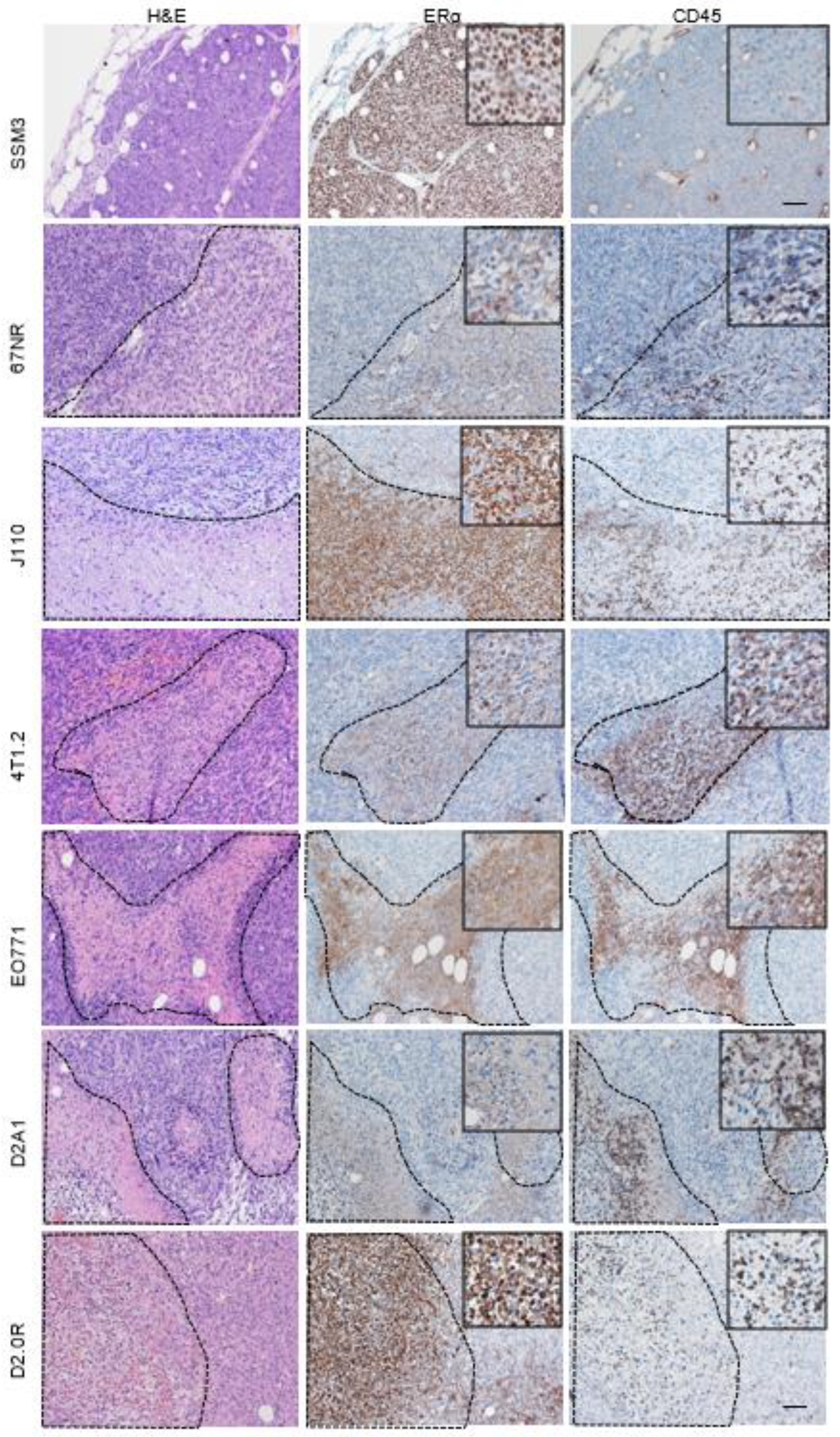
2.1. ERα Staining in Syngeneic Tumours
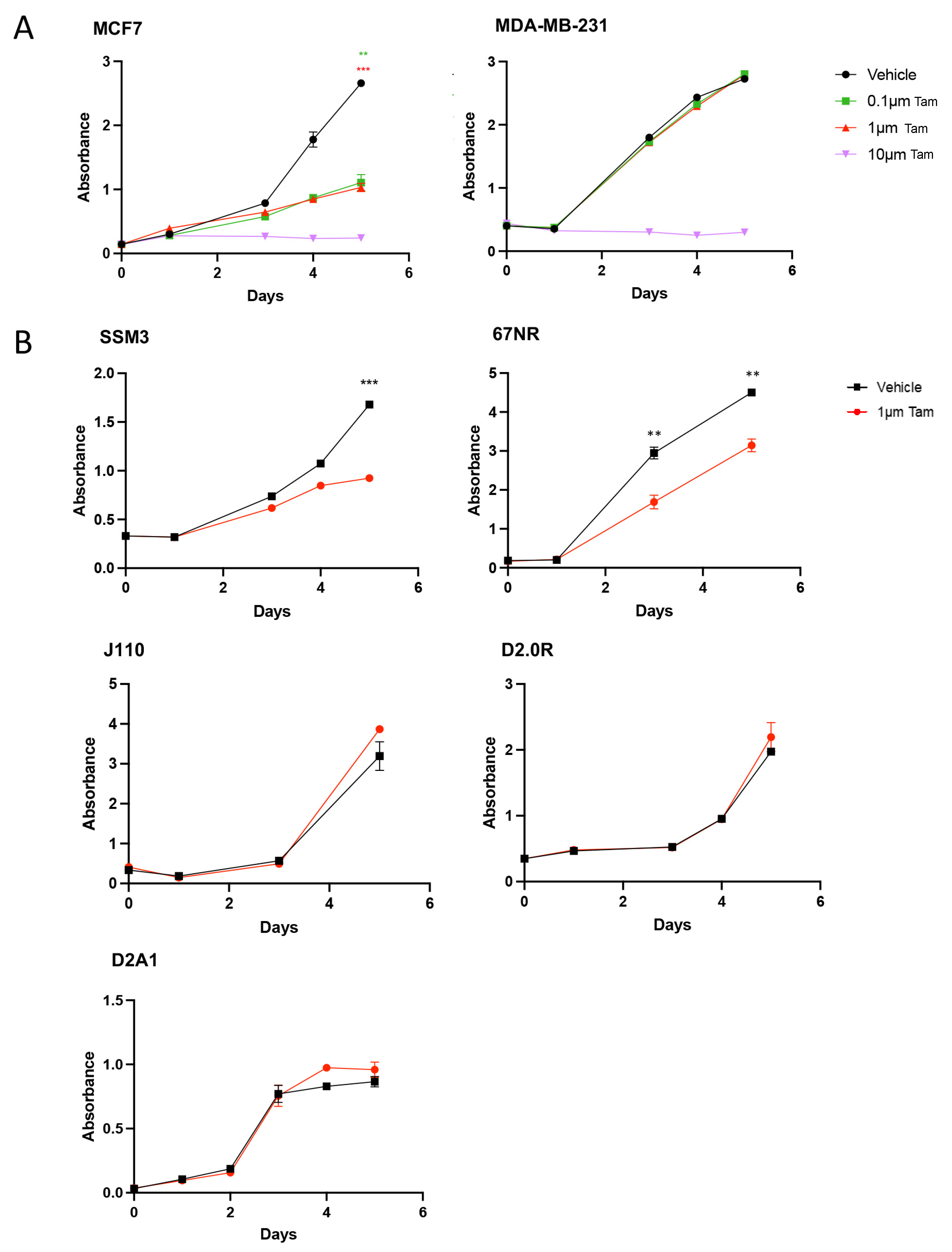
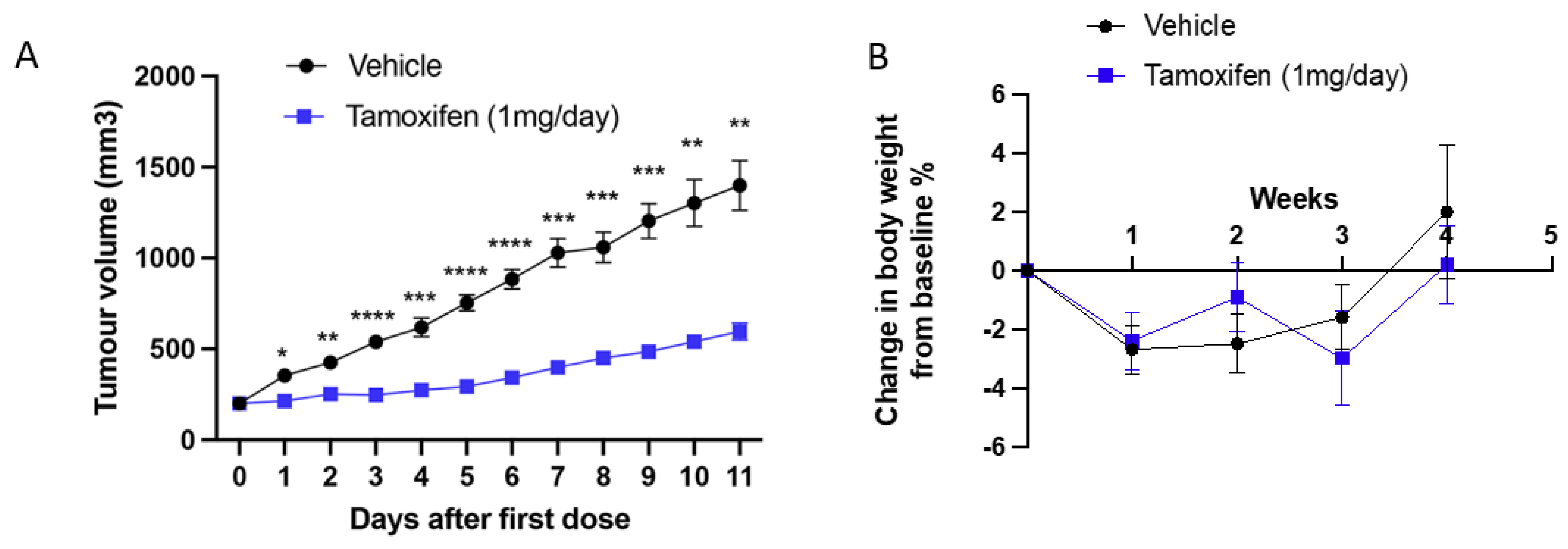
2.2. SSM3 Tumours Are the Only Syngeneic Model That Responds to Tamoxifen In Vitro and In Vivo
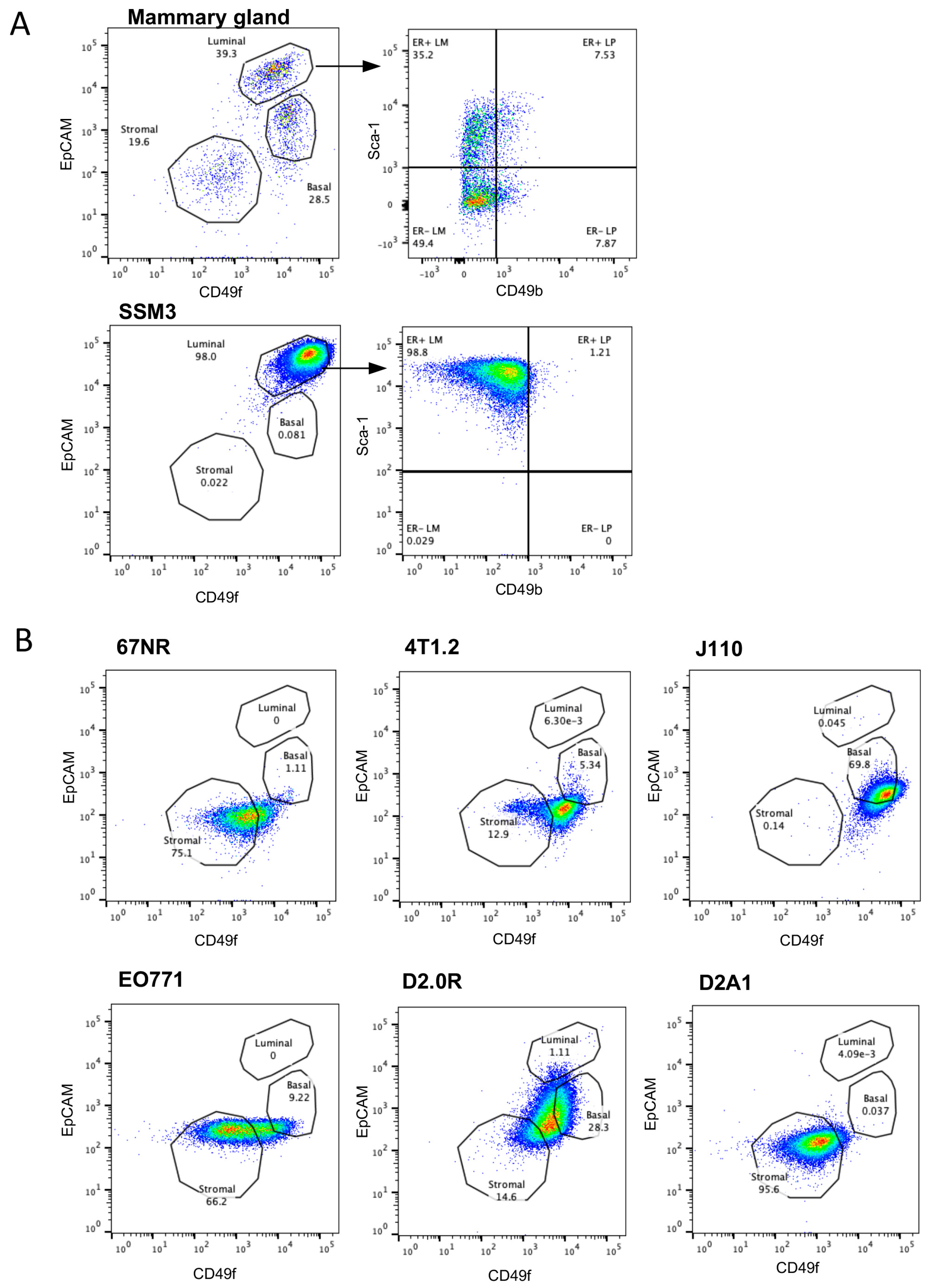
2.3. SSM3 Cells Are the Only Syngeneic Cell Line to Express Cell Surface Markers of Luminal Mammary Epithelial Cells
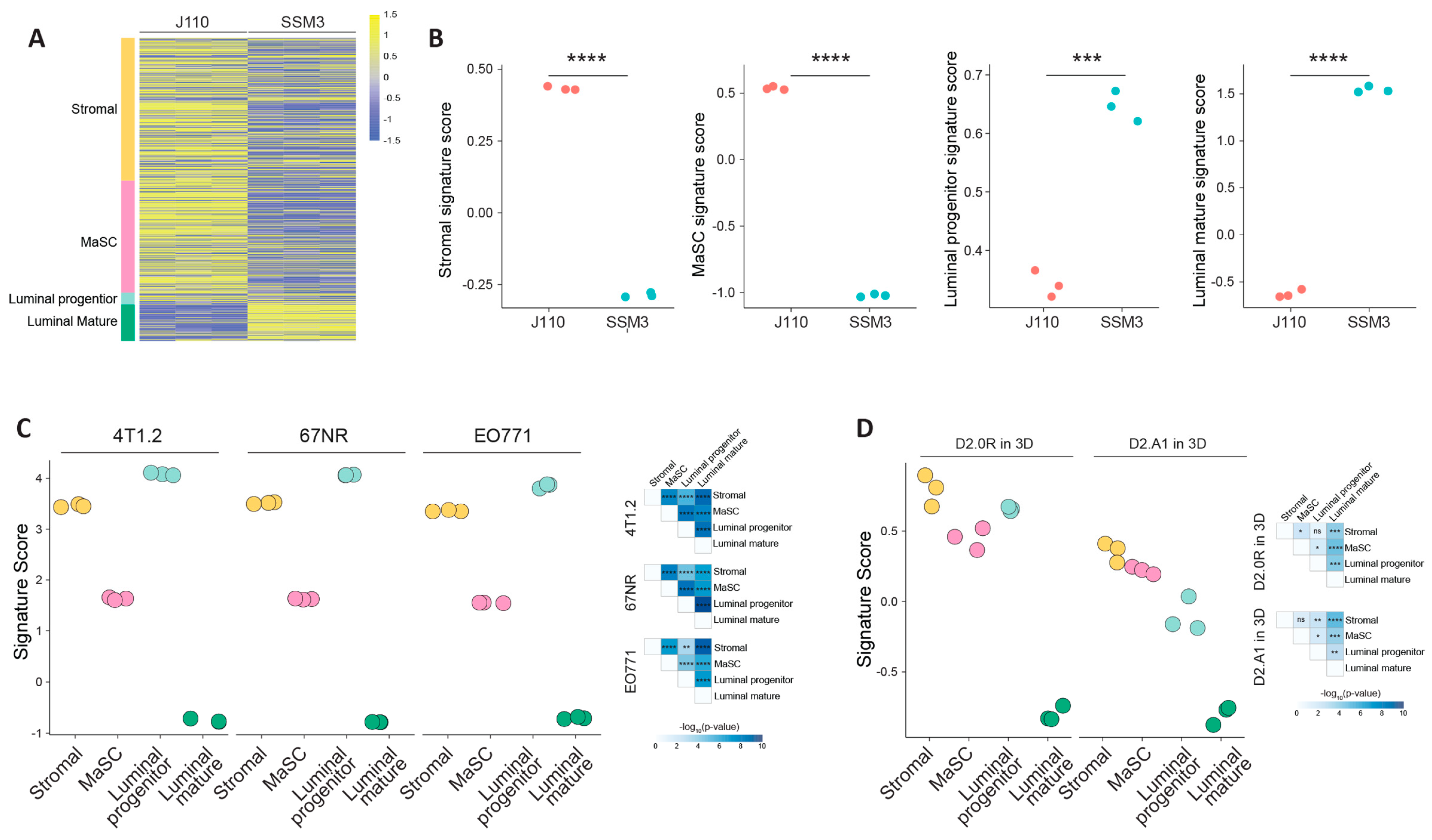
2.4. Molecular Characterisation of the Syngeneic Lines to Identify the Cell Lineage
3. Discussion
4. Materials and Methods
4.1. Cell Lines
4.2. Isolation, Staining and Flow Cytometric Analysis of Normal and Transformed Mouse Mammary Cells
4.3. Immunohistochemical Analysis
4.4. In Vitro Assessment of Tamoxifen Sensitivity
4.5. In Vivo Assessment of Tamoxifen Sensitivity in the SSM3 Mouse Model
4.6. RNAseq Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Cuzick, J.; Sestak, I.; Cawthorn, S.; Hamed, H.; Holli, K.; Howell, A.; Forbes, J.F.; Investigators, I.-I. Tamoxifen for prevention of breast cancer: Extended long-term follow-up of the IBIS-I breast cancer prevention trial. Lancet Oncol. 2015, 16, 67–75. [Google Scholar] [CrossRef] [PubMed]
- Normanno, N.; Di Maio, M.; De Maio, E.; De Luca, A.; De Matteis, A.; Giordano, A.; Perrone, F. Mechanisms of endocrine resistance and novel therapeutic strategies in breast cancer. Endocr.-Relat. Cancer 2005, 12, 721–747. [Google Scholar] [CrossRef] [Green Version]
- Smith, S.G.; Sestak, I.; Forster, A.; Partridge, A.; Side, L.; Wolf, M.; Horne, R.; Wardle, J.; Cuzick, J. Factors affecting uptake and adherence to breast cancer chemoprevention: A systematic review and meta-analysis. Ann. Oncol. 2016, 27, 575–590. [Google Scholar] [CrossRef]
