Macrophage Migration Inhibitory Factor in Psoroptes ovis: Molecular Characterization and Potential Role in Eosinophil Accumulation of Skin in Rabbit and Its Implication in the Host–Parasite Interaction
Abstract
:1. Introduction
2. Results
2.1. Sequence Comparison of Orthologous MIF from P. ovis and Other Organisms
2.2. Western Blotting Analysis of rPsoMIF
2.3. PsoMIF mRNA Expression Patterns in P. ovis Mites
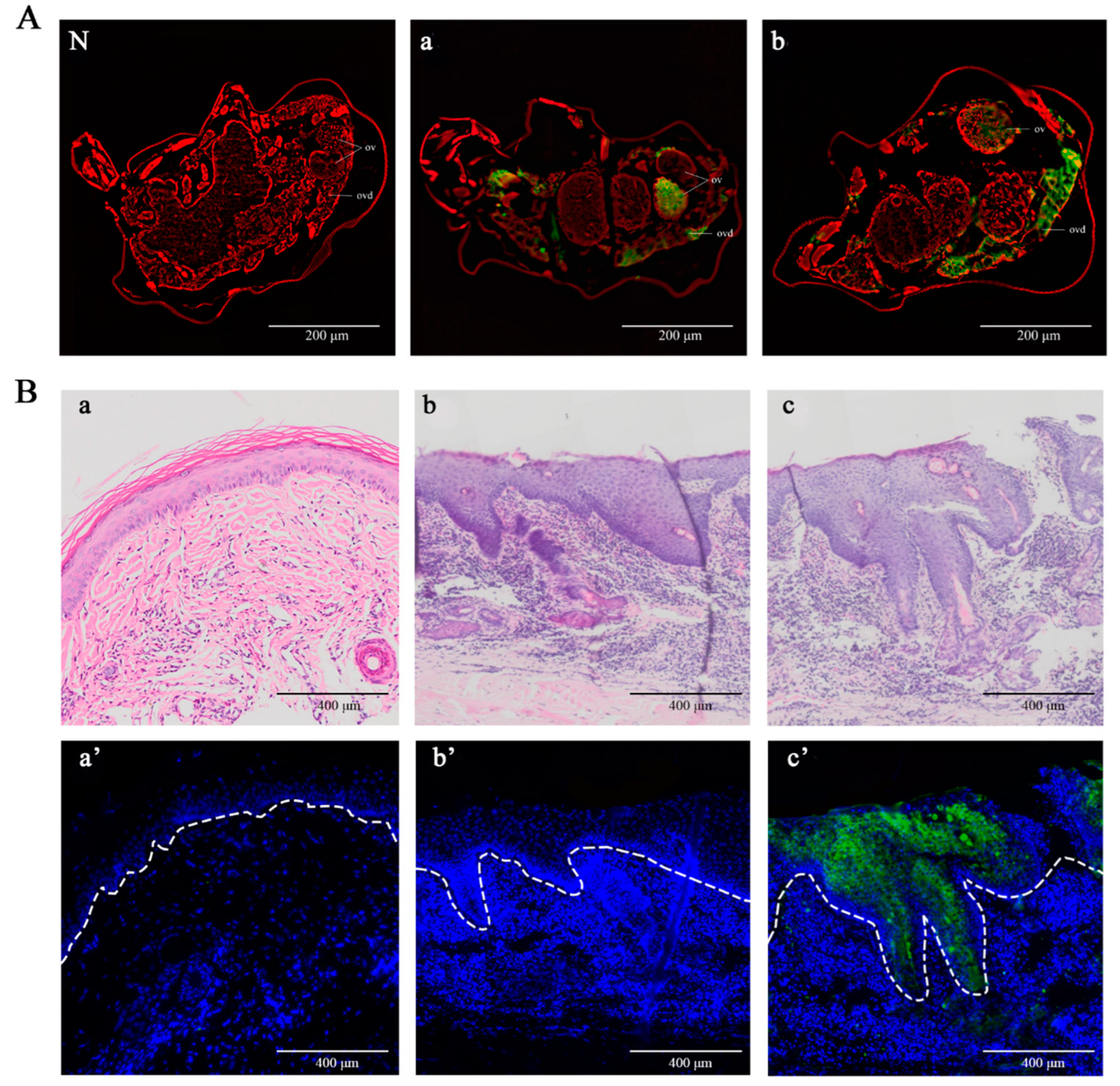
2.4. Detection of Native PsoMIF in Adult Female Mites and P. ovis Skin Lesions
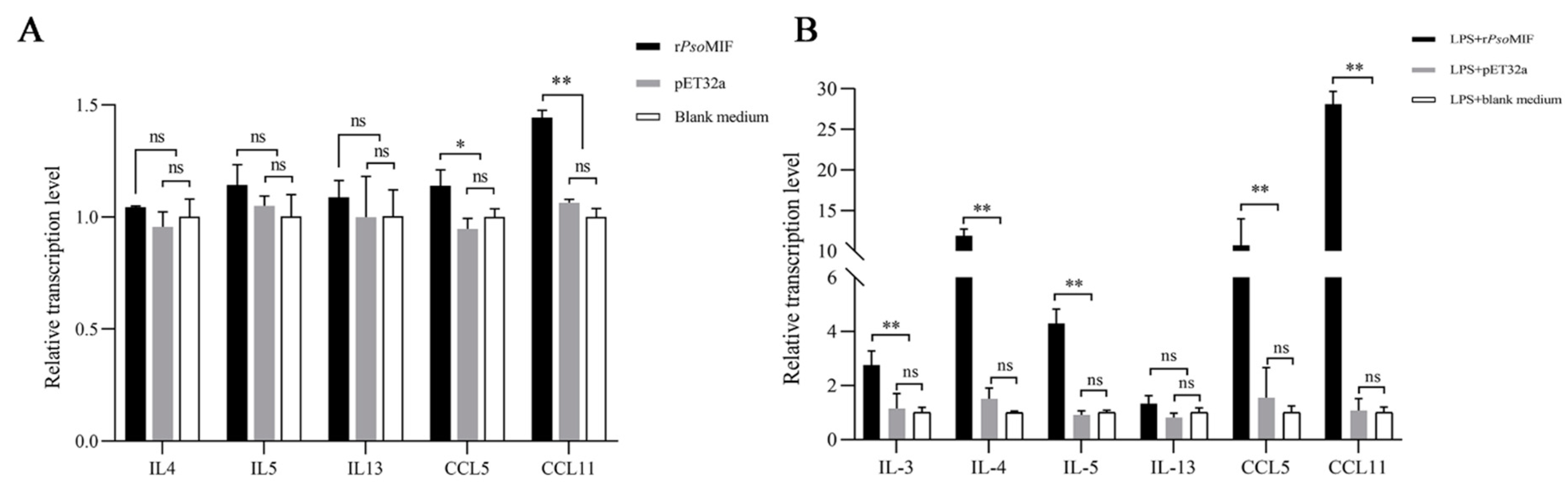
2.5. rPsoMIF Upregulates Eosinophil-Related Gene Expression in Allergic Rabbit PBMC
2.6. rPsoMIF Significantly Upregulates Eosinophil-Related Gene Expression in HaCaT Cells
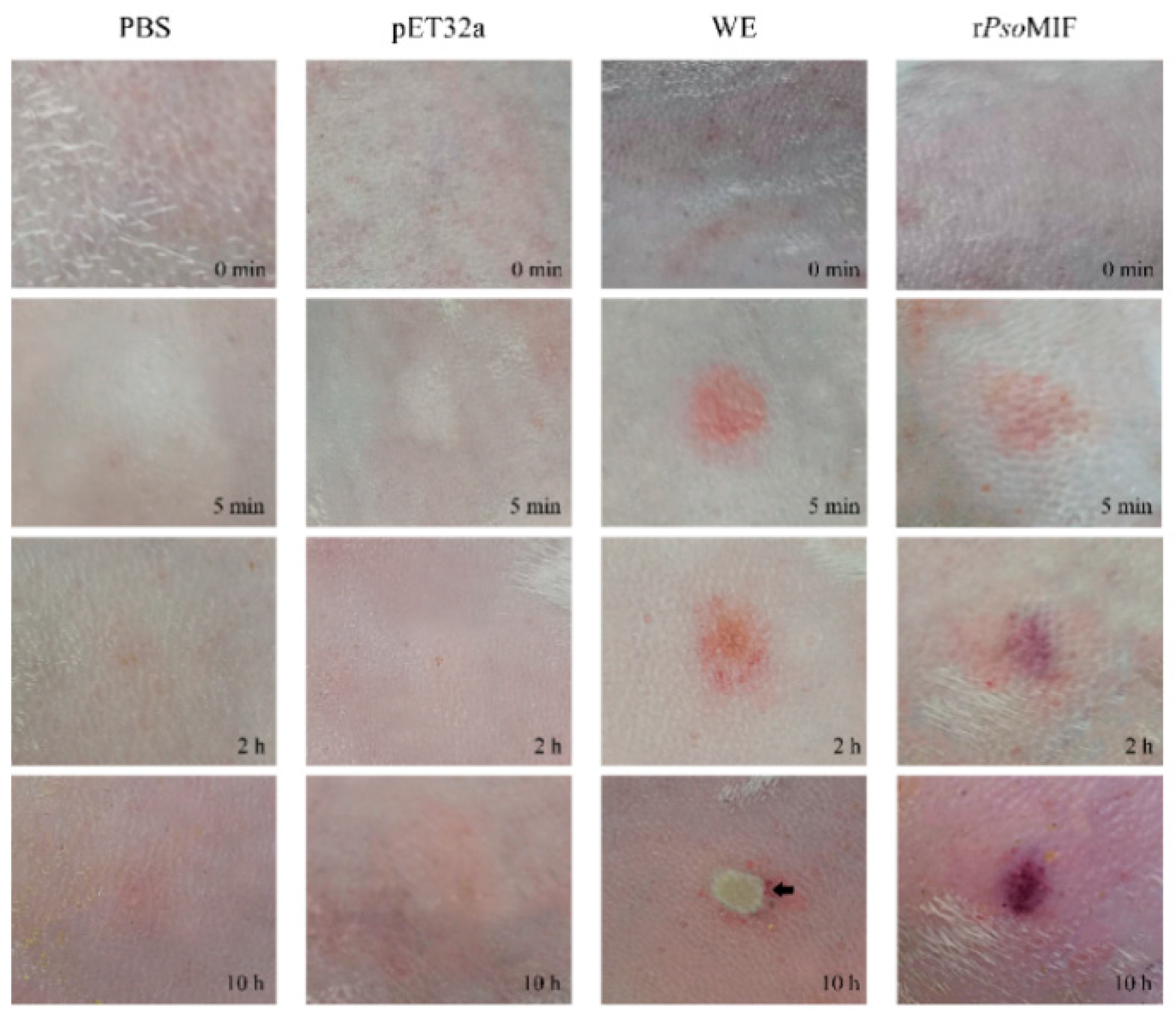
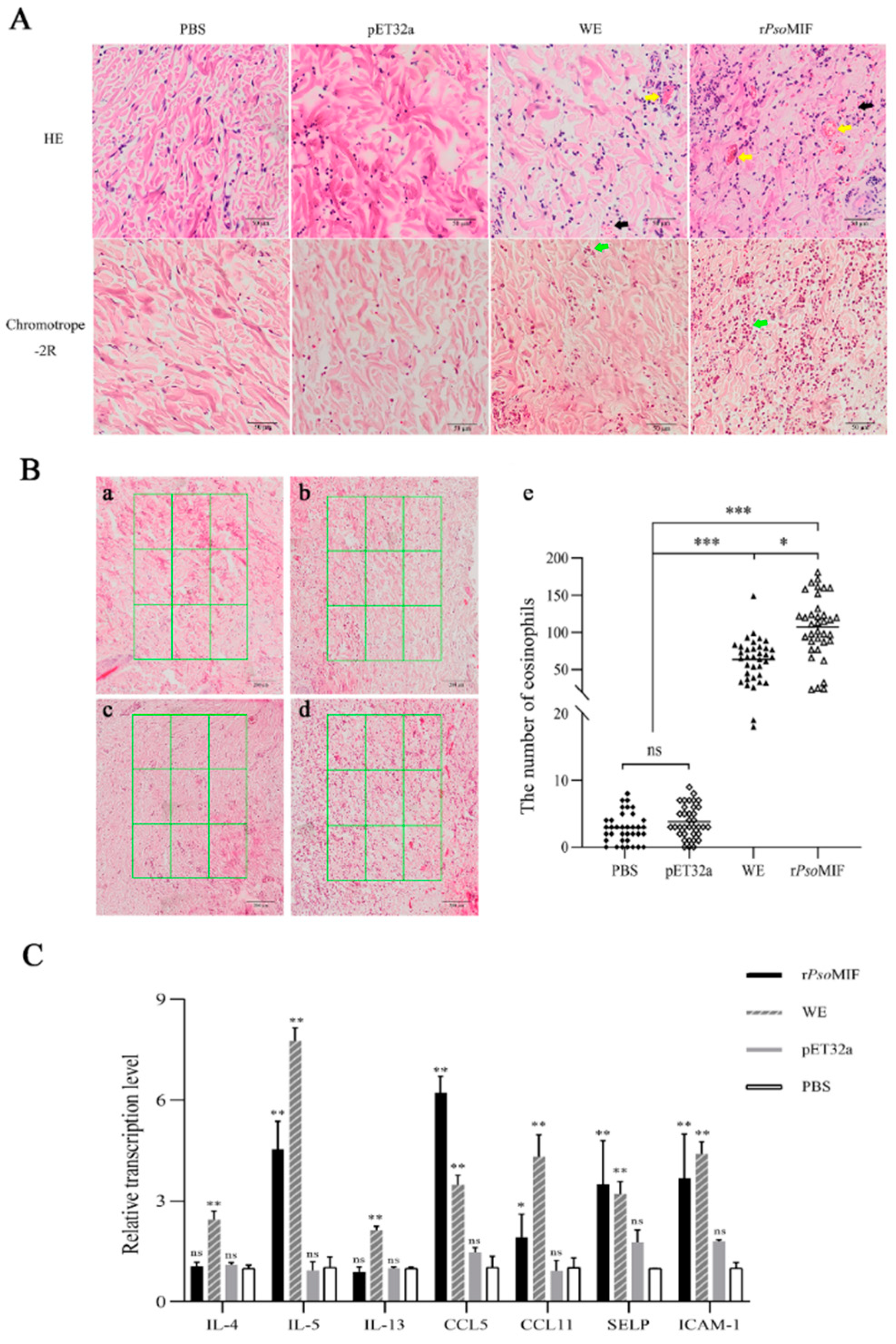
2.7. rPsoMIF Induces Skin Inflammation and Eosinophil Recruitment In Vivo
2.8. rPsoMIF Enhances Vascular Permeability
3. Discussion
4. Materials and Methods
4.1. Sequence Comparison of Orthologous MIF from Psoroptes ovis and Other Organisms
4.2. Mite Collection, Preparation of Recombinant PsoMIF (rPsoMIF), and Whole Mite Protein
4.3. Polyclonal Antibody Production and Western Blotting Analysis
4.4. Immunolocalization of MIF in Adult Female P. ovis var. cuniculi and Lesional Skin of Rabbit Infested with P. ovis var. cuniculi
4.5. In Vitro Treatment of Rabbit PBMC
4.6. In Vitro Treatment of HaCaT
4.7. In Vivo Treatment of New Zealand Rabbits
4.8. qRT-PCR
4.9. Skin Histopathology
4.10. Vascular Permeability Assay
4.11. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- He, M.L.; Xu, J.; He, R.; Shen, N.X. Preliminary analysis of Psoroptes ovis transcriptome in different developmental stages. Parasit Vector. 2016, 9, 570–581. [Google Scholar] [CrossRef] [Green Version]
