Gene Expression Profiling as a Novel Diagnostic Tool for Neurodegenerative Disorders
Abstract
1. Introduction
2. Results
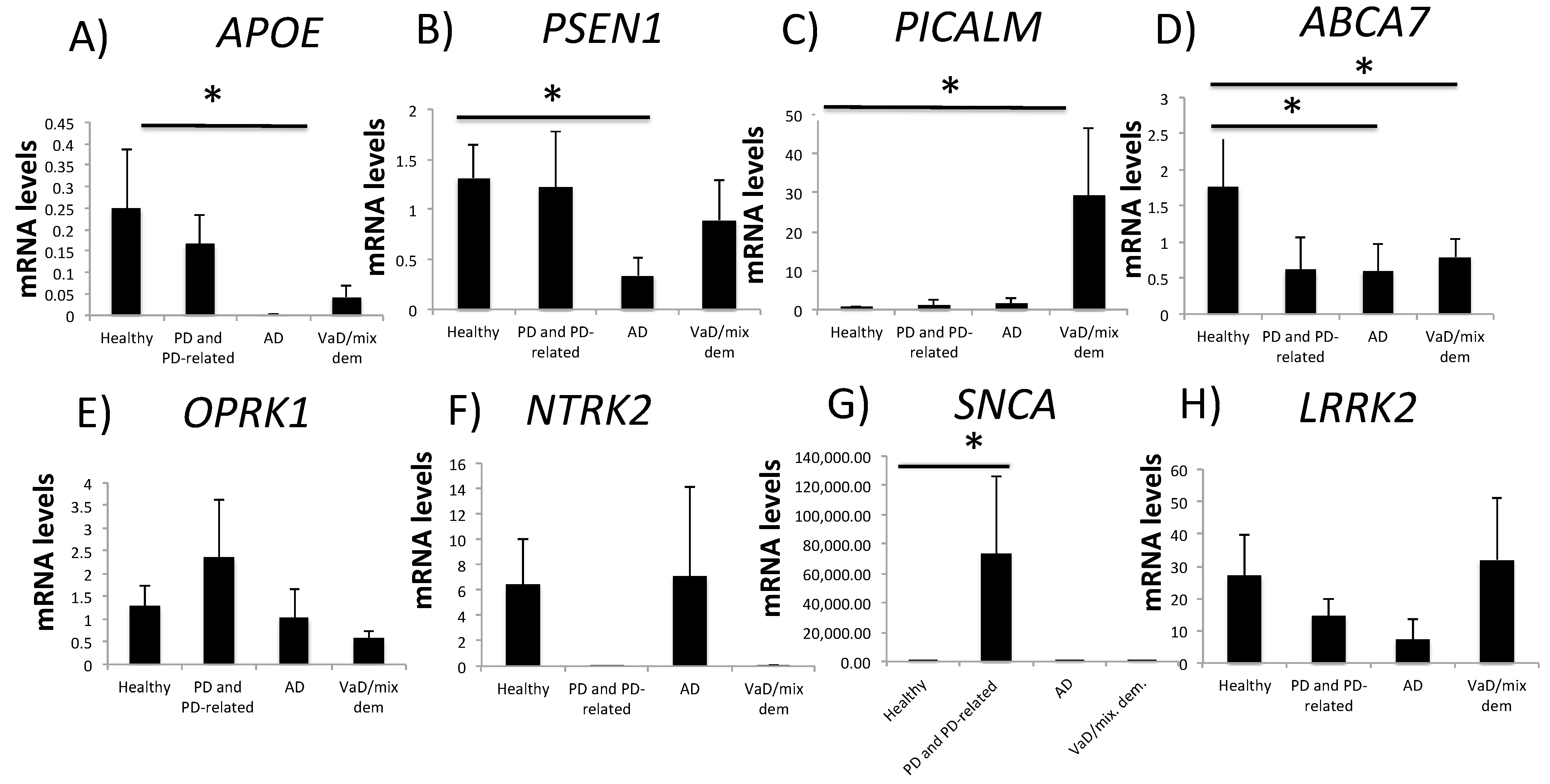
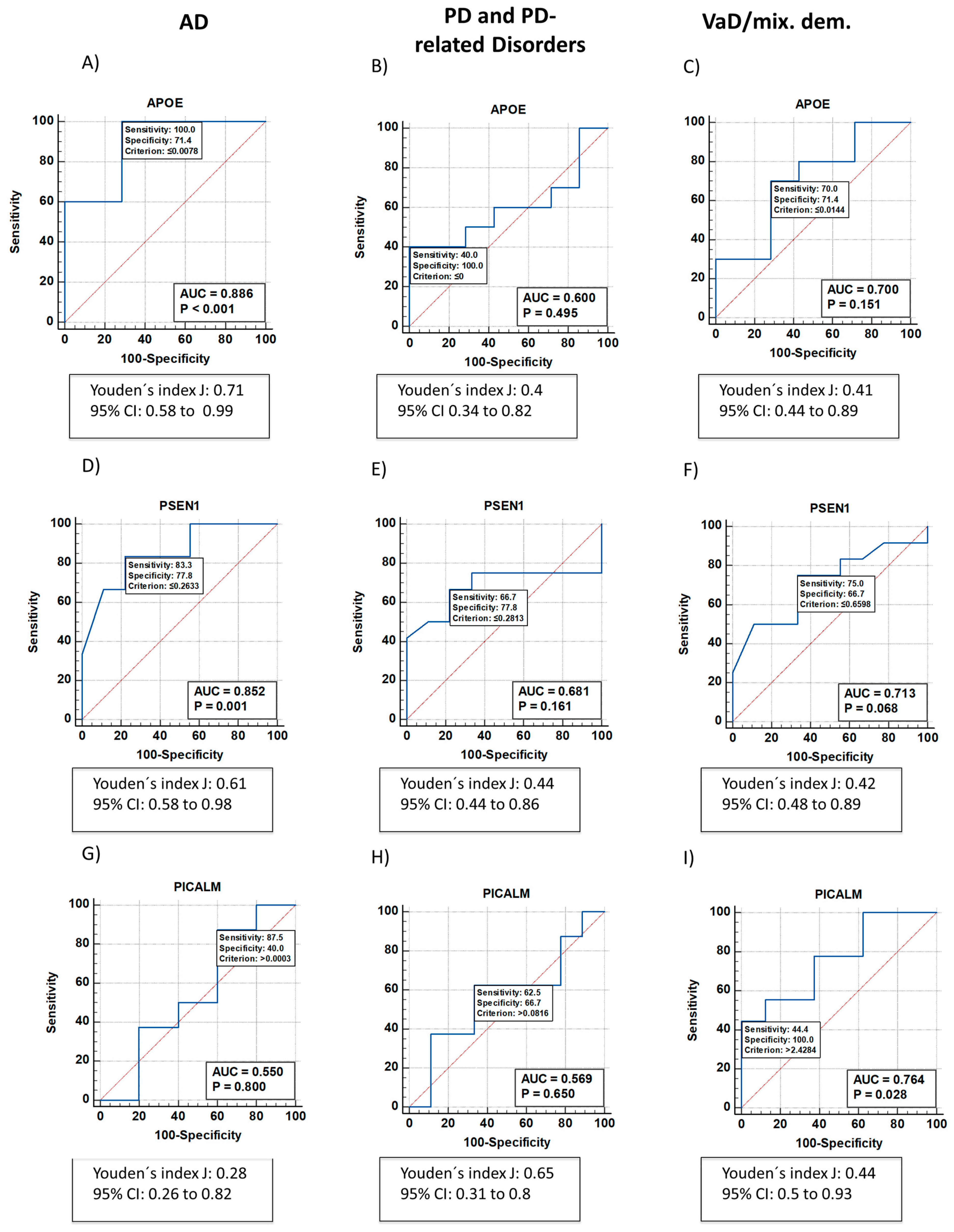
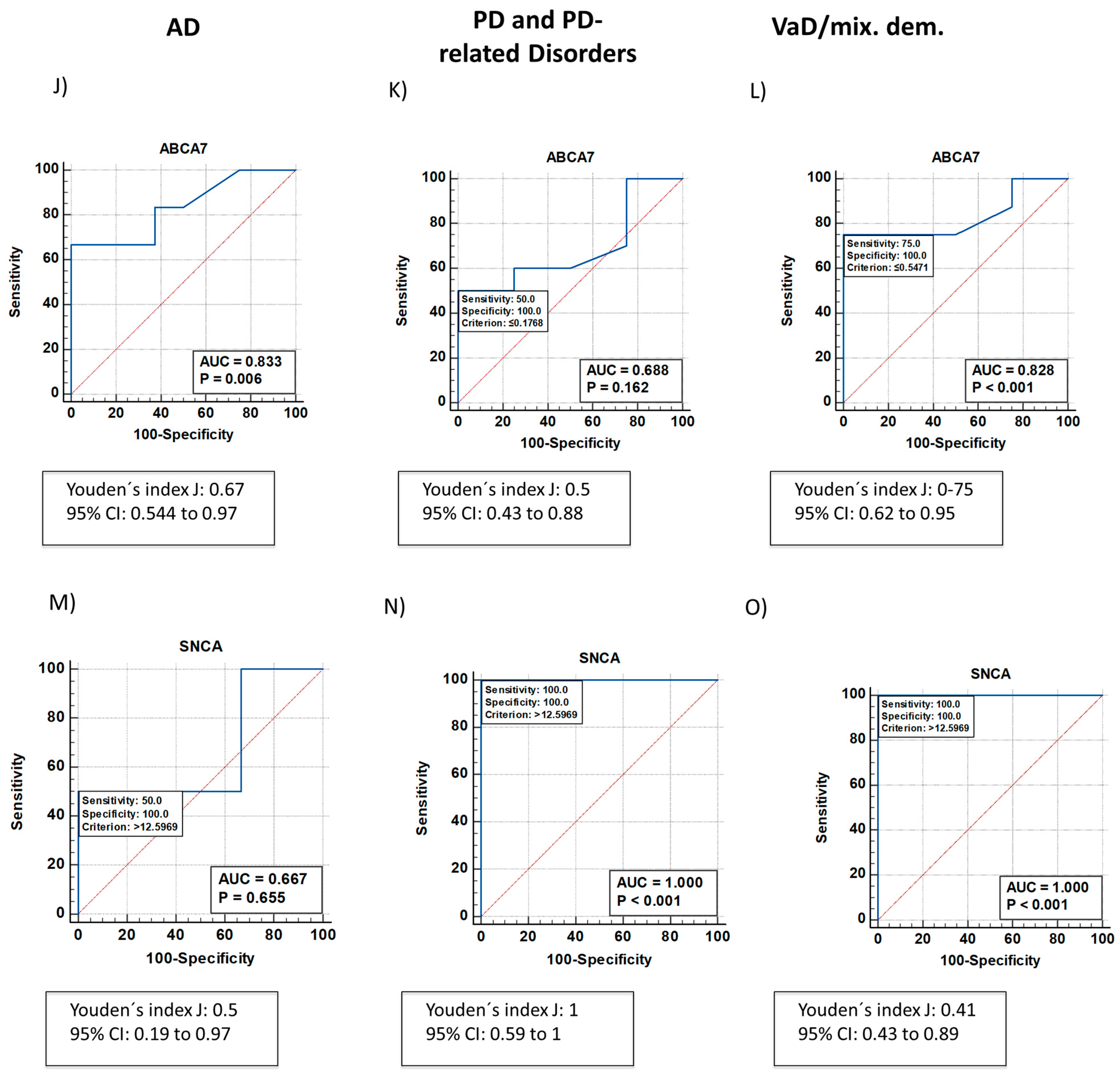
2.1. Neurodegeneration-Related Gene Expression Is Altered in Patients with NDDs
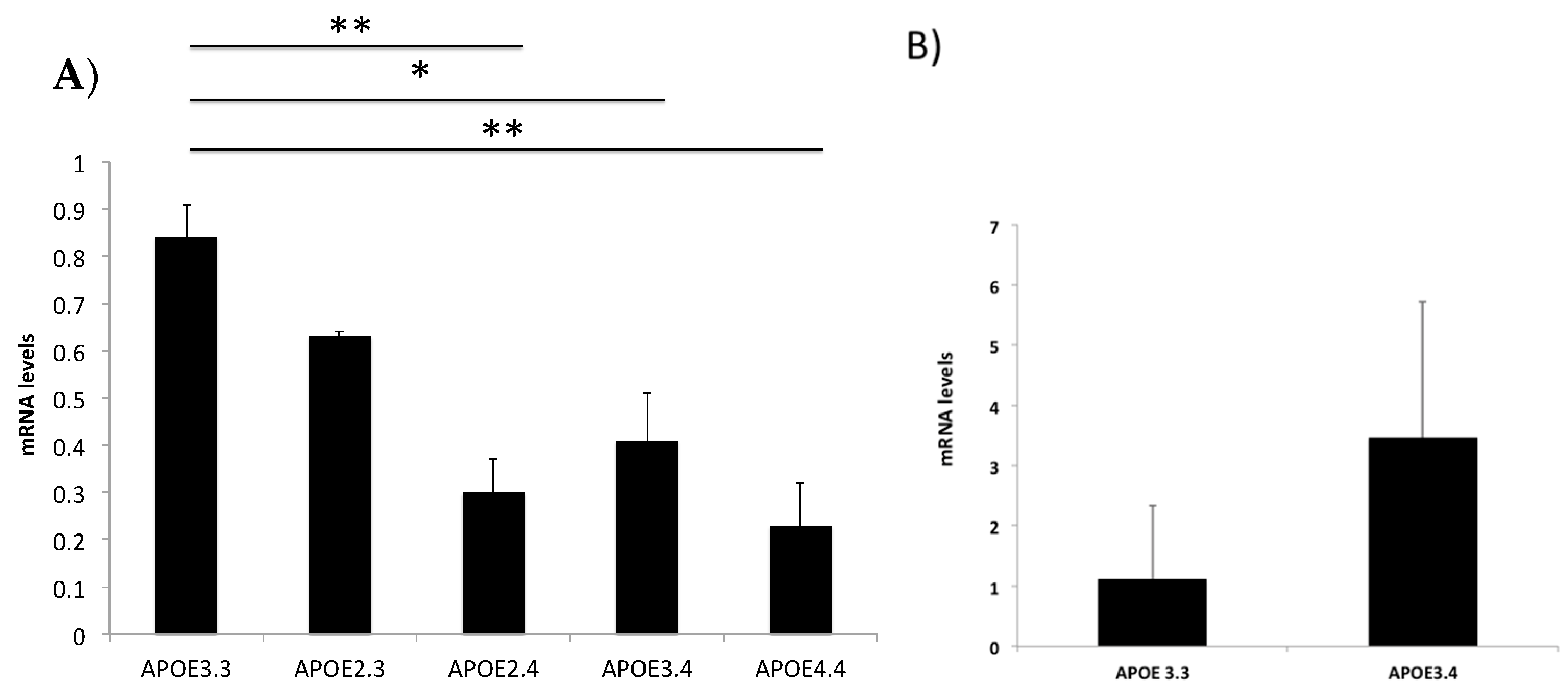
2.2. The ApoE4 Allele Alters APOE Expression Levels
2.3. PSEN1, PICALM, ABCA7 and SNCA Genotypes Do Not Affect Their Gene Expression
3. Discussion
4. Materials and Methods
4.1. Subjects
4.2. Sample Collection and Analysis
4.3. RNA Extraction
4.4. RT-qPCR
4.5. Genotyping
4.6. Statistical Analysis
5. Limitations of the Study
6. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Heemels, M.-T. Neurodegenerative disease. Nature 2016, 539, 179. [Google Scholar] [CrossRef]
- Gitler, A.; Dhillon, P.; Shorter, J. Neurodegenerative diseases:models, mechanisms, and a new hope. Dis. Model Mech. 2017, 10, 499–502. [Google Scholar] [CrossRef]
- Cacabelos, R.; Fernández-Novoa, L.; Lombardi, V.; Kubota, Y.; Takeda, M. Molecular genetics of Alzheiemr’s disease and aging. Methods Find. Exp. Clin. Pharm. 2005, 27, 1–573. [Google Scholar]
- Bloem, B.; Okun, M.; Klein, C. Parkinson’s Disease. Lancet 2021, 397, 2284–2303. [Google Scholar] [CrossRef]
- Spillantini, M.G.; Scmidt, M.L.; Lee, V.; Trojanowski, J.; Jakes, R.; Goedert, M. Alpha-synuclein in Lewy bodies. Nature 1997, 388, 839–840. [Google Scholar] [CrossRef]
- Avila, J. Common mechanisms in neurodegeneration. Nat. Med. 2010, 16, 1372. [Google Scholar] [CrossRef] [PubMed]
- Ciceri, F.; Rotllant, D.; Maes, T. Understanding epigenetic alterations in Alzheimer’s and Parkisnon’s Disease. Curr. Pharm. Des. 2017, 23, 839–857. [Google Scholar] [CrossRef] [PubMed]
- El Kadimiri, N.; Said, N.; Slassi, I.; El Moutawakil, B.; Dadifi, S. Biomarkers of Alzheimer Disease: Classical and novel candidates review. Neurosci 2018, 370, 181–190. [Google Scholar] [CrossRef]
- Janssens, J.; Vermeiren, Y.; Fransen, E.; Aerts, T.; Van Dam, D.; Engelborghs, S.; De Deyn, P. Cerebrospinal fluid and serum MHPG improve Alzheimer’s disease versus dementia with Lewy bodies differential diagnosis. Alzheimer’s Dement. 2018, 10, 172–181. [Google Scholar] [CrossRef] [PubMed]
- Hosoki, S.; Tanaka, T.; Ihara, M. Diagnostic and prognostic blood biomarkers in vascular dementia: From the viewpoint of ischemic stroke. Neurochem. Int. 2021, 146, 105015. [Google Scholar] [CrossRef]
- Li, Y.; Schindler, S.; Bollinger, J.; Ovod, V.; Mawuenyega, K.G.; Winer, M.W.; Shaw, L.M.; Masters, C.L.; Fowler, C.J.; Trojanowski, J.Q.; et al. Validation of plasma amyloid-b 42/40 for detecting Alzheimer Diseae amyloid plaques. Neurology 2022, 98, e688–e699. [Google Scholar] [CrossRef]
