Germline Variants in MLH1 and ATM Genes in a Young Patient with MSI-H in a Precancerous Colonic Lesion
Abstract
1. Introduction
2. Patient Report
3. Results
3.1. Analysis of MSI and Mutation Detection in MMR Genes
3.2. Characterization of the c.589-9_589-6 GTTT Variant in the MLH1 Gene
3.3. Next-Generation Sequencing Analysis
4. Discussion
5. Materials and Methods
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Conflicts of Interest
References
- Nolano, A.; Medugno, A.; Trombetti, S.; Liccardo, R.; De Rosa, M.; Izzo, P.; Duraturo, F. Hereditary Colorectal Cancer: State of the Art in Lynch Syndrome. Cancers 2022, 15, 75. [Google Scholar] [CrossRef] [PubMed]
- Duraturo, F.; Liccardo, R.; De Rosa, M.; Izzo, P. Genetics, diagnosis and treatment of Lynch syndrome: Old lessons and current challenges. Oncol. Lett. 2019, 17, 3048–3054. [Google Scholar] [CrossRef] [PubMed]
- Liccardo, R.; Della Ragione, C.; Mitilini, N.; De Rosa, M.; Izzo, P.; Duraturo, F. Novel variants of unknown significance in the PMS2 gene identified in patients with hereditary colon cancer. Cancer Manag. Res. 2019, 11, 6719–6725. [Google Scholar] [CrossRef] [PubMed]
- Liccardo, R.; De Rosa, M.; Rossi, G.B.; Carlomagno, N.; Izzo, P.; Duraturo, F. Incomplete Segregation of MSH6 Frameshift Variants with Phenotype of Lynch Syndrome. Int. J. Mol. Sci. 2017, 18, 999. [Google Scholar] [CrossRef]
- Duraturo, F.; Liccardo, R.; Izzo, P. Coexistence of MLH3 germline variants in colon cancer patients belonging to families with Lynch syndrome-associated brain tumors. J. Neuro-Oncol. 2016, 129, 577–578. [Google Scholar] [CrossRef]
- Duraturo, F.; Liccardo, R.; Cavallo, A.; De Rosa, M.; Grosso, M.; Izzo, P. Association of low-risk MSH3 and MSH2 variant alleles with Lynch syndrome: Probability of synergistic effects. Int. J. Cancer 2011, 129, 1643–1650. [Google Scholar] [CrossRef]
- Vasen, H.F.; Watson, P.; Mecklin, J.P.; Lynch, H.T. New clinical criteria for hereditary nonpolyposis colorectal cancer (HNPCC, Lynch syndrome) proposed by the International Collaborative group on HNPCC. Gastroenterology 1999, 116, 1453–1456. [Google Scholar] [CrossRef]
- Umar, A.; Boland, C.R.; Terdiman, J.P.; Syngal, S.; de la Chapelle, A.; Rüschoff, J.; Fishel, R.; Lindor, N.M.; Burgart, L.J.; Hamelin, R.; et al. Revised Bethesda guidelines for hereditary nonpolyposis colorectal cancer (Lynch syndrome) and microsatellite instability. J. Natl. Cancer Inst. 2004, 96, 261–268. [Google Scholar] [CrossRef]
- Shia, J. Immunohistochemistry versus microsatellite instability testing for screening colorectal cancer patients at risk for hereditary nonpolyposis colorectal cancer syndrome. Part. I. The utility of immunohistochemistry. J. Mol. Diagn. 2008, 10, 293–300. [Google Scholar] [CrossRef]
- Martin-Morales, L.; Rofes, P.; Diaz-Rubio, E.; Llovet, P.; Lorca, V.; Bando, I.; Perez-Segura, P.; de la Hoya, M.; Garre, P.; Garcia-Barberan, V.; et al. Novel genetic mutations detected by multigene panel are associated with hereditary colorectal cancer predisposition. PLoS ONE 2018, 13, e0203885. [Google Scholar] [CrossRef]
