Synthesis, Characterization and Biological Investigation of the Platinum(IV) Tolfenamato Prodrug–Resolving Cisplatin-Resistance in Ovarian Carcinoma Cell Lines
Abstract
:1. Introduction

2. Results and Discussion
2.1. Synthesis of the Active Esters 1–4
2.2. Synthesis of the Platinum(IV) Complexes 5–13
2.3. Anticancer Activity
2.4. Investigations on Stability Behavior
2.5. In Vitro Inhibition of COX Activity
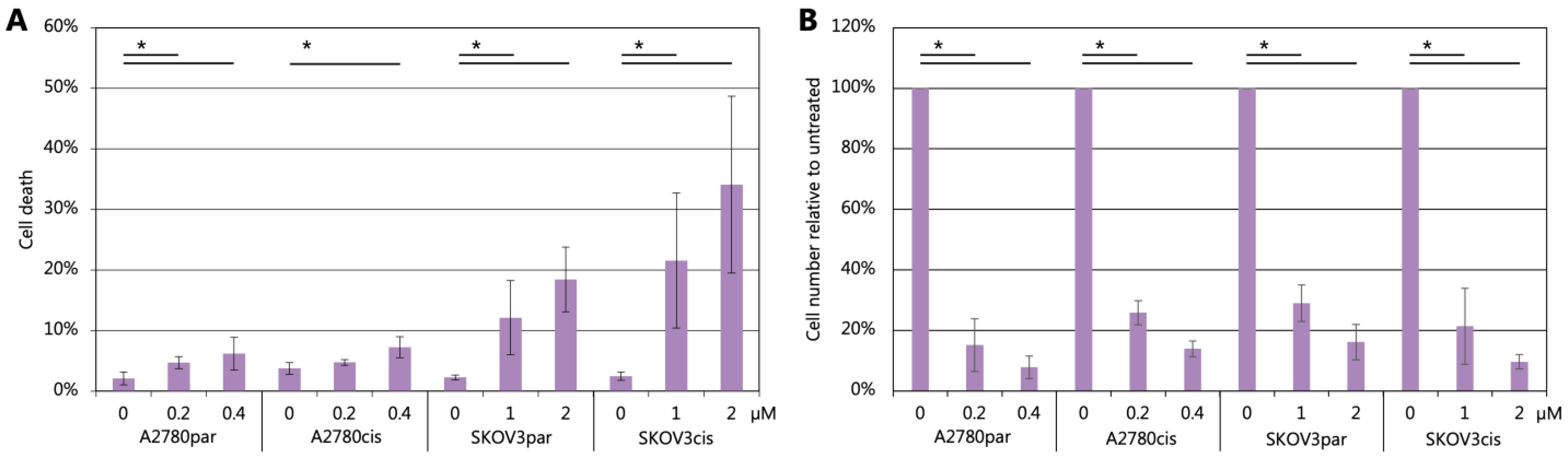
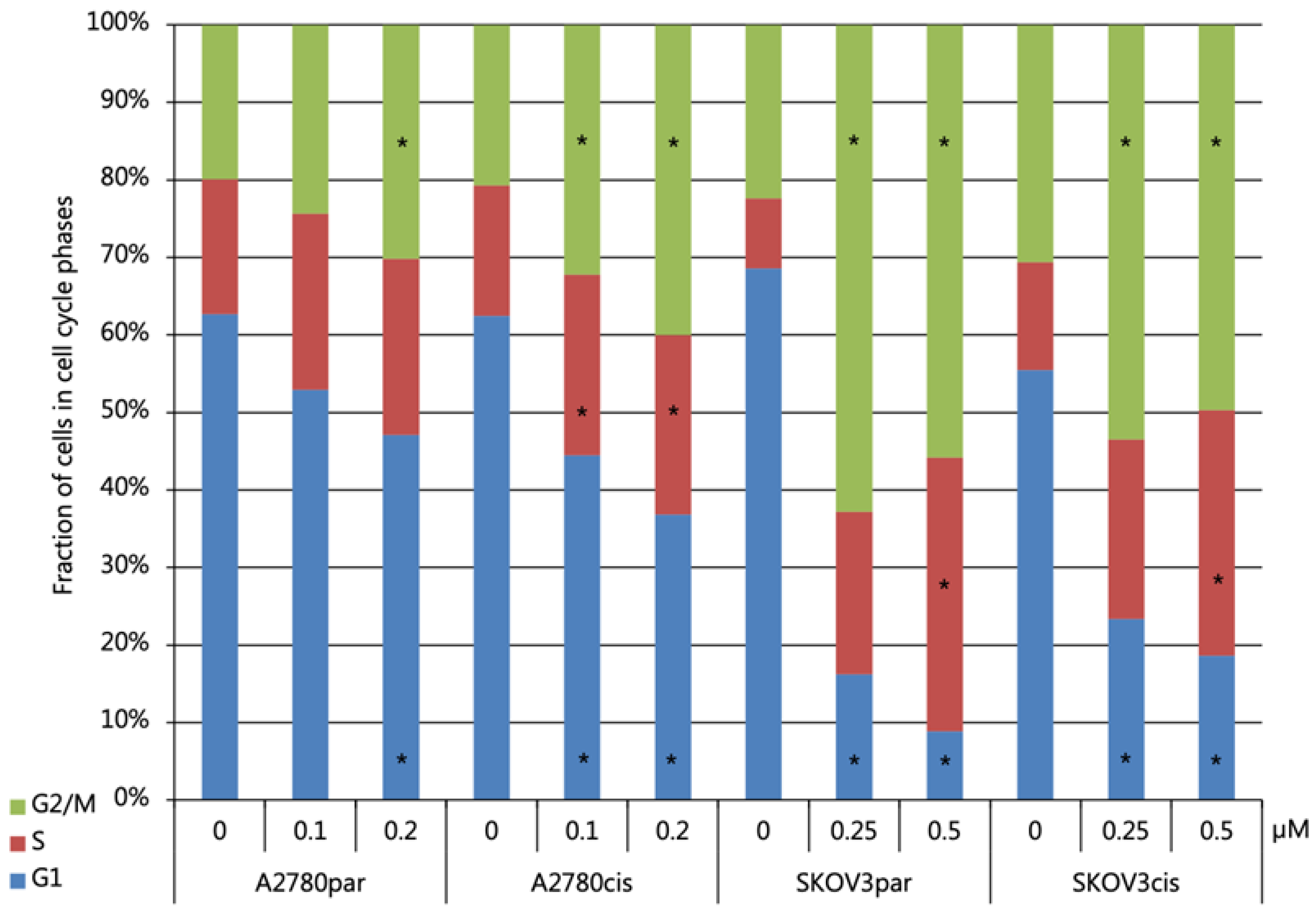
2.6. Cell Death and Cell Cycle Distribution Analyses
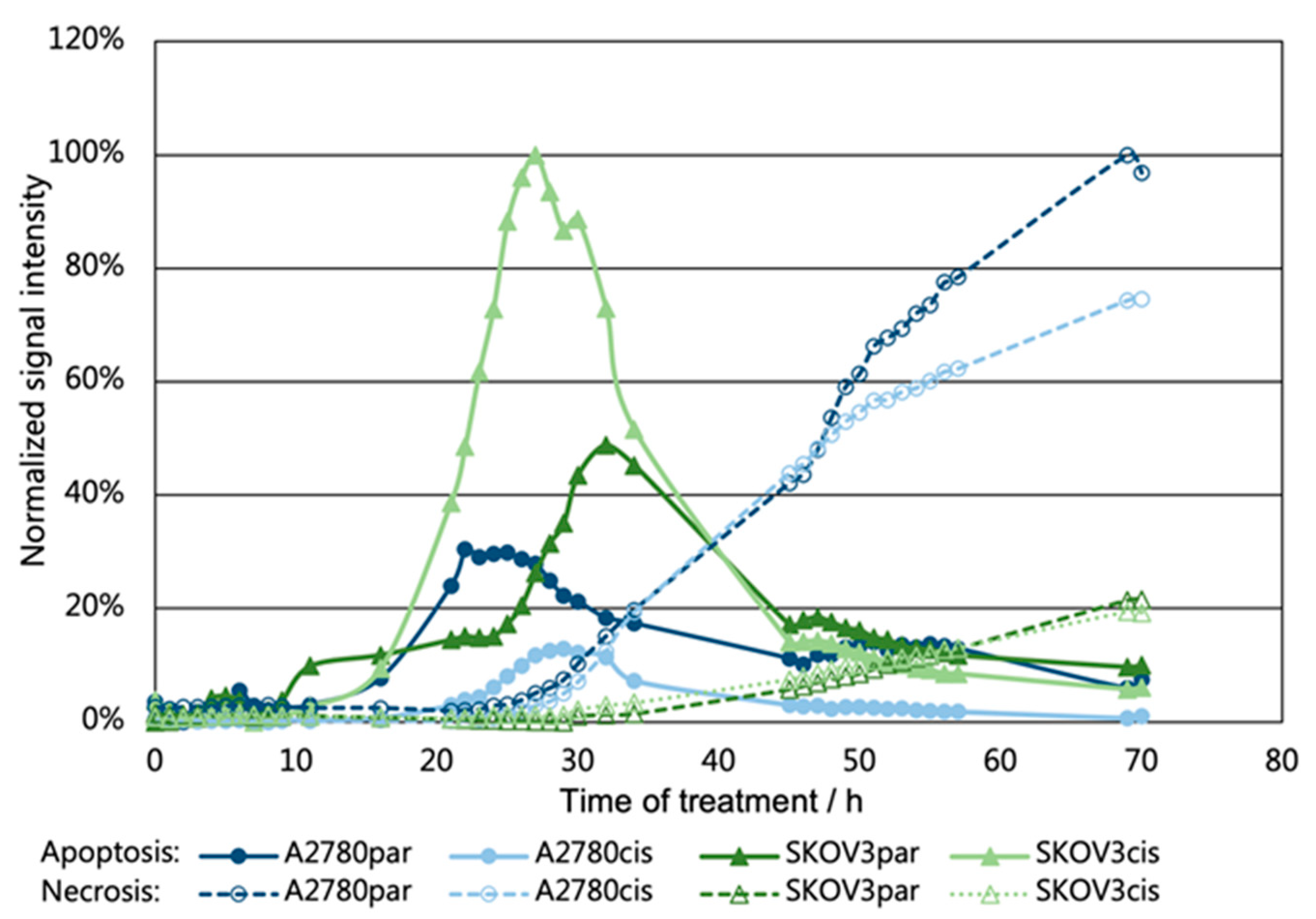
2.7. Investigation of Apoptosis/Necrosis
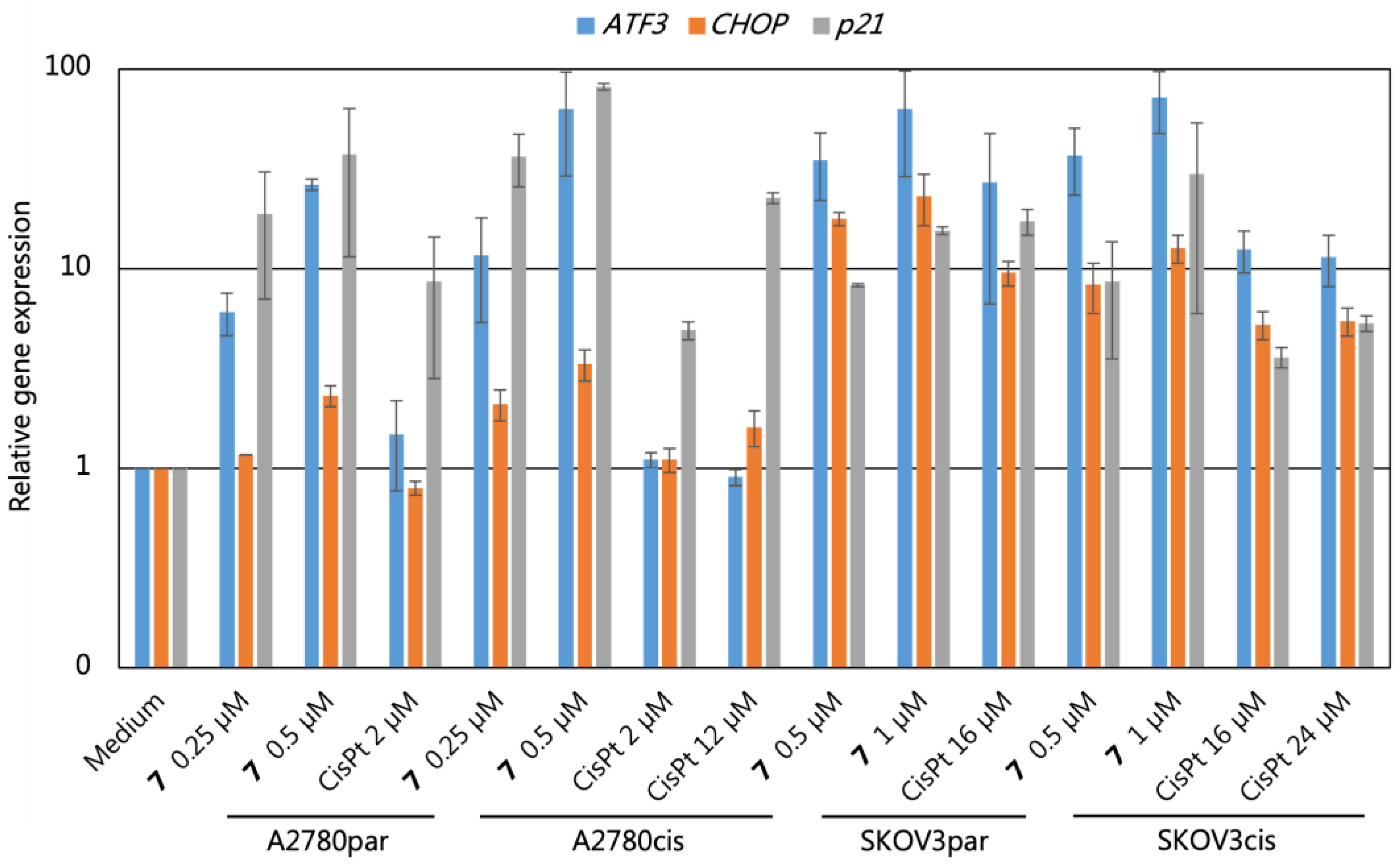
2.8. Gene Expression Analyses
3. Materials and Methods
3.1. Materials and Techniques
3.2. Syntheses
3.3. Stability Studies
3.4. Cell Culture Conditions
3.5. Determination of IC50 Values
3.6. Analyses with Flow-Cytometry
3.7. Real-Time-GloTM Annexin V Apoptosis and Necrosis Assay (Promega, Catalog no. JA1011)
3.8. COX (Ovine/Human) Inhibitor Screening Assay (Cayman Chemical, Item no. 560131)
3.9. Quantitative PCR
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Johnstone, T.C.; Suntharalingam, K.; Lippard, S.J. The Next Generation of Platinum Drugs: Targeted Pt(II) Agents, Nanoparticle Delivery, and Pt(IV) Prodrugs. Chem. Rev. 2016, 116, 3436–3486. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wheate, N.J.; Walker, S.; Craig, G.E.; Oun, R. The status of platinum anticancer drugs in the clinic and in clinical trials. Dalton Trans. 2010, 39, 8113–8127. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xu, Z.; Wang, Z.; Deng, Z.; Zhu, G. Recent advances in the synthesis, stability, and activation of platinum(IV) anticancer prodrugs. Coord. Chem. Rev. 2021, 442, 213991. [Google Scholar] [CrossRef]
- Peña, Q.; Wang, A.; Zaremba, O.; Shi, Y.; Scheeren, H.W.; Metselaar, J.M.; Kiessling, F.; Pallares, R.M.; Wuttke, S.; Lammers, T. Metallodrugs in cancer nanomedicine. Chem. Soc. Rev. 2022, 51, 2544–2582. [Google Scholar] [CrossRef] [PubMed]
- Dasari, S.; Tchounwou, P.B. Cisplatin in cancer therapy: Molecular mechanisms of action. Eur. J. Pharmacol. 2014, 740, 364–378. [Google Scholar] [CrossRef] [Green Version]
- Wang, L.; Zhao, X.; Fu, J.; Xu, W.; Yuan, J. The Role of Tumour Metabolism in Cisplatin Resistance. Front. Mol. Biosci. 2021, 8, 691795. [Google Scholar] [CrossRef]
- Fennell, D.A.; Summers, Y.; Cadranel, J.; Benepal, T.; Christoph, D.C.; Lal, R.; Das, M.; Maxwell, F.; Visseren-Grul, C.; Ferry, D. Cisplatin in the modern era: The backbone of first-line chemotherapy for non-small cell lung cancer. Cancer Treat. Rev. 2016, 44, 42–50. [Google Scholar] [CrossRef]
- Zhu, X.; Feng, J.; Fu, W.; Shu, X.; Wan, X.; Liu, J. Effects of cisplatin on the proliferation, invasion and apoptosis of breast cancer cells following β-catenin silencing. Int. J. Mol. Med. 2020, 45, 1838–1850. [Google Scholar] [CrossRef]
- Helm, C.W.; States, J.C. Enhancing the efficacy of cisplatin in ovarian cancer treatment—Could arsenic have a role. J. Ovarian Res. 2009, 2, 2. [Google Scholar] [CrossRef] [Green Version]
- Ghosh, S. Cisplatin: The first metal based anticancer drug. Bioorg. Chem. 2019, 88, 102925. [Google Scholar] [CrossRef]
- Martinez-Balibrea, E.; Martínez-Cardús, A.; Ginés, A.; de Porras, V.R.; Moutinho, C.; Layos, L.; Manzano, J.L.; Bugés, C.; Bystrup, S.; Esteller, M.; et al. Tumor-Related Molecular Mechanisms of Oxaliplatin Resistance. Mol. Cancer Ther. 2015, 14, 1767–1776. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Heinemann, V.; Haas, M.; Boeck, S. Systemic treatment of advanced pancreatic cancer. Cancer Treat. Rev. 2012, 38, 843–853. [Google Scholar] [CrossRef]