- Losson, B.J. Sheep psoroptic mange: An update. Vet. Parasitol. 2012, 189, 39–43. [Google Scholar] [CrossRef] [PubMed]
- Stoeckli, M.R.; Mcneilly, T.N.; Frew, D. The effect of Psoroptes ovis infestation on ovine epidermal barrier function. Vet. Res. 2013, 44, 11. [Google Scholar] [CrossRef] [Green Version]
- Broek, A.H.V.D.; Else, R.W.; Huntley, J.F. Early innate and longer-term adaptive cutaneous immunoinflammatory responses during primary infestation with the sheep scab mite, Psoroptes ovis. J. Comp. Pathol. 2004, 131, 318–329. [Google Scholar] [CrossRef] [PubMed]
- Broek, A.H.V.D.; Huntley, J.F. Sheep scab: The disease, patshogenesis and control. J. Comp. Pathol. 2003, 128, 79–91. [Google Scholar] [CrossRef] [PubMed]
- Chen, Z.; Claerebout, E.; Chiers, K. Dermal immune responses against Psoroptes ovis in two cattle breeds and effects of anti-inflammatory dexamethasone treatment on the development of psoroptic mange. Vet. Res. 2021, 52, 1–11. [Google Scholar] [CrossRef]
- Liao, X.X. Comparative Pathology and Evaluation of Characteristic Cytokines and Transcription Factors of Th1/Th2 Immune Response to Skin Lesion of Psoroptic Mange in Rabbits. Master’s Thesis, Sichuan Agricultural University, Yaan, China, 2017. [Google Scholar]
- Mathieson, B.R.F.; Lehane, M.J. Ultrastructure of the alimentary canal of the sheep scab mite, Psoroptes ovis (Acari: Psoroptidae). Vet. Parasitol. 2002, 104, 151–166. [Google Scholar] [CrossRef]
- Broek, A.H.V.D.; Huntley, J.F.; Machell, J. Cutaneous and systemic responses during primary and challenge infestations of sheep with the sheep scab mite, Psoroptes ovis. Parasite. Immunol. 2000, 22, 407–414. [Google Scholar] [CrossRef]