- Pascual-Lucas, M.; Allué, J.A.; Sarasa, L.; Fandos, N.; Castillo, S.; Terencio, J.; Sarasa, M.; Tartari, J.P.; Sanabria, Á.; Tárraga, L.; et al. Clinical performance of an antibody-free assay for plasma Aβ42/Aβ40 to detect early alterations of Alzheimer’s disease in individuals with subjective cognitive decline. Alzheimer’s Res. Ther. 2023, 15, 2. [Google Scholar] [CrossRef]
- Karikari, T.K.; Ashton, N.J.; Brinkmalm, G.; Brum, W.S.; Benedet, A.L.; Montoliu-Gaya, L.; Lantero-Rodriguez, J.; Pascoal, T.A.; Suárez-calvet, M.; Rosa-Neto, P.; et al. Blood phospho-tau in Alzheimer disease: Analysis, interpretation, and clinical utility. Nat. Rev. Neurol. 2022, 18, 400–418. [Google Scholar] [CrossRef]
- Palmqvist, S.; Janelidze, S.; Quiroz, Y.T.; Zetterberg, H.; Lopera, F.; Stomrud, E.; Su, Y.; Chen, Y.; Serrano, G.E.; Leuzy, A.; et al. Discriminative accuracy of plasma phospho-tau217 for Alzheimer disease vs other neurodegenerative disorders. JAMA 2020, 324, 772–781. [Google Scholar] [CrossRef] [PubMed]
- Palmqvist, S.; Tideman, P.; Cullen, N.; Zetterberg, H.; Blennow, K.; Alzheimer’s Disease Neuroimaging Initiative; Dage, J.J.; Stomrud, E.; Janelidze, S.; Mattsson-Carlgren, N.; et al. Prediction of future Alzheimer’s disease dementia using plasma phospho-tau combined with other accessible measures. Nat. Med. 2021, 27, 1034–1042. [Google Scholar] [CrossRef] [PubMed]
- Janelidze, S.; Berron, D.; Smith, R.; Strandburg, O.; Proctor, N.K.; Dage, J.L.; Stomrud, E.; Palmqvist, S.; Mattson-Calgren, N.; Hansson, O. Associations of plasma phospho-tau217 levels with tau positron emission tomography in early Alzheimer disease. JAMA Neurol. 2021, 78, 149–156. [Google Scholar] [CrossRef]
- Cullen, N.C.; Leuzy, A.; Janelidze, S.; Palmqvist, S.; Svenningsson, A.; Stomrud, E.; Dage, J.L.; Mattson-Carlgren, N.; Hansson, O. Plasma biomarkers of Alzheimer’s disease improve prediction of cognitive decline in cognitively unimpaired elderly populations. Nat. Commun. 2021, 12, 3555. [Google Scholar] [CrossRef] [PubMed]
- Porcu, E.; Sadler, M.; Lepik, K.; Auwerx, C.; Wood, A.; Weihs, A.; Sleiman, M.S.; Ribeiro, D.; Bandinelli, S.; Tanaka, T.; et al. Differentially expressed genes reflect disease-induced rather than disease-causing changes in the transcriptome. Nat. Commum. 2021, 12, 5647. [Google Scholar] [CrossRef]
- Yu, J.T.; Tan, L.; Hardy, J. Apolipoprotein E in Alzheimer’s Disease: An update. Ann. Rev. Neurosci. 2014, 37, 79–100. [Google Scholar] [CrossRef]
- Kabir, M.T.; Iddin, M.; Setu, J.; Ashraf, G.; Bin-Jumah, M.; Abdel-Daim, M. Exploring the role of PSEN mutations in the pathogenesis of Alzheimer’s Disease. Neurotox. Res. 2020, 38, 833–849. [Google Scholar] [CrossRef]
- Xu, W.; Tan, L.; Ju, J.-T. Rhe role of PICALM in Alzheiemr’s Disease. Mol. Neurobiol. 2015, 52, 399–413. [Google Scholar] [CrossRef] [PubMed]