- Inadomi, K.; Kusaba, H.; Matsushita, Y.; Tanaka, R.; Mitsugi, K.; Arimizu, K.; Hirano, G.; Makiyama, A.; Ohmura, H.; Uchino, K.; et al. Efficacy and Safety Analysis of Oxaliplatin-based Chemotherapy for Davanced Gastric Cancer. Anticancer Res. 2017, 37, 2663–2671. [Google Scholar]
- Comella, P.; Casaretti, R.; Sandomenico, C.; Avallone, A.; Franco, L. Role of oxaliplatin in the treatment of colorectal cancer. Ther. Clin. Risk Manag. 2009, 5, 229–238. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gibson, D. Multi-action Pt(IV) anticancer agents; do we understand how they work? J. Inorg. Biochem. 2019, 191, 77–84. [Google Scholar] [CrossRef] [PubMed]
- Gibson, D. Platinum(IV) anticancer agents; are we en route to the holy grail or to a dead end? J. Inorg. Biochem. 2021, 217, 111353. [Google Scholar] [CrossRef]
- Gibson, D. Pt(IV) Anticancer Prodrugs—A Tale of Mice and Men. ChemMedChem 2021, 16, 2188–2191. [Google Scholar] [CrossRef]
- Jung, Y.; Lippard, S.J. Direct Cellular Responses to Platinum-Induced DNA Damage. Chem. Rev. 2007, 107, 1387–1407. [Google Scholar] [CrossRef]
- Oun, R.; Moussa, Y.E.; Wheate, N.J. The side effects of platinum-based chemotherapy drugs: A review for chemists. Dalton Trans. 2018, 47, 6645–6653. [Google Scholar] [CrossRef]
- Wexselblatt, E.; Gibson, D. What do we know about the reduction of Pt(IV) prodrugs. J. Inorg. Biochem. 2012, 117, 220–229. [Google Scholar] [CrossRef]
- Gibson, D. Platinum(IV) anticancer prodrugs—Hypotheses and facts. Dalton Trans. 2016, 45, 12983–12991. [Google Scholar] [CrossRef]
- Zhou, J.; Kang, Y.; Chen, L.; Wang, H.; Liu, J.; Zeng, S.; Yu, L. The Drug-Resistance Mechanisms of Five Platinum-Based Antitumor Agents. Front. Pharmacol. 2020, 11, 343. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rottenberg, S.; Disler, C.; Perego, P. The rediscovery of platinum-based cancer therapy. Nature Rev. 2021, 21, 37–50. [Google Scholar] [CrossRef]
- Kenny, R.G.; Chuah, S.W.; Crawford, A.; Marmion, C.J. Platinum(IV) Prodrugs—A Step Closer to Ehrlich’s Vision. Eur. J. Inorg. Chem. 2017, 2017, 1596–1612. [Google Scholar] [CrossRef] [Green Version]
- Ravera, M.; Gabano, E.; McGlinchey, M.J.; Osella, D. A view on multi-action Pt(IV) antitumor prodrugs. Inorg. Chim. Acta 2019, 492, 32–47. [Google Scholar] [CrossRef]
- Spector, D.; Krasnovskaya, O.; Pavlov, K.; Erofeev, A.; Gorelkin, P.; Beloglazkina, E.; Majouga, A. Pt(IV) Prodrugs with NSAIDs as Axial Ligands. Int. J. Mol. Sci. 2021, 22, 3817. [Google Scholar] [CrossRef] [PubMed]
- Ravera, M.; Gabano, E.; McGlinchey, M.J.; Osella, D. Pt(IV) antitumor prodrugs: Dogmas, paradigms, and realities. Dalton Trans. 2022, 51, 2121–2134. [Google Scholar] [CrossRef] [PubMed]
- Kenny, R.G.; Marmion, J. Toward Multi-Targeted Platinum and Ruthenium Drugs—A New Paradigm in Cancer Drug Treatment Regimens? Chem. Rev. 2019, 119, 1058–1137. [Google Scholar] [CrossRef]
- Pathak, R.K.; Marrache, S.; Choi, J.H.; Berding, T.B.; Dhar, S. The Prodrug Platin—A: Simultaneous Release of Cisplatin and Aspirin. Angew. Chem. Int. Ed. 2014, 53, 1963–1967. [Google Scholar] [CrossRef]
- Neumann, W.; Crews, B.C.; Marnett, L.J.; Hey-Hawkins, E. Conjugates of Cisplatin and Cyclooxygenase Inhibitors as Potent Antitumor Agents Overcoming Cisplatin Resistance. ChemMedChem 2014, 9, 1150–1153. [Google Scholar] [CrossRef] [Green Version]
- Basu, U.; Banik, B.; Wen, R.; Pathak, R.K.; Dhar, S. The Platin-X series: Activation, targeting, and delivery. Dalton Trans. 2016, 45, 12992–13004. [Google Scholar] [CrossRef] [Green Version]