- Wildblood, L.A.; Jones, D.G. Stimulation of the in vitro migration of ovine eosinophils by factors derived from the sheep scab mite, Psoroptes ovis. Vet. Res. 2007, 31, 197–206. [Google Scholar] [CrossRef]
- Sparkes, A.; Baetselier, P.; Roelants, K. The non-mammalian MIF superfamily. Immunobiology 2017, 222, 473–482. [Google Scholar] [CrossRef] [Green Version]
- Sumaiya, K.; Langford, D.; Natarajaseenivasan, K. Macrophage migration inhibitory factor (MIF): A multifaceted cytokine regulated by genetic and physiological strategies. Pharmacol. Ther. 2022, 233, 108024. [Google Scholar] [CrossRef]
- Falcone, F.H.; Loke, P.; Zang, X.; MacDonald, A.S.; Maizels, R.M.; Allen, J.E. A Brugia malayi homolog of macrophage migration inhibitory factor reveals an important link between macrophages and eosinophil recruitment during nematode infection. J. Immunol. 2001, 167, 5348–5354. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Prieto-Lafuente, L.; Gregory, W.F.; Allen, J.E.; Maizels, R.M. MIF homologues from a filarial nematode parasite synergize with IL-4 to induce alternative activation of host macrophages. J. Leukoc. Biol. 2009, 85, 844–854. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Baumann, R.; Casaulta, C.; Simon, D.; Conus, S.; Yousefi, S.; Simon, H.U. Macrophage migration inhibitory factor delays apoptosis in neutrophils by inhibiting the mitochondria-dependent death pathway. FASEB J. 2003, 17, 2221–2230. [Google Scholar] [CrossRef] [Green Version]
- Magalhães, E.S.; Paiva, C.N.; Souza, H.S.; Pyrrho, A.S.; Mourao-Sá, D.; Figueiredo, R.T.; Vieira-de-Abreu, A.; Dutra, H.S.; Silveira, M.S.; Gaspar-Elsas, M.I.C.; et al. Macrophage migration inhibitory factor is critical to interleukin-5-driven eosinophilopoiesis and tissue eosinophilia triggered by Schistosoma mansoni infection. FASEB J. 2009, 23, 1262–1271. [Google Scholar] [CrossRef] [PubMed]