- De Roeck, A.; Van Broeckhoven, C.; Sleegers, K. The role of ABCA7 in Alzheiemr’s disease: Evidence grom genomics, transcriptomics and methylomics. Acta Neuropathol. 2019, 138, 201–220. [Google Scholar] [CrossRef]
- Siddiqui, I.; Pervaiz, N.; Ali Abbasi, A. The Parkisnon Disease gene SNCA: Evolutionary and structural insights with pathological implication. Sci. Rep. 2016, 6, 24475. [Google Scholar] [CrossRef] [PubMed]
- Ji, H.; Wang, Y.; Liu, G.; Xu, X.; Dai, D.; Chen, Z.; Zhou, D.; Zhou, X.; Han, L.; Li, Y.; et al. OPRK1 promoter hypermethylation increases the risk of Alzheimer’s disease. Neurosci. Lett. 2015, 606, 24–29. [Google Scholar] [CrossRef] [PubMed]
- Sunderland, T.; Berrettini, W.H.; Molchan, S.E.; Lawlor, B.A.; Martinez, R.A.; Vitiello, B.; Tariot, P.N.; Cohen, R.M. Reduced cerebrospinal fluid dynorphin A1-8 in Alzheimer’s disease. Biol. Psychiatry 1991, 30, 81–87. [Google Scholar] [CrossRef]
- Chen, Z.; Simons, M.; Perry, R.; Wiener, H.; Harrell, L.; Go, R. Genetic association of neurotrophic tyrosine kinase receptor type 2 (NTRK2) with Alzheimer’s Disease. Am. J. Med. Genet. 2007, 147B, 363–369. [Google Scholar] [CrossRef]
- Santpere, G.; Ferrer, I. LRRK2 and neurodegeneration. Acta Neuropathol. 2009, 117, 227–246. [Google Scholar] [CrossRef]
- Angelpoulou, E.; Paudel, Y.; Papageorgiou, S.; Piperi, C. APOE genotype and Alzheimer’s disease:the influence of lifestyle and environmental factors. ACS Chem. Neurosci. 2021, 12, 2749–2764. [Google Scholar] [CrossRef]
- Husain, A.; Subramaniyan, K.; Ahmed, S.; Ramakrishnan, V. Association of PSEN1 rs165932 polymorphism with Alzheimer’s disease susceptibility: An extensive meta-analysis. Meta Gene 2019, 19, 123–133. [Google Scholar] [CrossRef]
- Xu, W.; Tan, C.-C.; Cao, X.-P.; Tan, L. Association of Alzheimer’s disease risk variants on the PICALM gene with PICALM expression, core biomarkers, and feature neurodegenration. Aging 2020, 12, 21202–21219. [Google Scholar] [CrossRef]
- Hollingworth, P.; Harld, D.; Sims, R.; Gerrish, A.; Lambert, J.-C.; Carrasquillo, M.; Abraham, R.; Hanshere, M.L.; Pahwa, J.S.; Moskvina, V.; et al. Common variants at ABCA7, MS4A6A/MS4A4E, EPHA1, CD33 and CD2AP are associated with Alzheiemer’s Disease. Nat. Genet. 2011, 43, 429–435. [Google Scholar] [CrossRef]
- Nordestgaard, L.; Christoffersen, M.; Frikke-Schmidt, R. Shared risk factors between dementia and atherosclerotic cardiovascular disease. Int. J. Mol. Sci. 2022, 23, 9777. [Google Scholar] [CrossRef] [PubMed]
- Hernandez, D.G.; Reed, X.; Singleton, A. Genetics in parkinson disease: Mendelian versus non-mendelian inheritance. J. Neurochem. 2016, 139, 59–74. [Google Scholar] [CrossRef] [PubMed]
- de la Torre, J.C. Vascular basis of Alzheimer’s pathogenesis. Ann. N. Y. Acad. Sci. 2002, 977, 196–215. [Google Scholar] [CrossRef]