- Neumann, W.; Crews, B.C.; Sárosi, M.B.; Daniel, C.M.; Ghebreselasie, K.; Scholz, M.S.; Marnett, L.J.; Hey-Hawkins, E. Conjugation of Cisplatin Analogues and Cyclooxygenase Inhibitors to Overcome Cisplatin Resistance. ChemMedChem. 2015, 10, 183–192. [Google Scholar] [CrossRef] [Green Version]
- Curci, A.; Denora, N.; Iacobazzi, R.M.; Ditaranto, N.; Hoeschele, J.D.; Margiotta, N.; Natile, G. Synthesis, characterization, and in vitro cytotoxicity of a Kiteplatin-Ibuprofen Pt(IV) prodrug. Inorg. Chim. Acta 2018, 472, 221–228. [Google Scholar] [CrossRef]
- Tan, J.; Li, C.; Wang, Q.; Li, S.; Chen, S.; Zhang, J.; Wang, P.C.; Ren, L.; Liang, X.J. A Carrier-Free Nanostructure Based on Platinum(IV) Prodrug Enhances Cellular Uptake and Cytotoxicity. Mol. Pharm. 2018, 15, 1724–1728. [Google Scholar] [CrossRef]
- Ravera, M.; Zanellato, I.; Gabano, E.; Perin, E.; Rangone, B.; Coppola, M.; Osella, D. Antiproliferative Activity of Pt(IV) Conjugates Containing the Non-Steroidal Anti-Inflammatory Drugs (NSAIDs) Ketoprofen and Naproxen. Int. J. Mol. Sci. 2019, 20, 3074. [Google Scholar] [CrossRef] [Green Version]
- Tolan, D.A.; Abdel-Monem, Y.K.; El-Nagar, M.A. Anti-tumor platinum (IV) complexes bearing the anti-inflammatory drug naproxen in the axial position. Appl. Organomet. Chem. 2019, 33, e4763. [Google Scholar] [CrossRef]
- Chen, Y.; Wang, Q.; Li, Z.; Liu, Z.; Zhao, Y.; Zhang, J.; Liu, M.; Wang, Z.; Li, D.; Han, J. Naproxen platinum(IV) hybrids inhibiting cycloxygenases and matrix metalloproteinases and causing DNA damage: Synthesis and biological evaluation as antitumor agents in vitro and in vivo. Dalton Trans. 2020, 49, 5192–5204. [Google Scholar] [CrossRef] [PubMed]
- Jin, S.; Muhammad, N.; Sun, Y.; Tan, Y.; Yuan, H.; Song, D.; Guo, Z.; Wang, X. Multispecific Platinum(IV) Complex Deters Breast Cancer via Interposing Inflammation and Immunosuppression as an Inhibitor of COX-2 and PD-L1. Angew. Chem. Int. Ed. 2020, 59, 23313–23321. [Google Scholar] [CrossRef]
- Song, X.Q.; Ma, Z.Y.; Wu, Y.G.; Dai, M.L.; Wang, D.B.; Xu, J.Y.; Liu, Y. New NSAID-Pt(IV) prodrugs to suppress metastasis and invasion of tumor cells and enhance anti-tumor effect in vitro and in vivo. Eur. J. Med. Chem. 2019, 167, 377–387. [Google Scholar] [CrossRef]
- Hu, W.; Fang, L.; Hua, W.; Gou, S. Biotin-Pt (IV)-indomethacin hybrid: A targeting anticancer prodrugproviding enhanced cancer cellular uptake and reversing cisplatin resistance. J. Inorg. Biochem. 2017, 175, 47–57. [Google Scholar] [CrossRef] [PubMed]
- Petruzzella, E.; Sirota, R.; Solazzo, I.; Gandin, V.; Gibson, D. Triple action Pt(IV) derivatives of cisplatin: A new class of potent anticancer agents that overcome resistance. Chem. Sci. 2018, 9, 4299–4307. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Karmakar, S.; Kostrhunova, H.; Ctvrtlikova, T.; Novohradsky, V.; Gibson, D.; Brabec, V. Platinum(IV)-Estramustine Multiaction Prodrugs Are Effective Antiproliferative Agents against Prostate Cancer Cells. J. Med. Chem. 2020, 63, 13861–13877. [Google Scholar] [CrossRef]
- Predarska, I.; Saoud, M.; Morgan, I.; Eichhorn, T.; Kaluđerović, G.N.; Hey-Hawkins, E. Cisplatin-cyclooxygenase inhibitor conjugates, free and immobilised in mesoporous silica SBA-15, prove highly potent against triple-negative MDA-MB-468 breast cancer cell line. Dalton Trans. 2022, 51, 857–869. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.; Chen, F.; Shang, L. Advances in antitumor effects of NSAIDs. Cancer Manag. Res. 2018, 10, 4631–4640. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wong, R.S.Y. Role of Nonsteroidal Anti-Inflammatory Drugs (NSAIDs) in Cancer Prevention and Cancer Promotion. Adv. Pharmacol. Sci. 2019, 2019, 3418975. [Google Scholar] [CrossRef] [Green Version]