- Bozza, M.T.; Lintomen, L.; Kitoko, J.Z.; CN Paiva, P.C. Olsen, The role of MIF on eosinophil biology and eosinophilic inflammation. Clin. Rev. Allergy Immunol. 2020, 58, 15–24. [Google Scholar] [CrossRef] [PubMed]
- Zheng, Y.L.; Chen, Y.H.; Xie, Y.; Yang, G.Y.; He, R.; Gu, X.B. The effects of recombinant macrophage migration inhibitory factor of Psoroptes ovis on the characteristic molecules of Th1/Th2 and Th17/Treg type immune responses in rabbit peripheral blood mononuclear cells in vitro. Acta Vet. Zootech. Sin. 2020, 51, 594–601. (In Chinese) [Google Scholar]
- Bendrat, K.; Al-Abed, Y.; Callaway, D.J. Biochemical and mutational investigations of the enzymatic activity of macrophage migration inhibitory factor. Biochemistry 1997, 36, 15356–15362. [Google Scholar] [CrossRef]
- Lubetsky, J.B.; Swope, M.; Dealwis, C. Pro-1 of macrophage migration inhibitory factor functions as a catalytic base in the phenylpyruvate tautomerase activity. Biochemistry 1999, 38, 7346–7354. [Google Scholar] [CrossRef]
- Xu, L.; Li, Y.Y.; Sun, H.Y. Current developments of macrophage migration inhibitory factor (MIF) inhibitors. Drug Discov. Today. 2013, 18, 592–600. [Google Scholar] [CrossRef]
- Nguyen, M.T.; Beck, J.; Lue, H. A 16-residue peptide fragment of macrophage migration inhibitory factor, MIF-(50–65), exhibits redox activity and has MIF-like biological functions. J. Biol. Chem. 2003, 278, 33654–33671. [Google Scholar] [CrossRef] [Green Version]