- Fernandez-Calle, R.; Konings, S.; Frontiñan-Rubio, J.; García-Revilla, J.; Camprubí-Ferrer, L.; Svensson, M.; Martisnon, I.; Boza-Serrano, A.; Venero, J.L.; Nielsen, H.M.; et al. APOE in the bullseye of neurodegenerative diseases: Impact of the APOE genotype in Alzheimer’s disease pathology and brain diseases. Mol. Neurodeg. 2022, 17, 62. [Google Scholar] [CrossRef]
- Rasmussen, K. Plasma levels of apolipoprotein E, APOE genotype and risk of dementia and ischemic heart disease: A review. Atherosclerosis 2016, 255, 145–155. [Google Scholar] [CrossRef] [PubMed]
- Soreide, K. Receiver-operating characteristic curve analysis in diagnostic, prognostic and predictive biomarker research. J. Clin. Pathol. 2009, 62, 1–5. [Google Scholar] [CrossRef]
- Hsu, M.-J.; Chang, Y.-C.; Hsueh, H.-M. Biomarker selection for medical diagnosis using the partial area under the ROC curve. BMC Res. Notes 2014, 7, 25. [Google Scholar] [CrossRef]
- Rasmussen, K.; Tybjaerg-Hansen, A.; Nordestgaard, B.; Frikke-Scmidt, R. Plasma levels of apolipoprotein E and risk of dementia in the general population. Ann. Neurol. 2014, 77, 301–311. [Google Scholar] [CrossRef]
- Landén, M.; Fredman, C.; Regland, B.; Wallin, A.; Blennow, K. Apolipoprotein E in cerebrospinal fluid from patients with Alzheimer’s disease and other forms of dementia is reduced but without any correlation to the apoE4 isoform. Dementia 1996, 7, 5. [Google Scholar] [CrossRef]
- Villa, C.; Stoccoro, A. Epigenetic peropheral biomarkers for early diagnosis of Alzheimer’s Disease. Genes 2022, 13, 1308. [Google Scholar] [CrossRef] [PubMed]
- Jagtap, A.; Gawande, S.; Sharma, S. Biomarkers in vascular dementia: A recent update. Biomark. Genom. Med. 2015, 7, 43–56. [Google Scholar] [CrossRef]
- Ridell, D.; Zhou, H.; Atchison, K.; Warwick, H.; Atkinsons, P.; Jefferson, J.; Xu, L.; Aschmies, S.; Kirksey, Y.; Hu, Y.; et al. Impact of Apolipoprotein E (ApoE) polymorphism on brain ApoE levels. J. Neurosci. 2008, 28, 11445–11453. [Google Scholar] [CrossRef]
- Makin, S. The amyloid hypothesis on trial. Nature 2018, 559, S4–S7. [Google Scholar] [CrossRef]
- Solis, E.; Hascup, K.; Hascup, E. Alzheimer’s Disease: The Link Between Amyloid-β and Neurovascular Dysfunction. J. Alzheimers Dis. 2020, 76, 1179–1198. [Google Scholar] [CrossRef] [PubMed]
- Xiao, X.; Liu, H.; Liu, X.; Zhang, W.; Zhang, S.; Jiao, B. APP, PSEN1, and PSEN2 Variants in Alzheimer’s Disease: Systematic Re-evaluation According to ACMG Guidelines. Front. Aging Neurosci. 2021, 13, 695808. [Google Scholar] [CrossRef] [PubMed]
- Cacquevel, M.; Aeschbach, L.; Houacine, J.; Fraering, P. Alzheimer’s disease-linked mutations in presenilin-1 result in a drastic loss of activity in purified c-secretase complexes. PLoS ONE 2012, 7, e35133. [Google Scholar] [CrossRef]