- Donnelly, L.; Evans, D.; Wiseman, J.; Fox, J.; Greenhalgh, R.; Affen, J.; Juraskova, I.; Stavrinos, P.; Dawe, S.; Cuzick, J. Uptake of tamoxifen in consecutive premenopausal women under surveillance in a high-risk breast cancer clinic. Br. J. Cancer 2014, 110, 1681–1687. [Google Scholar] [CrossRef] [PubMed]
- DeCensi, A.; Thorat, M.A.; Bonanni, B.; Smith, S.G.; Cuzick, J. Barriers to preventive therapy for breast and other major cancers and strategies to improve uptake. Ecancermedicalscience 2015, 9, 595. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Crew, K.D.; Albain, K.S.; Hershman, D.L.; Unger, J.M.; Lo, S.S. How do we increase uptake of tamoxifen and other anti-estrogens for breast cancer prevention? NPJ Breast Cancer 2017, 3, 1–7. [Google Scholar] [CrossRef] [Green Version]
- Tikoo, A.; Roh, V.; Montgomery, K.G.; Ivetac, I.; Waring, P.; Pelzer, R.; Hare, L.; Shackleton, M.; Humbert, P.; Phillips, W.A. Physiological levels of Pik3ca H1047R mutation in the mouse mammary gland results in ductal hyperplasia and formation of ERα-positive tumors. PLoS ONE 2012, 7, e36924. [Google Scholar] [CrossRef] [PubMed]
- Chan, S.R.; Vermi, W.; Luo, J.; Lucini, L.; Rickert, C.; Fowler, A.M.; Lonardi, S.; Arthur, C.; Young, L.J.; Levy, D.E. STAT1-deficient mice spontaneously develop estrogen receptor α-positive luminal mammary carcinomas. Breast Cancer Res. 2012, 14, 1–21. [Google Scholar] [CrossRef] [Green Version]
- Torres-Arzayus, M.I.; de Mora, J.F.; Yuan, J.; Vazquez, F.; Bronson, R.; Rue, M.; Sellers, W.R.; Brown, M. High tumor incidence and activation of the PI3K/AKT pathway in transgenic mice define AIB1 as an oncogene. Cancer Cell 2004, 6, 263–274. [Google Scholar] [CrossRef] [Green Version]