- Dodaro, C.; Grifasi, C.; Florio, J.; Santangelo, M.L.; Duraturo, F.; De Rosa, M.; Izzo, P.; Renda, A. The role of mutation analysis of the APC gene in the management of FAP patients. A controversial issue. Ann. Ital. Chir. 2016, 87, 321–325. [Google Scholar]
- Rijcken, F.E.M.; Hollema, H.; Kleibeuker, J.H. Proximal Adenomas in Hereditary Non-Polyposis Colorectal Cancer Are Prone to Rapid Malignant Transformation. Gut 2002, 50, 382–386. [Google Scholar] [CrossRef]
- Ahadova, A.; Gallon, R.; Gebert, J.; Ballhausen, A.; Endris, V.; Kirchner, M.; Stenzinger, A.; Burn, J.; von Knebel Doeberitz, M.; Bläker, H.; et al. Three Molecular Pathways Model Colorectal Carcinogenesis in Lynch Syndrome. Int. J. Cancer 2018, 143, 139–150. [Google Scholar] [CrossRef] [PubMed]
- Sekine, S.; Mori, T.; Ogawa, R.; Tanaka, M.; Yoshida, H.; Taniguchi, H.; Nakajima, T.; Sugano, K.; Yoshida, T.; Kato, M.; et al. Mismatch Repair Deficiency Commonly Precedes Adenoma Formation in Lynch Syndrome-Associated Colorectal Tumorigenesis. Mod. Pathol. 2017, 30, 1144–1151. [Google Scholar] [CrossRef]
- Møller, P. The Prospective Lynch Syndrome Database Reports Enable Evidence-Based Personal Precision Health Care. Hered. Cancer Clin. Pract. 2020, 18, 6. [Google Scholar] [CrossRef] [PubMed]
- Toyota, M.; Ahuja, N.; Ohe-Toyota, M.; Herman, J.G.; Baylin, S.B.; Issa, J.P. CpG Island Methylator Phenotype in Colorectal Cancer. Proc. Natl. Acad. Sci. USA 1999, 96, 8681–8686. [Google Scholar] [CrossRef] [PubMed]
- Valo, S.; Kaur, S.; Ristimäki, A.; Renkonen-Sinisalo, L.; Järvinen, H.; Mecklin, J.-P.; Nyström, M.; Peltomäki, P. DNA Hypermethylation Appears Early and Shows Increased Frequency with Dysplasia in Lynch Syndrome-Associated Colorectal Adenomas and Carcinomas. Clin. Epigenet. 2015, 7, 71. [Google Scholar] [CrossRef] [PubMed]
- De Jong, A.E.; Morreau, H.; Van Puijenbroek, M.; Eilers, P.H.; Wijnen, J.; Nagengast, F.M.; Griffioen, G.; Cats, A.; Menko, F.H.; Kleibeuker, J.H.; et al. The Role of Mismatch Repair Gene Defects in the Development of Adenomas in Patients with HNPCC. Gastroenterology 2004, 126, 42–48. [Google Scholar] [CrossRef] [PubMed]
- Rondagh, E.J.A.; Gulikers, S.; Gómez-García, E.B.; Vanlingen, Y.; Detisch, Y.; Winkens, B.; Vasen, H.F.A.; Masclee, A.A.M.; Sanduleanu, S. Nonpolypoid Colorectal Neoplasms: A Challenge in Endoscopic Surveillance of Patients with Lynch Syndrome. Endoscopy 2013, 45, 257–264. [Google Scholar] [CrossRef]
- Remo, A.; Fassan, M.; Lanza, G. Immunohistochemical Evaluation of Mismatch Repair Proteins in Colorectal Carcinoma: The AIFEG/GIPAD Proposal. Pathologica 2016, 108, 104–109. [Google Scholar]
- Lanspa, S.J.; Jenkins, J.X.; Cavalieri, R.J.; Smyrk, T.C.; Watson, P.; Lynch, J.; Lynch, H.T. Surveillance in Lynch Syndrome: How Aggressive? Am. J. Gastroenterol. 1994, 89, 1978–1980. [Google Scholar] [PubMed]