- Sommerville, C.; Richardson, J.M.; Williams, R.A. Henriquez, Biochemical and immunological characterization of Toxoplasma gondii macrophage migration inhibitory factor. J. Biol. Chem. 2013, 288, 12733–12741. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Miller, J.L.; Harupa, A.; Kappe, S.H. Plasmodium yoelii macrophage migration inhibitory factor is necessary for efficient liver-stage development. Infect. Immun. 2012, 80, 1399–1407. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Qu, G.; Fetterer, R.; Leng, L. Ostertagia ostertagi macrophage migration inhibitory factor is present in all developmental stages and may cross-regulate host functions through interaction with the host receptor. Int. J. Parasitol. 2014, 44, 355–367. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Naessens, E.; Dubreuil, G.; Giordanengo, P. A secreted MIF cytokine enables aphid feeding and represses plant immune responses. Curr. Biol. 2015, 25, 1898–1903. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, Y.P.; Twu, O.; Johnson, P.J. Trichomonas vaginalis macrophage migration inhibitory factor mediates parasite survival during nutrient stress. mBio 2018, 9, e00910-18. [Google Scholar] [CrossRef] [Green Version]
- Mcnair, C.M.; Billingsley, P.F.; Nisbet, A.J. Feeding-associated gene expression in sheep scab mites (Psoroptes ovis). Vet. Res. 2010, 41, 16. [Google Scholar] [CrossRef] [Green Version]
- Qu, G.; Fetterer, R.; Jenkins, M. Characterization of Neospora caninum macrophage migration inhibitory factor. Exp. Parasitol. 2013, 135, 246–256. [Google Scholar] [CrossRef] [Green Version]
- Han, C.; Lin, Y.; Shan, G. Plasma concentration of malaria parasite-derived macrophage migration inhibitory factor in uncomplicated malaria patients correlates with parasitemia and disease severity. Clin. Vaccine Immunol. 2010, 17, 1524–1532. [Google Scholar] [CrossRef] [Green Version]