- Chulpanova, D.S.; Kitaeva, K.V.; Rutland, C.S.; Rizvanov, A.A.; Solovyeva, V.V. Mouse tumor models for advanced cancer immunotherapy. Int. J. Mol. Sci. 2020, 21, 4118. [Google Scholar] [CrossRef]
- Torres-Arzayus, M.I.; Yuan, J.; DellaGatta, J.L.; Lane, H.; Kung, A.L.; Brown, M. Targeting the AIB1 oncogene through mammalian target of rapamycin inhibition in the mammary gland. Cancer Res. 2006, 66, 11381–11388. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hammond, M.E.H.; Hayes, D.F.; Dowsett, M.; Allred, D.C.; Hagerty, K.L.; Badve, S.; Fitzgibbons, P.L.; Francis, G.; Goldstein, N.S.; Hayes, M. American Society of Clinical Oncology/College of American Pathologists guideline recommendations for immunohistochemical testing of estrogen and progesterone receptors in breast cancer (unabridged version). Arch. Pathol. Lab. Med. 2010, 134, e48–e72. [Google Scholar] [CrossRef]
- Bogina, G.; Zamboni, G.; Sapino, A.; Bortesi, L.; Marconi, M.; Lunardi, G.; Coati, F.; Massocco, A.; Molinaro, L.; Pegoraro, C. Comparison of Anti–Estrogen Receptor Antibodies SP1, 6F11, and 1D5 in Breast Cancer: Lower 1D5 Sensitivity but Questionable Clinical Implications. Am. J. Clin. Pathol. 2012, 138, 697–702. [Google Scholar] [CrossRef] [Green Version]
- Johnstone, C.N.; Smith, Y.E.; Cao, Y.; Burrows, A.D.; Cross, R.S.; Ling, X.; Redvers, R.P.; Doherty, J.P.; Eckhardt, B.L.; Natoli, A.L.; et al. Functional and molecular characterisation of EO771.LMB tumours, a new C57BL/6-mouse-derived model of spontaneously metastatic mammary cancer. Dis. Model Mech. 2015, 8, 237–251. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lim, E.; Vaillant, F.; Wu, D.; Forrest, N.C.; Pal, B.; Hart, A.H.; Asselin-Labat, M.L.; Gyorki, D.E.; Ward, T.; Partanen, A.; et al. Aberrant luminal progenitors as the candidate target population for basal tumor development in BRCA1 mutation carriers. Nat. Med. 2009, 15, 907–913. [Google Scholar] [CrossRef] [PubMed]