- Day, R.O.; Graham, G.G. The vascular effects of COX-2 selective inhibitors. Aust. Prescr. 2004, 27, 142–145. [Google Scholar] [CrossRef] [Green Version]
- Attiq, A.; Jalil, J.; Husain, K.; Ahmad, W. Raging the War Against Inflammation with Natural Products. Front. Pharmacol. 2018, 9, 976. [Google Scholar] [CrossRef] [Green Version]
- Williams, C.S.; Mann, M.; Dubois, R.N. The role of cyclooxygenases in inflammation, cancer, and development. Oncogene 1999, 18, 7908–7916. [Google Scholar] [CrossRef] [Green Version]
- Nørregaard, R.; Kwon, T.H.; Frøkiær, J. Physiology and pathophysiology of cyclooxygenase-2 and prostaglandin E2 in the kidney. Kidney Res. Clin. Pract. 2015, 34, 194–200. [Google Scholar] [CrossRef] [Green Version]
- Zappavigna, S.; Cossu, A.M.; Grimaldi, A.; Bocchetti, M.; Ferraro, G.A.; Nicoletti, G.F.; Filosa, R.; Caraglia, M. Advances in Pharmacological Sciences. Int. J. Mol. Sci. 2020, 21, 2605. [Google Scholar] [CrossRef] [Green Version]
- Rawat, C.; Kukal, S.; Dahiya, U.R.; Kukreti, R. Cyclooxygenase-2 (COX-2) inhibitors: Future therapeutic strategies for epilepsy management. J. Neuroinflammation 2019, 16, 197. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gosh, N.; Chaki, R.; Mandal, V.; Mandal, S.C. COX-2 as a target for cancer chemotherapy. Pharmacol. Rep. 2010, 62, 233–244. [Google Scholar] [CrossRef]
- Goradel, N.H.; Najafi, M.; Salehi, E.; Farhood, B.; Mortezaee, K. Cyclooxygenase-2 in cancer: A review. J. Cell Physiol. 2019, 234, 5683–5699. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Zhang, R.; Yu, L.; Xiao, J.; Zhou, X.; Li, X.; Song, P.; Li, X. Aspirin Use and Common Cancer Risk: A Meta-Analysis of Cohort Studies and Randomized Controlled Trials. Front. Oncol. 2021, 11, 690219. [Google Scholar] [CrossRef]
- Ng, K.; Meyerhardt, J.A.; Chan, A.T.; Sato, K.; Chan, J.A.; Niedzwiecki, D.; Saltz, L.B.; Mayer, R.J.; Benson, A.B., III; Schaefer, P.L.; et al. Aspirin and COX-2 Inhibitor Use in Patients With Stage III Colon Cancer. J. Natl. Cancer Inst. 2015, 107, 345. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Okamoto, K.; Saito, Y.; Narumi, K.; Furugen, A.; Iseki, K.; Kobayashi, M. Anticancer effects of non-steroidal anti-inflammatory drugs against cancer cells and cancer stem cells. Toxicol. Vitr. 2021, 74, 105155. [Google Scholar] [CrossRef]
- Gurpinar, E.; Grizzle, W.E.; Piazza, G.A. NSAIDs Inhibit Tumorigenesis, but How? Clin. Cancer Res. 2014, 20, 1104–1113. [Google Scholar] [CrossRef] [Green Version]
- Khan, S.A.; Imam, S.M.; Ahmad, A.; Basha, S.H.; Husain, A. Synthesis, molecular docking with COX 1& II enzyme, ADMET screening and in vivo anti-inflammatory activity of oxadiazole, thiadiazole and triazole analogs of felbinac. Bioorg Med. Chem 2018, 22, 469–484. [Google Scholar]
- Hosie, G.; Bird, H. The topical NSAID felbinac versus oral NSAIDs: A critical review. Eur. J. Rheumatol. Inflamm. 1994, 14, 21–28. [Google Scholar]
- El-Sheikh, A.; Khired, Z. Interactions of Analgesics with Cisplatin: Modulation of Anticancer Efficacy and Potential Organ Toxicity. Medicina 2022, 58, 46. [Google Scholar] [CrossRef]