- Baroni, A.; Buommino, E.; De Gregorio, V. Structure and function of the epidermis related to barrier properties. Clin. Dermatol. 2012, 30, 257–262. [Google Scholar] [CrossRef]
- Watkins, C.A.; Mackellar, A.; Frew, D. Gene expression profiling of ovine keratinocytes stimulated with Psoroptes ovis mite antigen--a preliminary study. Parasite Immunol. 2009, 31, 304–311. [Google Scholar] [CrossRef]
- Siegfried, E.; Ochs, H.; Deplazes, P. Clinical development and serological antibody responses in sheep and rabbits experimentally infested with Psoroptes ovis and Psoroptes cuniculi. Vet. Parasitol. 2004, 124, 109–124. [Google Scholar] [CrossRef] [Green Version]
- Kato, M.; Kephart, G.M.; Morikawa, A. Eosinophil infiltration and degranulation in normal human tissues: Evidence for eosinophil degranulation in normal gastrointestinal tract. Int. Arch. Allergy Immunol. 2001, 125, 55–58. [Google Scholar] [CrossRef] [PubMed]
- Kovalszki, A.; Weller, P.F. Eosinophilia. Prim. Care 2016, 43, 607–617. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ravin, A.K.; Loy, M. The eosinophil in infection. Clin. Rev. Allergy Immunol. 2016, 50, 214–227. [Google Scholar] [CrossRef] [PubMed]
- Fulkerson, P.C.; Schollaert, K.L.; Bouffi, C. IL-5 triggers a cooperative cytokine network that promotes eosinophil precursor maturation. J. Immunol. 2014, 193, 4043–4052. [Google Scholar] [CrossRef] [Green Version]
- Hogan, S.P.; Rosenberg, H.F.; Moqbel, R. Eosinophils: Biological properties and role in health and disease. Clin. Exp. Allergy 2008, 38, 709–750. [Google Scholar] [CrossRef] [PubMed]
- Ahrens, R.; Waddell, A.; Seidu, L. Intestinal macrophage/epithelial cell-derived CCL11/eotaxin-1 mediates eosinophil recruitment and function in pediatric ulcerative colitis. J. Immunol. 2008, 181, 7390–7399. [Google Scholar] [CrossRef] [Green Version]