- Lim, E.; Wu, D.; Pal, B.; Bouras, T.; Asselin-Labat, M.L.; Vaillant, F.; Yagita, H.; Lindeman, G.J.; Smyth, G.K.; Visvader, J.E. Transcriptome analyses of mouse and human mammary cell subpopulations reveal multiple conserved genes and pathways. Breast Cancer Res. 2010, 12, R21. [Google Scholar] [CrossRef] [Green Version]
- Nolan, E.; Vaillant, F.; Branstetter, D.; Pal, B.; Giner, G.; Whitehead, L.; Lok, S.W.; Mann, G.B.; Kathleen Cuningham Foundation Consortium for Research into Familial Breast Cancer; Rohrbach, K.; et al. RANK ligand as a potential target for breast cancer prevention in BRCA1-mutation carriers. Nat. Med. 2016, 22, 933–939. [Google Scholar] [CrossRef]
- Dall, G.V.; Vieusseux, J.; Seyed-Razavi, Y.; Godde, N.; Ludford-Menting, M.; Russell, S.M.; Ashworth, A.; Anderson, R.L.; Risbridger, G.P.; Shackleton, M.; et al. Parity reduces mammary repopulating activity but does not affect mammary stem cells defined as CD24 + CD29/CD49fhi in mice. Breast Cancer Res. Treat. 2020, 183, 565–575. [Google Scholar] [CrossRef]
- Dall, G.V.; Vieusseux, J.L.; Korach, K.S.; Arao, Y.; Hewitt, S.C.; Hamilton, K.J.; Dzierzak, E.; Boon, W.C.; Simpson, E.R.; Ramsay, R.G.; et al. SCA-1 Labels a Subset of Estrogen-Responsive Bipotential Repopulating Cells within the CD24(+) CD49f(hi) Mammary Stem Cell-Enriched Compartment. Stem Cell Rep. 2017, 8, 417–431. [Google Scholar] [CrossRef]
- Shehata, M.; Teschendorff, A.; Sharp, G.; Novcic, N.; Russell, I.A.; Avril, S.; Prater, M.; Eirew, P.; Caldas, C.; Watson, C.J.; et al. Phenotypic and functional characterisation of the luminal cell hierarchy of the mammary gland. Breast Cancer Res. 2012, 14, R134. [Google Scholar] [CrossRef] [Green Version]