- Engel, C.; Ahadova, A.; Seppälä, T.T.; Aretz, S.; Bigirwamungu-Bargeman, M.; Bläker, H.; Bucksch, K.; Büttner, R.; de Vos Tot Nederveen Cappel, W.T.; Endris, V.; et al. Associations of Pathogenic Variants in MLH1, MSH2, and MSH6 with Risk of Colorectal Adenomas and Tumors and with Somatic Mutations in Patients with Lynch Syndrome. Gastroenterology 2020, 158, 1326–1333. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Jiang, K.; Hu, Y.; Zhao, X.; Yin, L.; Diao, X.; Ma, X.; Xu, Y.; Bai, Y.; Zhang, Y.; et al. An exploration of gastric cancer with heterogeneous mismatch repair status. Virchows Arch. 2023. [Google Scholar] [CrossRef] [PubMed]
- Aaltonen, L.A.; Salovaara, R.; Kristo, P.; Canzian, F.; Hemminki, A.; Peltomäki, P.; Chadwick, R.B.; Kääriäinen, H.; Eskelinen, M.; Järvinen, H.; et al. Incidence of Hereditary Nonpolyposis Colorectal Cancer and the Feasibility of Molecular Screening for the Disease. N. Engl. J. Med. 1998, 338, 1481–1487. [Google Scholar] [CrossRef] [PubMed]
- Takayama, T.; Miyanishi, K.; Hayashi, T.; Sato, Y.; Niitsu, Y. Colorectal Cancer: Genetics of Development and Metastasis. J. Gastroenterol. 2006, 41, 185–192. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Li, R.; He, Y.; Yi, Y.; Wu, H.; Liang, Z. Next-Generation Sequencing Reveals Heterogeneous Genetic Alterations in Key Signaling Pathways of Mismatch Repair Deficient Colorectal Carcinomas. Mod. Pathol. 2020, 33, 2591–2601. [Google Scholar] [CrossRef] [PubMed]
- Ahadova, A.; von Knebel Doeberitz, M.; Bläker, H.; Kloor, M. CTNNB1-Mutant Colorectal Carcinomas with Immediate Invasive Growth: A Model of Interval Cancers in Lynch Syndrome. Fam. Cancer 2016, 15, 579–586. [Google Scholar] [CrossRef] [PubMed]
- Liccardo, R.; Lambiase, M.; Nolano, A.; De Rosa, M.; Izzo, P.; Duraturo, F. Significance of rare variants in genes involved in the pathogenesis of Lynch syndrome. Int. J. Mol. Med. 2022, 49, 81. [Google Scholar] [CrossRef]
- Richards, S.; Aziz, N.; Bale, S.; Bick, D.; Das, S.; Gastier-Foster, J.; Grody, W.W.; Hegde, M.; Lyon, E.; Spector, E.; et al. ACMG Laboratory Quality Assurance Committee. Standards and guidelines for the interpretation of sequence variants: A joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet. Med. 2015, 17, 405–424. [Google Scholar] [CrossRef]
- Lin, E.I.; Tseng, L.H.; Gocke, C.D.; Reil, S.; Le, D.T.; Azad, N.S.; Eshleman, J.R. Mutational profiling of colorectal cancers with microsatellite instability. Oncotarget 2015, 6, 42334–42344. [Google Scholar] [CrossRef]
- Liu, C.; Wang, Q.S.; Wang, Y.J. The CHEK2 I157T variant and colorectal cancer susceptibility: A systematic review and meta-analysis. Asian Pac. J. Cancer Prev. 2012, 13, 2051–2055. [Google Scholar] [CrossRef]
- Alim, I.; Loke, J.; Yam, S.; Templeton, A.S.; Newcomb, P.; Lindor, N.M.; Pai, R.K.; Jenkins, M.A.; Buchanan, D.D.; Gallinger, S.; et al. Cancer Risk C (CR-C), a functional genomics test is a sensitive and rapid test for germline mismatch repair deficiency. Genet. Med. 2022, 24, 1821–1830. [Google Scholar] [CrossRef]