- Davidsson, P.; Bogdanovic, N.; Lannfelt, L.; Blennow, K. Reduced expressio of amyloid precursor protein, Presenilin-1 and rab3a in cortical brain regions in Alzheimer’s disease. Dem. Geriatr. Cogn. Disord. 2001, 12, 243–250. [Google Scholar] [CrossRef]
- Carboni, L.; Lattanzio, F.; Candeletti, S.; Porcellini, E.; Raschi, E.; Licastro, F.; Romualdi, P. Peripheral leukocyte expression of the potential biomatker proteins Bdnf, Sirt1, and Psen1 is not regulated by promother methylation in Alzheiemr’s disease patients. Neurosci. Lett. 2015, 605, 44–48. [Google Scholar] [CrossRef]
- de Franca, J.; Talib, L.; Joaquim, H.; Sarno, T.; Gattaz, W.; Forlenza, O. Protein levels of ADAm10, BACE1, and PSEN1 in platelets and leukocytes of Alzheimer’s disease patients. Eur. Arch. Psych. Clin. Neurosci. 2018, 269, 963–972. [Google Scholar]
- Pluta, R. Alzheimer’s Disease conncected genes in the post-ischemic hippocampus and temporal cortex. Genes 2022, 13, 1059. [Google Scholar] [CrossRef] [PubMed]
- Jansen, I.E.; Savage, J.E.; Watanabe, K.; Bryois, J.; Williams, D.M.; Steinberg, S.; Sealock, J.; Karlsson, I.K.; Hägg, S.; Athanasiu, L.; et al. Genome-Wide Meta-Analysis Identifies New Loci and Functional Pathways Influencing Alzheimer’s Disease Risk. Nat. Genet. 2019, 51, 404–413. [Google Scholar] [CrossRef] [PubMed]
- Bellenguez, C.; Kucukali, F.; Jansen, I.E.; Kleineidam, L.; Moreno-Grau, S.; Amin, N.; Naj, A.C.; Campos-Martin, R.; Grenier-Boley, B.; Andrade, V.; et al. New Insights into the Genetic Etiology of Alzheimer’s Disease and Related Dementias. Nat. Genet. 2022, 54, 412–436. [Google Scholar] [CrossRef] [PubMed]
- Ando, K.; Nagaraj, S.; Kucukali, F.; de Fisenne, M.-A.; Kosa, A.-C.; Doeraene, E.; lópez Gutierrez, L.; Brion, J.-P.; Leroy, K. PICALM and Alzheiemr’s Disease: An update and perspectives. Cells 2022, 11, 3994. [Google Scholar] [CrossRef]
- Kumon, H.; Yoshino, Y.; Funahashi, Y.; Mori, H.; Ueno, M.; Ozaki, Y.; Yamazaki, K.; Ochi, S.; Mori, T.; Iga, J.-I.; et al. PICALM mRNA expression in the blood of patients with neurodegenerative disease and geriatric depression. J. Alzheimers Dis. 2021, 79, 1055–1062. [Google Scholar] [CrossRef] [PubMed]
- Tasaki, S.; Xu, J.; Avey, D.; Johnson, L.; Petjuk, V.; Dawe, R.; Bennett, D.; Wang, Y.; Gaiteri, C. Inferring protein expression changes from mRNA in Alzheimer’s dementia using deep natural networks. Nat. Commun. 2022, 13, 655. [Google Scholar] [CrossRef]
- Rasmussen, I.; Tybjaerg-Hansen, A.; Rasmussen, K.; Nordestgaard, B.; Frikke-Scmidt, R. Blood.brain barrier transcytosis genes, risk of dementia and stroke: A prospective cohort study of 74,754 individuals. Eur. J. Epidemiol. 2019, 34, 579–590. [Google Scholar] [CrossRef]