- Vera-Ramirez, L.; Vodnala, S.K.; Nini, R.; Hunter, K.W.; Green, J.E. Autophagy promotes the survival of dormant breast cancer cells and metastatic tumour recurrence. Nat. Commun. 2018, 9, 1–12. [Google Scholar] [CrossRef] [Green Version]
- Prunier, C.; Alay, A.; Van Dijk, M.; Ammerlaan, K.L.; Van Gelderen, S.; Marvin, D.L.; Teunisse, A.; Slieker, R.C.; Szuhai, K.; Jochemsen, A. Breast cancer dormancy is associated with a 4NG1 state and not senescence. NPJ Breast Cancer 2021, 7, 140. [Google Scholar] [CrossRef] [PubMed]
- Roberto, M.; Astone, A.; Botticelli, A.; Carbognin, L.; Cassano, A.; D’auria, G.; Fabbri, A.; Fabi, A.; Gamucci, T.; Krasniqi, E. CDK4/6 inhibitor treatments in patients with hormone receptor positive, Her2 negative advanced breast cancer: Potential molecular mechanisms, clinical implications and future perspectives. Cancers 2021, 13, 332. [Google Scholar] [CrossRef]
- Le Naour, A.; Koffi, Y.; Diab, M.; Le Guennec, D.; Rougé, S.; Aldekwer, S.; Goncalves-Mendes, N.; Talvas, J.; Farges, M.-C.; Caldefie-Chezet, F. EO771, the first luminal B mammary cancer cell line from C57BL/6 mice. Cancer Cell Int. 2020, 20, 328. [Google Scholar] [CrossRef] [PubMed]
- Bendau, E.; Smith, J.; Zhang, L.; Ackerstaff, E.; Kruchevsky, N.; Wu, B.; Koutcher, J.A.; Alfano, R.; Shi, L. Distinguishing metastatic triple-negative breast cancer from nonmetastatic breast cancer using second harmonic generation imaging and resonance Raman spectroscopy. J. Biophotonics 2020, 13, e202000005. [Google Scholar] [CrossRef] [PubMed]
- Smart, E.; Alejo, L.H.; Frasor, J. Cytoplasmic ERα and NFκB promote cell survival in mouse mammary cancer cell lines. Horm. Cancer 2020, 11, 76–86. [Google Scholar] [CrossRef] [PubMed]