- Takamoto, M.; Sugane, K. Synergism of IL-3, IL-5, and GM-CSF on eosinophil differentiation and its application for an assay of murine IL-5 as an eosinophil differentiation factor. Immunol. Lett. 1995, 45, 43–46. [Google Scholar] [CrossRef]
- Lampinen, M.; Carlson, M.; Håkansson, L.D. Cytokine-regulated accumulation of eosinophils in inflammatory disease. Allergy 2004, 59, 793–805. [Google Scholar] [CrossRef]
- Bullard, D.C.; Qin, L.; Lorenzo, I.; Quinlin, W.M. P-selectin/ICAM-1 double mutant mice: Acute emigration of neutrophils into the peritoneum is completely absent but is normal into pulmonary alveoli. J. Clin. Investig. 1995, 95, 1782–1788. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Subramaniam, M.; Saffaripour, S.; Watson, S.R. Reduced recruitment of inflammatory cells in a contact hypersensitivity response in P-selectin-deficient mice. J. Exp. Med. 1995, 181, 2277–2282. [Google Scholar] [CrossRef]
- Collins, D.S.; Dupuis, R.; Gleich, G.J. Immunoglobulin E-mediated increase in vascular permeability correlates with eosinophilic inflammation. Am. Rev. Respir. Dis. 1993, 147, 677–683. [Google Scholar] [CrossRef] [PubMed]
- Deloach, J.R.; Wright, F.C. Ingestion of rabbit erythrocytes containing 51Cr-labeled hemoglobin by Psoroptes spp. (Acari: Psoroptidae) that originated on cattle, mountain sheep, or rabbits. J. Med. Entomol. 1981, 18, 345–348. [Google Scholar] [CrossRef] [PubMed]
- Mathieson, B.R.F. An Investigation of Psoroptes ovis, the Sheep Scab Mite with a View to Developing an In Vitro Feeding System. Ph.D Thesis, University of Wales, Bangor, Wales, 1995. [Google Scholar]