- Scotland, P.; Heath, J.; Conwy, A.; Porter, N.; Armstrong, M.; Walker, J.; Klebig, M.; Lavau, C.; Wechsler, D. The PICALM protein plays a key role in iron homeostasi and cell proliferation. PLoS ONE 2012, 7, e44252. [Google Scholar] [CrossRef]
- Mercer, J.L.; Argus, J.; Crabtree, D.; Keenan, M.; Wilks, M.; Ashley Chi, J.-T.; Bensinger, S.; Lavau, C.; Wechsle, D. Modulation of PICALM levels perturbs cellular cholesterol homeostais. PLoS ONE 2015, 10, e0129776. [Google Scholar] [CrossRef]
- Bjorkhem, I.; Meaney, S. Brain cholesterol: Long secret life behind a barrier. Arterioscler. Thromb. Vasc. Biol. 2004, 24, 805–815. [Google Scholar] [CrossRef]
- Duong, M.; Nasrallah, I.; Wolk, D.; Chang, C.; Chang, T.-Y. Cholesterol, atherosclerosis, and APOE in vascular contributions to cognitive impairment and Dementia (VCID): Potential mechanisms and therapy. Front. Aging Neurosci. 2021, 13, 2021. [Google Scholar] [CrossRef]
- Cacabelos, R.; López-Múñoz, F. The ABCB1 transporter in Alzheimer’s disease. Clin. Exp. Pharmacol. 2014, 4, e128. [Google Scholar] [CrossRef]
- Yamazaki, K.; Yoshino, Y.; Mori, T.; Yoshida, T.; Ozaki, Y.; Sao, T.; Mori, Y.; Ochi, S.; Iga, J.-I.; Ueno, S.-I. Gene expression and methylation analysis of ABCA7 in patients with Alzheiemr’s Disease. J. Alzheimer’s Dis. 2017, 57, 171–181. [Google Scholar] [CrossRef]
- Vasquez, J.; Fardo, D.; Estus, S. ABCA7 expression is associated with alzheimer’s disease polymorphism and disease status. Neurosci. Lett. 2013, 556, 58–62. [Google Scholar] [CrossRef] [PubMed]
- Chiba-Falek, O.; Lopez, G.; Nussbaum, R. Levels of alpha-synuclein mRNA in sporadic Parkisnon disease patients. Mov. Disord. 2006, 21, 1703–1708. [Google Scholar] [CrossRef] [PubMed]
- Takahashi, M.; Suzuki, M.; Fukuoka, M.; Fujikake, N.; Watanabe, S.; Murata, M.; Wada, K.; Nagai, Y.; Hohjoh, H. Normalization of overexpresses a-synuclein causing Parkinson’s Disease by a moderate gene silencing with rna interference. Mol. Therapy 2015, 4, e241. [Google Scholar]
- Marsal-Garcia, L.; Urbizu, A.; Arnaldo, L.; Campdekacreu, J.; Vilas, D.; Ispierto, L.; Gascón-Bayarri, J.; Reñé, R.; Álvarez, R.; Beyer, K. Expression levels of an alpha-synuclein transcript in blood may distingish between early dementia with Lewy bodies and Parkinson’s Disease. Int. J. Mol. Sci. 2021, 22, 775. [Google Scholar] [CrossRef]
- Philstrom, L.; Blauwendraat, C.; Cappelleti, C.; Berge-Seidl, V.; Langmyhr, M.; Hernriksen, S.P.; van de Berg, W.; Gibbs, R.; Cookson, M.; the Int Parkinson Disease Genomics Consortium; et al. A comprehensive analysis of SNCA-related genetic risk in sporadic parkinson disease. Ann. Neurol. 2018, 84, 117–129. [Google Scholar] [CrossRef]
- Cheng, F.; Zheng, W.; Liu, C.; Barbuty, P.; Yu-Taeger, L.; Casadei, N.; Huebener-Schmid, J.; Admard, J.; Boldt, K.; Junger, K.; et al. Intronic enhancers of the human SNCA gene predominantly regulate its expression in brain in vivo. Sci. Adv. 2022, 8, 47. [Google Scholar] [CrossRef] [PubMed]