- Torres-Arzayus, M.I.; Zhao, J.; Bronson, R.; Brown, M. Estrogen-dependent and estrogen-independent mechanisms contribute to AIB1-mediated tumor formation. Cancer Res. 2010, 70, 4102–4111. [Google Scholar] [CrossRef] [Green Version]
- Meraz, M.A.; White, J.M.; Sheehan, K.C.; Bach, E.A.; Rodig, S.J.; Dighe, A.S.; Kaplan, D.H.; Riley, J.K.; Greenlund, A.C.; Campbell, D. Targeted disruption of the Stat1 gene in mice reveals unexpected physiologic specificity in the JAK–STAT signaling pathway. Cell 1996, 84, 431–442. [Google Scholar] [CrossRef] [Green Version]
- Bach, E.A.; Aguet, M.; Schreiber, R.D. The IFNγ receptor: A paradigm for cytokine receptor signaling. Annu. Rev. Immunol. 1997, 15, 563–591. [Google Scholar] [CrossRef]
- Cancer Genome Atlas Network. Comprehensive molecular portraits of human breast tumours. Nature 2012, 490, 61. [Google Scholar] [CrossRef] [Green Version]
- Griffith, O.L.; Spies, N.C.; Anurag, M.; Griffith, M.; Luo, J.; Tu, D.; Yeo, B.; Kunisaki, J.; Miller, C.A.; Krysiak, K. The prognostic effects of somatic mutations in ER-positive breast cancer. Nat. Commun. 2018, 9, 1–16. [Google Scholar] [CrossRef]
- Casey, A.E.; Laster Jr, W.R.; Ross, G.L. Sustained enhanced growth of carcinoma EO771 in C57 black mice. Proc. Soc. Exp. Biol. Med. 1951, 77, 358–362. [Google Scholar] [CrossRef] [PubMed]
- Aslakson, C.J.; Miller, F.R. Selective events in the metastatic process defined by analysis of the sequential dissemination of subpopulations of a mouse mammary tumor. Cancer Res. 1992, 52, 1399–1405. [Google Scholar] [PubMed]
- Lelekakis, M.; Moseley, J.M.; Martin, T.J.; Hards, D.; Williams, E.; Ho, P.; Lowen, D.; Javni, J.; Miller, F.R.; Slavin, J. A novel orthotopic model of breast cancer metastasis to bone. Clin. Exp. Metastasis 1999, 17, 163–170. [Google Scholar] [CrossRef]