- Matsumoto, R.; Tsuda, M.; Yoshida, K.; Tanino, M.; Kimura, T.; Nishihara, H.; Abe, T.; Shinohara, N.; Nonomura, K.; Tanaka, S. Aldo-keto reductase 1C1 induced by interleukin-1β mediates the invasive potential and drug resistance of metastatic bladder cancer cells. Sci. Rep. 2016, 6, 34625. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shiiba, M.; Yamagami, H.; Yamamoto, A.; Minakawa, Y.; Okamoto, A.; Kasamatsu, A.; Sakamoto, Y.; Uzawa, K.; Takiguchi, Y.; Tanzawa, H. Mefenamic acid enhances anticancer drug sensitivity via inhibition of aldo-keto reductase 1C enzyme activity. Oncol. Rep. 2017, 37, 2025–2032. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xing, L.; Yang, C.X.; Zhao, D.; Shen, L.J.; Zhou, T.J.; Bi, Y.Y.; Huang, Z.J.; Weid, Q.; Li, L.; Li, F.; et al. A carrier-free anti-inflammatory platinum (II) self-delivered nanoprodrug for enhanced breast cancer therapy. J. Control. Release 2021, 331, 460–471. [Google Scholar] [CrossRef] [PubMed]
- Yang, C.X.; Xing, L.; Chang, X.; Zhou, T.J.; Bi, Y.Y.; Yu, Z.Q.; Zhang, Z.Q.; Jiang, H.L. Synergistic Platinum(II) Prodrug Nanoparticles for Enhanced Breast Cancer Therapy. Mol. Pharm. 2020, 17, 1300–1309. [Google Scholar] [CrossRef]
- Totta, X.; Papadopoulou, A.A.; Hatzidimitriou, A.G.; Papadopoulos, A.; Psomas, G. Synthesis, structure and biological activity of nickel(II) complexes with mefenamato and nitrogen-donor ligands. J. Inorg. Biochem. 2015, 145, 79–93. [Google Scholar] [CrossRef]
- Kovala-Demertzi, D.; Hadjipavlou-Litina, D.; Primikiri, A.; Staninska, M.; Kotoglou, C.; Demertzis, M.A. Anti-Inflammatory, Antiproliferative, and Radical-Scavenging Activities of Tolfenamic Acid and Its Metal Complexes. Chem. Biodivers. 2009, 6, 948–960. [Google Scholar] [CrossRef]
- Gacki, M.; Kafarska, K.; Pietrzak, A.; Szczesio, M.; Korona-Głowniak, I.; Wolf, W.M. Transition Metal Complexes with Flufenamic Acid for Pharmaceutical Applications—A Novel Three-Centered Coordination Polymer of Mn(II) Flufenamate. Materials 2020, 13, 3705. [Google Scholar] [CrossRef]
- Hurtado, M.; Sankpal, U.T.; Chhabra, J.; Brown, D.T.; Maram, R.; Patel, R.; Gurung, R.K.; Simecka, J.; Holder, A.A.; Basha, R. Copper-tolfenamic acid: Evaluation of stability and anti-cancer activity. Invest. New Drugs 2019, 37, 27–34. [Google Scholar] [CrossRef]
- Mazumder, M.M.U.; Sukul, A.; Saha, S.K.; Chowdhury, A.A.; Mamun, Y. A comprehensive in vitro biological investigation of metal complexes of tolfenamic acid. Alexandria Med. J. 2018, 54, 23–26. [Google Scholar] [CrossRef] [Green Version]
- Hudecova, L.; Jomova, K.; Lauro, P.; Simunkova, M.; Alwasel, S.H.; Alhazza, I.M.; Moncol, J.; Valko, M. Antimicrobial and antifungal activities of bifunctional copper(II) complexes with non-steroidal anti-inflammatory drugs, flufenamic, mefenamic and tolfenamic acids and 1,10-phenanthroline. Open Chem. J. 2020, 18, 1444–1451. [Google Scholar] [CrossRef]
- Smolko, L.; Smolková, R.; Samoľová, E.; Morgan, I.; Saoud, M.; Kaluđerović, G.N. Two isostructural Co(II) flufenamato and niflumato complexes with bathocuproine: Analogues with a different cytotoxic activity. J. Inorg. Biochem. 2020, 210, 111160. [Google Scholar] [CrossRef] [PubMed]
- Yang, L.; Xie, H.-J.; Li, Y.-Y.; Wang, X.; Liu, X.-X.; Mai, J. Molecular mechanisms of platinum-based chemotherapy resistance in ovarian cancer (Review). Oncol. Rep. 2022, 47, 82. [Google Scholar] [CrossRef] [PubMed]
- Guo, C.; Song, C.; Zhang, J.; Gao, Y.; Qi, Y.; Zhao, Z.; Yuan, C. Revisiting chemoresistance in ovarian cancer: Mechanism, biomarkers, and precision medicine. Genes Dis. 2022, 9, 668–681. [Google Scholar] [CrossRef] [PubMed]