- Gong, X.; Zhang, H.; Liu, X.; Liu, Y.; Liu, J.; Fapohunda, F.; Lü, P.; Wang, K.; Tang, M. Is liquid biopsy mature enough for the diagnosis of Alzheimer’s Disease? Front. Aging Neurosci. 2022, 14, 2022. [Google Scholar] [CrossRef] [PubMed]
- Manzine, P.R.; Vatanabe, I.; Grigoli, M.; Pedroso, R.; Monteiro, M.P.; Oliveira, D.; Nascimento, C.; Peron, R.; Orlandi, F.; Cominetti, M. Potential protein in blood-based biomarkers indifferent types of Dementia: A therapeutic overview. Curr. Pharm. Des. 2022, 28, 1170–1186. [Google Scholar] [CrossRef] [PubMed]
- Thijssen, E.H.; Verberk, I.; Kindermans, J.; Abramian, A.; Vanbrabant, J.; Ball, A.; Pijjenburg, Y.; Lemstra, A.; van der Flier, W.; Stoops, E.; et al. Differential diagnostic performance of a panel of plasma biomarkers for different types of Dementia. Alzheimers 2022, 14, e12285. [Google Scholar] [CrossRef] [PubMed]
- Mayeux, R.; Honig, L.S.; Tang, M.-X.; Manly, J.; Stern, Y.; Schupf, N.; Mehta, P.D. Plasma Ab40 and Ab42 and Alzheiemr’s Disease: Relation to age, mortality and risk. Neurology 2003, 62, 1219–1222. [Google Scholar]
- Moscoso, A.; Grothe, M.; Ashton, N.; Karikari, T.; Lantero Rodriguez, J.; Snellman, A.; Súarez-Calvet, M.; Zetterberg, H.; Blennow, K.; Schöll, M.; et al. Time course of phosphorilated-tau181 in blood across the Alzheimer’s disease spectrum. Brain 2021, 144, 325–339. [Google Scholar] [CrossRef] [PubMed]
- Pereira, J.B.; Janelidze, S.; Stomrud, E.; Palmqvist, S.; van Westen, D.; Dage, J.; Mattson-Carlgren, N.; Hansson, O. Plasma markers predict changes in amyloid, tau, atrophy and cognition in nin-dementia subjects. Brain 2021, 144, 2826–2836. [Google Scholar] [CrossRef]
- Martínez-Iglesias, O.; Carrera, I.; Carril, J.C.; Fernandez-Novoa, L.; Cacabelos, N.; Cacabelos, R. DNA methylation in Neurodegenerative and cerebrovascular Disorders. Int. J. Mol. Sci. 2020, 21, 2220. [Google Scholar] [CrossRef] [PubMed]
- Martínez-Iglesias, O.; Naidoo, V.; Cacbelos, N.; Cacabelos, R. Epigenetic biomarkers as diagnostic tools for Neurodegenrative Disorders. Int. J. Mol. Sci. 2022, 23, 13. [Google Scholar] [CrossRef]
- Mandrekar, J.N. Receiver operating characteristic curve in diagnostic test assessment. Journal of thoracic oncology: Official publication of the International Association for the Study of Lung Cancer. Int. Assoc. Study Lung Cancer 2010, 5, 1315–1316. [Google Scholar]

| Group | Clinical Diagnosis | MMSE | Age (Years) |
|---|---|---|---|
| Healthy | Healthy (n = 9) | 29.18 ± 1.18 | 61.36 ± 8.58 |
| AD | AD (n = 8) | 14.92 ± 4.94 | 69.76 ± 7.9 |
| VaD/mixed dementia | Vascular encephalopathy multi-infarction Binswanger-like (n = 4) | 18.2 ± 4.8 | 77.6 ± 5.84 |
| Stroke (n = 2) | |||