- Reitsam, N.G.; Märkl, B.; Dintner, S.; Waidhauser, J.; Vlasenko, D.; Grosser, B. Concurrent loss of MLH1, PMS2 and MSH6 immunoexpression in digestive system cancers indicating a widespread dysregulation in DNA repair processes. Front. Oncol. 2022, 12, 1019798. [Google Scholar] [CrossRef] [PubMed]
- Sriramulu, S.; Ramachandran, M.; Subramanian, S.; Kannan, R.; Gopinath, M.; Sollano, J.; Bissi, L.; Banerjee, A.; Marotta, F.; Pathak, S. A review on role of ATM gene in hereditary transfer of colorectal cancer. Acta Biomed. 2019, 89, 463. [Google Scholar] [PubMed]
- Hall, M.J.; Bernhisel, R.; Hughes, E.; Larson, K.; Rosenthal, E.T.; Singh, N.A.; Lancaster, J.M.; Kurian, A.W. Germline Pathogenic Variants in the Ataxia Telangiectasia Mutated (ATM) Gene are Associated with High and Moderate Risks for Multiple Cancers. Cancer Prev. Res. 2021, 14, 433–440. [Google Scholar] [CrossRef] [PubMed]
- Maillet, P.; Chappuis, P.O.; Vaudan, G.; Dobbie, Z.; Müller, H.; Hutter, P.; Sappino, A.P. A poly-morphism in the ATM gene modulates the penetrance of hereditary non-polyposis colo-rectal cancer. Int. J. Cancer 2000, 88, 928–931. [Google Scholar] [CrossRef]
- van Leerdam, M.E.; Roos, V.H.; van Hooft, J.E.; Balaguer, F.; Dekker, E.; Kaminski, M.F.; Latchford, A.; Neumann, H.; Ricciardiello, L.; Rupińska, M.; et al. Endoscopic management of Lynch syndrome and of familial risk of colorectal cancer: European Society of Gastrointestinal Endoscopy (ESGE) Guideline. Endoscopy 2019, 51, 1082–1093. [Google Scholar] [CrossRef]
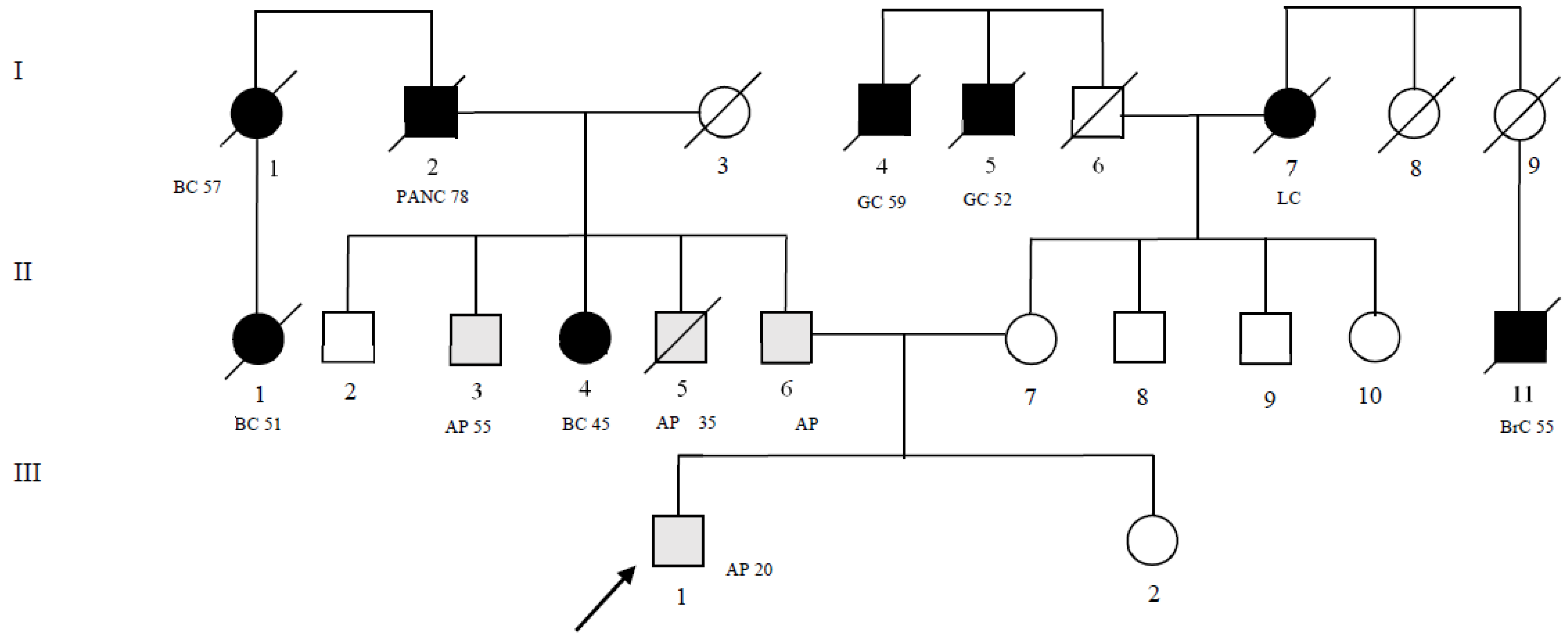

| Patient Pedigree ID (Figure 1) | MLH1 Gene | ATM Gene |
|---|---|---|
| II-4 | c.589-9_589-6delgttt | c.5975A>C |
| II-6 | c.589-9_589-6delgttt | c.5975A>C |
| II-7 | - | c.8734A>G |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Nolano, A.; Rossi, G.B.; D’Angelo, V.; Liccardo, R.; Rosa, M.D.; Izzo, P.; Duraturo, F. Germline Variants in MLH1 and ATM Genes in a Young Patient with MSI-H in a Precancerous Colonic Lesion. Int. J. Mol. Sci. 2023, 24, 5970. https://doi.org/10.3390/ijms24065970
Nolano A, Rossi GB, D’Angelo V, Liccardo R, Rosa MD, Izzo P, Duraturo F. Germline Variants in MLH1 and ATM Genes in a Young Patient with MSI-H in a Precancerous Colonic Lesion. International Journal of Molecular Sciences. 2023; 24(6):5970. https://doi.org/10.3390/ijms24065970
Chicago/Turabian StyleNolano, Antonio, Giovanni Battista Rossi, Valentina D’Angelo, Raffaella Liccardo, Marina De Rosa, Paola Izzo, and Francesca Duraturo. 2023. "Germline Variants in MLH1 and ATM Genes in a Young Patient with MSI-H in a Precancerous Colonic Lesion" International Journal of Molecular Sciences 24, no. 6: 5970. https://doi.org/10.3390/ijms24065970
APA StyleNolano, A., Rossi, G. B., D’Angelo, V., Liccardo, R., Rosa, M. D., Izzo, P., & Duraturo, F. (2023). Germline Variants in MLH1 and ATM Genes in a Young Patient with MSI-H in a Precancerous Colonic Lesion. International Journal of Molecular Sciences, 24(6), 5970. https://doi.org/10.3390/ijms24065970