- Allemani, C.; Weir, H.K.; Carreira, H.; Harewood, R.; Spika, D.; Wang, X.-S.; Bannon, F.; Ahn, J.V.; Johnson, C.J.; Bonaventure, A.; et al. Global surveillance of cancer survival 1995–2009: Analysis of individual data for 25,676,887 patients from 279 population-based registries in 67 countries (CONCORD-2). Lancet 2015, 385, 977–1010. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, J.; Kuang, Y.; Shi, J.; Gao, Y.; Zhou, J.; Xu, B. The conjugation of nonsteroidal anti-inflammatory drugs (NSAID) to small peptides for generating multifunctional supramolecular nanofibers/hydrogels. Beilstein, J. Org. Chem. 2013, 9, 908–917. [Google Scholar] [CrossRef] [Green Version]
- Yempala, T.; Babu, T.; Karmakar, S.; Nemirovski, A.; Ishan, M.; Gandin, V.; Gibson, D. Expanding the Arsenal of Pt(IV) Anticancer Agents: Multi-action Pt(IV) Anticancer Agents with Bioactive Ligands Possessing a Hydroxy Functional Group. Angew. Chem. Int. Ed. Engl. 2019, 58, 18218–18223. [Google Scholar] [CrossRef]
- Ermondi, G.; Caron, G.; Ravera, M.; Gabano, E.; Bianco, S.; Platts, J.A.; Osella, D. Molecular interaction fields vs. quantum-mechanical-based descriptors in the modelling of lipophilicity of platinum(IV) complexes. Dalton Trans. 2013, 42, 3482–3489. [Google Scholar] [CrossRef] [Green Version]
- Shi, Y.; Liu, S.A.; Kerwood, D.J.; Goodisman, J.; Dabrowiak, J.C. Pt(IV) complexes as prodrugs for cisplatin. J. Inorg. Biochem. 2012, 107, 6–14. [Google Scholar] [CrossRef] [Green Version]
- Raveendran, R.; Braude, J.P.; Wexselblatt, E.; Novohradsky, V.; Stuchlikova, O.; Brabec, V.; Gandin, V.; Gibson, D. Pt(IV) derivatives of cisplatin and oxaliplatin with phenylbutyrate axial ligands are potent cytotoxic agents that act by several mechanisms of action. Chem. Sci. 2016, 7, 2381–2391. [Google Scholar] [CrossRef] [Green Version]
- Petruzzella, E.; Braude, J.P.; Aldrich-Wright, J.R.; Gandin, V.; Gibson, D. A Quadruple-Action Platinum(IV) Prodrug with Anticancer Activity Against KRAS Mutated Cancer Cell Lines. Angew. Chem. Int. Ed. 2017, 56, 11539–11544. [Google Scholar] [CrossRef]
- Li, X.; Liu, Y.; Tian, H. Current Developments in Pt(IV) Prodrugs Conjugated with Bioactive Ligands. Bioinorg. Chem. Appl. 2018, 2018, 8276139. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kritsch, D.; Hoffmann, F.; Steinbach, D.; Jansen, L.; Photini, S.M.; Gajda, M.; Mosig, A.S.; Sonnemann, J.; Peters, S.; Melnikova, M.; et al. Tribbles 2 mediates cisplatin sensitivity and DNA damage response in epithelial ovarian cancer. Int. J. Cancer 2017, 141, 1600–1613. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Verheijen, M.; Lienhard, M.; Schrooders, Y.; Clayton, O.; Nudischer, R.; Boerno, S.; Timmermann, B.; Selevsek, N.; Schlapbach, R.; Gmuender, H.; et al. DMSO induces drastic changes in human cellular processes and epigenetic landscape in vitro. Sci. Rep. 2019, 9, 4641. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kuete, V.; Karaosmanoğlu, O.; Sivas, H. Anticancer Activities of African Medicinal Spices and Vegetables. In Medicinal Spices and Vegetables from Africa; Elsevier: Amsterdam, The Netherlands, 2017. [Google Scholar]
- Riss, T.L.; Moravec, R.A.; Niles, A.L.; Duellman, S.; Benink, H.A.; Worzella, T.J.; Minor, L. Cell Viability Assays; Eli Lilly & Company and the National Center for Advancing Translational Sciences: Bethesda, MD, USA, 2004. [Google Scholar]
- Ammar, A.A.; Raveendran, R.; Gibson, D.; Nassar, T.; Benita, S. A Lipophilic Pt(IV) Oxaliplatin Derivative Enhances Antitumor Activity. J. Med. Chem. 2016, 59, 9035–9046. [Google Scholar] [CrossRef]
- Moeller, N.; Kangarloo, B.S.; Puscasu, I.; Mock, C.; Krebs, B.; Wolff, J.E. Rational design of platinum chemotherapeutic drugs. Anticancer Res. 2000, 20, 4435–4439. [Google Scholar] [PubMed]