| Mixed Dementia (Vascular-hypovitaminosis) | |||
| Vascular encephalopathy (n = 2) | |||
| Ischemic vascular encephalopathy (n = 4) | |||
| PD and PD-related disorders | Parkinson’s (n = 7) | 26.4 ± 2.26 | 67.8 ± 8.15 |
| Left vascular hemiparkinsonism | |||
| Incipient Parkinsonism (n = 2) | |||
| Parkinson’s with VaD (n = 2) | |||
| Familial Parkinson’s |
| GENOTYPE | DIAGNOSIS | AGE (Years) | SEX |
|---|---|---|---|
| APOE 3.3 | NCD mixed dementia | 80 | F |
| PD | 65 | F | |
| NCD mixed dementia | 80 | M | |
| NCD mixed dementia | 71 | F | |
| Healthy (n = 4) | 68 ± 8.12 | 2M, 2F | |
| APOE 2.3 | PD | 69 | F |
| NCD | 65 | M | |
| BIPOLAR DISORDER | 64 | M | |
| NCD | 61 | M | |
| APOE 2.4 | NCD mixed dementia | 78 | M |
| PD | 75 | F | |
| Depression | 74 | F | |
| Maturational delay | 22 | M | |
| NCD | 75 | M | |
| APOE 3.4 | NCD | 76 | M |
| NCD | 71 | F | |
| NCD mixed dementia | 65 | M | |
| Depression | 40 | F | |
| Stroke | 60 | M | |
| Healthy (n = 4) | 66.25 ± 12.5 | 2M, 2F | |
| APOE 4.4 | Encephalopathy | 63 | M |
| NCD | 69 | F | |
| NCD | 68 | M | |
| NCD-AD | 70 | F | |
| NCD | 70 | M |
| APOE | PICALM | OPRK1 | LRRK2 | ABCA7 | PSEN1 | NTRK2 | SNCA | |
|---|---|---|---|---|---|---|---|---|
| AD | low | - | - | - | low | low | - | - |
| VaD/mixed dementia | - | high | - | - | low | - | - | - |
| PD and PD-related disorders | - | - | - | - | - | - | high |
| GENE | ID |
|---|---|
| APOE | Hs00171160_m1 |
| PSEN1 | Hs00240518_m1 |
| PICALM | Hs00300318_m1 |
| ABCA7 | Hs1105094_m1 |
| OPRK1 | Hs00175127_m1 |
| NTRK2 | Hs00178811_m1 |
| SNCA | Hs00240906_m1 |
| LRRK2 | Hs00968192_m1 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Martínez-Iglesias, O.; Naidoo, V.; Carril, J.C.; Seoane, S.; Cacabelos, N.; Cacabelos, R. Gene Expression Profiling as a Novel Diagnostic Tool for Neurodegenerative Disorders. Int. J. Mol. Sci. 2023, 24, 5746. https://doi.org/10.3390/ijms24065746
Martínez-Iglesias O, Naidoo V, Carril JC, Seoane S, Cacabelos N, Cacabelos R. Gene Expression Profiling as a Novel Diagnostic Tool for Neurodegenerative Disorders. International Journal of Molecular Sciences. 2023; 24(6):5746. https://doi.org/10.3390/ijms24065746
Chicago/Turabian StyleMartínez-Iglesias, Olaia, Vinogran Naidoo, Juan Carlos Carril, Silvia Seoane, Natalia Cacabelos, and Ramón Cacabelos. 2023. "Gene Expression Profiling as a Novel Diagnostic Tool for Neurodegenerative Disorders" International Journal of Molecular Sciences 24, no. 6: 5746. https://doi.org/10.3390/ijms24065746
APA StyleMartínez-Iglesias, O., Naidoo, V., Carril, J. C., Seoane, S., Cacabelos, N., & Cacabelos, R. (2023). Gene Expression Profiling as a Novel Diagnostic Tool for Neurodegenerative Disorders. International Journal of Molecular Sciences, 24(6), 5746. https://doi.org/10.3390/ijms24065746