- Jungwirth, U.; van Weverwijk, A.; Melake, M.J.; Chambers, A.F.; Gao, Q.; Fivaz, M.; Isacke, C.M. Generation and characterisation of two D2A1 mammary cancer sublines to model spontaneous and experimental metastasis in a syngeneic BALB/c host. Dis. Model. Mech. 2018, 11, dmm031740. [Google Scholar] [CrossRef] [Green Version]
- Miller, F.R.; McEachern, D.; Miller, B.E. Growth regulation of mouse mammary tumor cells in collagen gel cultures by diffusible factors produced by normal mammary gland epithelium and stromal fibroblasts. Cancer Res. 1989, 49, 6091–6097. [Google Scholar] [PubMed]
- Britt, K.L.; Kendrick, H.; Regan, J.L.; Molyneux, G.; Magnay, F.A.; Ashworth, A.; Smalley, M.J. Pregnancy in the mature adult mouse does not alter the proportion of mammary epithelial stem/progenitor cells. Breast Cancer Res. 2009, 11, R20. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sleeman, K.E.; Kendrick, H.; Ashworth, A.; Isacke, C.M.; Smalley, M.J. CD24 staining of mouse mammary gland cells defines luminal epithelial, myoepithelial/basal and non-epithelial cells. Breast Cancer Res. 2006, 8, R7. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vichai, V.; Kirtikara, K. Sulforhodamine B colorimetric assay for cytotoxicity screening. Nat. Protoc. 2006, 1, 1112–1116. [Google Scholar] [CrossRef]
- Martin, M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet. J. 2011, 17, 10–12. [Google Scholar] [CrossRef]
- Kim, D.; Langmead, B.; Salzberg, S.L. HISAT: A fast spliced aligner with low memory requirements. Nat. Methods 2015, 12, 357–360. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liao, Y.; Smyth, G.K.; Shi, W. featureCounts: An efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics 2014, 30, 923–930. [Google Scholar] [CrossRef] [Green Version]