- Tallen, G.; Mock, C.; Gangopadhyay, S.B.; Kangarloo, B.; Krebs, B.; Wolff, J.E. Overcoming cisplatin resistance: Design of novel hydrophobic platinum compounds. Anticancer Res. 2000, 20, 445–449. [Google Scholar]
- Kim, H.J.; Bae, S.C. Histone deacetylase inhibitors: Molecular mechanisms of action and clinical trials as anti-cancer drugs. Am. J. Transl. Res. 2011, 3, 166–179. [Google Scholar]
- Kusaczuk, M.; Krętowski, R.; Bartoszewicz, M.; Cechowska-Pasko, M. Phenylbutyrate—A pan-HDAC inhibitor—Suppresses proliferation of glioblastoma LN-229 cell line. Tumor Biol. 2016, 37, 931–942. [Google Scholar] [CrossRef] [Green Version]
- Barbosa, M.A.G.; Xavier, C.P.R.; Pereira, R.F.; Petrikaitė, V.; Vasconcelos, M.H. 3D Cell Culture Models as Recapitulators of the Tumor Microenvironment for the Screening of Anti-Cancer Drugs. Cancers 2022, 14, 190. [Google Scholar] [CrossRef]
- Melnikova, M.; Wauer, U.S.; Mendus, D.; Hilger, R.A.; Oliver, T.G.; Mercer, K.; Gohlke, B.O.; Erdmann, K.; Niederacher, D.; Neubauer, H.; et al. Diphenhydramine increases the therapeutic window for platinum drugs by simultaneously sensitizing tumor cells and protecting normal cells. Mol. Oncol. 2020, 14, 686–703. [Google Scholar] [CrossRef]
- Khoury, A.; Sakoff, J.A.; Gilbert, J.; Scott, K.F.; Karan, S.; Gordon, C.P.; Aldrich-Wright, J.R. Cyclooxygenase-Inhibiting Platinum(IV) Prodrugs with Potent Anticancer Activity. Pharmaceutics 2022, 14, 787. [Google Scholar] [CrossRef] [PubMed]
- Crowley, L.C.; Scott, A.P.; Marfell, B.J.; Boughaba, J.A.; Chojnowski, G.; Waterhouse, N.J. Measuring Cell Death by Propidium Iodide Uptake and Flow Cytometry. Cold Spring Harb. Protoc. 2016, 2016, prot087163. [Google Scholar] [CrossRef]
- Shen, Y.; Vignali, P.; Wang, R. Rapid Profiling Cell Cycle by Flow Cytometry Using Concurrent Staining of DNA and Mitotic Markers. Bio Protoc. 2017, 7, e2517. [Google Scholar] [CrossRef] [Green Version]
- Galluzzi, L.; Vitale, I.; Senovilla, L.; Eisenberg, T.; Carmona-Gutierrez, D.; Vacchelli, E.; Robert, T.; Ripoche, H.; Jägemann, N.; Paccard, C.; et al. Independent transcriptional reprogramming and apoptosis induction by cisplatin. Cell Cycle 2012, 11, 3472–3480. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schoch, S.; Gajewski, S.; Rothfuß, J.; Hartwig, A.; Köberle, B. Comparative Study of the Mode of Action of Clinically Approved Platinum-Based Chemotherapeutics. Int. J. Mol. Sci. 2020, 21, 6928. [Google Scholar] [CrossRef] [PubMed]
- Liggett, J.L.; Zhang, X.; Eling, T.E.; Baek, S.J. Anti-tumor activity of non-steroidal anti-inflammatory drugs: Cyclooxygenase-independent targets. Cancer Lett. 2014, 346, 217–224. [Google Scholar] [CrossRef] [Green Version]
- Min, D.; Zhao, Y.; Monks, A.; Palmisano, A.; Hose, C.; Teicher, B.A.; Doroshow, J.H.; Simon, R.M. Identification of pharmacodynamic biomarkers and common molecular mechanisms of response to genotoxic agents in cancer cell lines. Cancer Chemother. Pharmacol. 2019, 84, 771–780. [Google Scholar] [CrossRef]
- Ohoka, N.; Yoshii, S.; Hattori, T.; Onozaki, K.; Hayashi, H. TRB3, a novel ER stress-inducible gene, is induced via ATF4–CHOP pathway and is involved in cell death. EMBO J. 2005, 24, 1243–1255. [Google Scholar] [CrossRef]
- Lee, K.; Tirasophon, W.; Shen, X.; Michalak, M.; Prywes, R.; Okada, T.; Yoshida, H.; Mori, K.; Kaufman, R.J. IRE1-mediated unconventional mRNA splicing and S2P-mediated ATF6 cleavage merge to regulate XBP1 in signaling the unfolded protein response. Genes Dev. 2002, 16, 452–466. [Google Scholar] [CrossRef] [Green Version]
- Bottone, F.G.; Martinez, J.M.; Alston-Mills, B.; Eling, T.E. Gene modulation by Cox-1 and Cox-2 specific inhibitors in human colorectal carcinoma cancer cells. Carcinogenesis 2004, 25, 349–357. [Google Scholar] [CrossRef] [PubMed]