- Ritchie, M.E.; Phipson, B.; Wu, D.; Hu, Y.; Law, C.W.; Shi, W.; Smyth, G.K. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015, 43, e47. [Google Scholar] [CrossRef]
- McCarthy, D.J.; Chen, Y.; Smyth, G.K. Differential expression analysis of multifactor RNA-Seq experiments with respect to biological variation. Nucleic Acids Res. 2012, 40, 4288–4297. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dobin, A.; Davis, C.A.; Schlesinger, F.; Drenkow, J.; Zaleski, C.; Jha, S.; Batut, P.; Chaisson, M.; Gingeras, T.R. STAR: Ultrafast universal RNA-seq aligner. Bioinformatics 2013, 29, 15–21. [Google Scholar] [CrossRef] [PubMed]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lambouras, M.; Roelofs, C.; Pereira, M.; Gruber, E.; Vieusseux, J.L.; Lanteri, P.; Johnstone, C.N.; Muntz, F.; O’Toole, S.; Ooms, L.M.; et al. Functional and Phenotypic Characterisations of Common Syngeneic Tumour Cell Lines as Estrogen Receptor-Positive Breast Cancer Models. Int. J. Mol. Sci. 2023, 24, 5666. https://doi.org/10.3390/ijms24065666
Lambouras M, Roelofs C, Pereira M, Gruber E, Vieusseux JL, Lanteri P, Johnstone CN, Muntz F, O’Toole S, Ooms LM, et al. Functional and Phenotypic Characterisations of Common Syngeneic Tumour Cell Lines as Estrogen Receptor-Positive Breast Cancer Models. International Journal of Molecular Sciences. 2023; 24(6):5666. https://doi.org/10.3390/ijms24065666
Chicago/Turabian StyleLambouras, Maria, Charlotte Roelofs, Melrine Pereira, Emily Gruber, Jessica L. Vieusseux, Patrick Lanteri, Cameron N. Johnstone, Fenella Muntz, Sandra O’Toole, Lisa M. Ooms, and et al. 2023. "Functional and Phenotypic Characterisations of Common Syngeneic Tumour Cell Lines as Estrogen Receptor-Positive Breast Cancer Models" International Journal of Molecular Sciences 24, no. 6: 5666. https://doi.org/10.3390/ijms24065666