- Bottone, F.G.; Martinez, J.M.; Collins, J.B.; Afshari, C.A.; Eling, T.E. Gene Modulation by the Cyclooxygenase Inhibitor, Sulindac Sulfide, in Human Colorectal Carcinoma Cells: Possible Link to Apoptosis. J. Biol. Chem. 2003, 278, 25790–25801. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fu, D.; Wang, C.; Yu, L.; Yu, R. Induction of ferroptosis by ATF3 elevation alleviates cisplatin resistance in gastric cancer by restraining Nrf2/Keap1/xCT signaling. Cell. Mol. Biol. Lett. 2021, 26, 26. [Google Scholar] [CrossRef] [PubMed]
| Compound | A2780par | A2780cis | SKOV3par | SKOV3cis |
|---|---|---|---|---|
| 5 | 0.15 ± 0.06 | 0.14 ± 0.04 | 1.19 ± 0.12 | 0.39 ± 0.04 |
| RF (5) | 0.9 | 0.3 | ||
| 6 | 0.88 ± 0.55 | 0.32 ± 0.14 | 3.95 ± 1.33 | 1.37 ± 0.57 |
| RF (6) | 0.4 | 0.3 | ||
| 7 | 0.22 ± 0.12 | 0.12 ± 0.02 | 1.07 ± 0.25 | 0.38 ± 0.03 |
| RF (7) | 0.5 | 0.3 | ||
| 8 | 7.08 ± 3.40 | 6.07 ± 1.14 | 55.95 ± 12.34 | 65.35 ± 14.35 |
| RF (8) | 0.9 | 1.2 | ||
| 10 | 5.60 ± 3.11 | 4.88 ± 0.61 | 59.98 ± 12.94 | 66.92 ± 12.89 |
| RF (10) | 0.9 | 1.1 | ||
| 11 | 42.40 ± 9.04 | 21.86 ± 0.69 | >100 a | >100 a |
| RF (11) | 0.5 | - | ||
| 12 | 13.31 ± 8.16 | 8.33 ± 0.61 | >100 a | >100 a |
| RF (12) | 0.6 | - | ||
| 13 | 1.58 ± 0.78 | 1.05 ± 0.14 | 10.64 ± 2.04 | 9.82 ± 2.51 |
| RF (13) | 0.7 | 0.9 | ||
| MEF | >100 a | >100 a | >100 a | >100 a |
| RF (MEF) | - | - | ||
| FLU | >100 a | >100 a | >100 a | >100 a |
| RF (FLU) | - | - | ||
| TOLF | >100 a | >100 a | >100 a | >100 a |
| RF (TOLF) | - | - | ||
| FEL | >100 a | >100 a | >100 a | >100 a |
| RF (FEL) | - | - | ||
| CisPt | 3.41 ± 0.95 | 11.43 ± 2.53 | 16.97 ± 4.48 | 25.33 ± 8.76 |
| RF (CisPt) | 3.4 | 1.5 | ||
| CisPt 0.1% DMSO | 2.47 ± 0.16 | 10.20 ± 2.00 | 15.81 ± 3.47 | 27.77 ± 11.57 |
| RF (CisPt 0.1% DMSO) | 4.1 | 1.7 | ||
| CisPt + TOLF (1:1) | 1.64 ± 0.57 | 8.25 ± 1.60 | 13.98 ± 4.29 | 21.37 ± 8.18 |
| RF (CisPt + TOLF, 1:1) | 5.0 | 1.5 | ||
| OxaPt | 1.06 ± 0.27 | 1.49 ± 0.85 | 39.23 ± 8.75 | 113.98 ± 28.39 |
| RF (OxaPt) | 1.4 | 2.9 |
| Gene | Sequence | Ta | |
|---|---|---|---|
| GAPDH | F | GCGACACCCACTCCTCCACC | 60 °C |
| R | GAGGTCCACCACCCTGTTGC | ||
| HPRT | F | ACGAAGTGTTGGATATAAGC | 56 °C |
| R | ATAATTTTACTGGCGATGTC | ||
| ATF3 | F | GCCATCCAGAACAAGCACCT | 60 °C |
| R | TTCTCGTCGCCTCTTTTTCC | ||
| CHOP | F | GCACCTCCCAGAGCCCTCACTCTCC | 60 °C |
| R | GTCTACTCCAAGCCTTCCCCCTGCG | ||
| p21 | F | TCGACTTTGTCACCGAGACACCAC | 60 °C |
| R | CAGGTCCACATGGTCTTCCTCTG |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Barth, M.-C.; Häfner, N.; Runnebaum, I.B.; Weigand, W. Synthesis, Characterization and Biological Investigation of the Platinum(IV) Tolfenamato Prodrug–Resolving Cisplatin-Resistance in Ovarian Carcinoma Cell Lines. Int. J. Mol. Sci. 2023, 24, 5718. https://doi.org/10.3390/ijms24065718
Barth M-C, Häfner N, Runnebaum IB, Weigand W. Synthesis, Characterization and Biological Investigation of the Platinum(IV) Tolfenamato Prodrug–Resolving Cisplatin-Resistance in Ovarian Carcinoma Cell Lines. International Journal of Molecular Sciences. 2023; 24(6):5718. https://doi.org/10.3390/ijms24065718
Chicago/Turabian StyleBarth, Marie-Christin, Norman Häfner, Ingo B. Runnebaum, and Wolfgang Weigand. 2023. "Synthesis, Characterization and Biological Investigation of the Platinum(IV) Tolfenamato Prodrug–Resolving Cisplatin-Resistance in Ovarian Carcinoma Cell Lines" International Journal of Molecular Sciences 24, no. 6: 5718. https://doi.org/10.3390/ijms24065718