- Huang, L.; Beiting, D.P.; Gebreselassie, N.G. Eosinophils and IL-4 Support Nematode Growth Coincident with an Innate Response to Tissue Injury. PLoS Pathog. 2015, 11, e1005347. [Google Scholar] [CrossRef]
- Yoshimura, K. Mechanism of parasite killing by eosinophils in parasitic infections. Jpn J. Clin. Med. 1993, 51, 657–663. [Google Scholar]
- Zhang, Y.C.; Gu, X.B.; Chen, Y.H. Molecular characterization of four novel serpins in Psoroptes ovis var. cuniculi and their implications in the host-parasite interaction. Int. J. Biol. Macromol. 2021, 182, 1399–1408. [Google Scholar]
- Sanders, A.; Froggatt, P.; Wall, R. Life-cycle stage morphology of Psoroptes mange mites. Med. Vet. Entomol. 2000, 14, 131–141. [Google Scholar] [CrossRef]
- Gu, X.B.; Chen, Y.H.; Zhang, C.Y. Molecular characterization and serodiagnostic potential of two serpin proteins in Psoroptes ovis var. cuniculi. Parasite Vector. 2020, 13, 620. [Google Scholar] [CrossRef]
- Shen, N.X.; Chen, Y.H.; Wei, W.R. Comparative analysis of the allergenic characteristics and serodiagnostic potential of recombinant chitinase-like protein-5 and -12 from Sarcoptes scabiei. Parasite Vector. 2021, 14, 148. [Google Scholar] [CrossRef]
- Lanzafame, M.; Botta, E.; Teson, M. Reference genes for gene expression analysis in proliferating and differentiating human keratinocytes. J. Exp. Dermatol. 2015, 24, 314–316. [Google Scholar] [CrossRef] [PubMed]
- Song, Y.; Yin, Y.S.; Chang, H. Comparison of four staining methods for detecting eosinophils in nasal polyps. Sci. Rep. 2018, 8, 17718. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Brash, J.T.; Ruhrberg, C.; Fantin, A. Evaluating vascular hyperpermeability-inducing agents in the skin with the miles assay. J. Vis. Exp. 2018, 136, e57524. [Google Scholar]
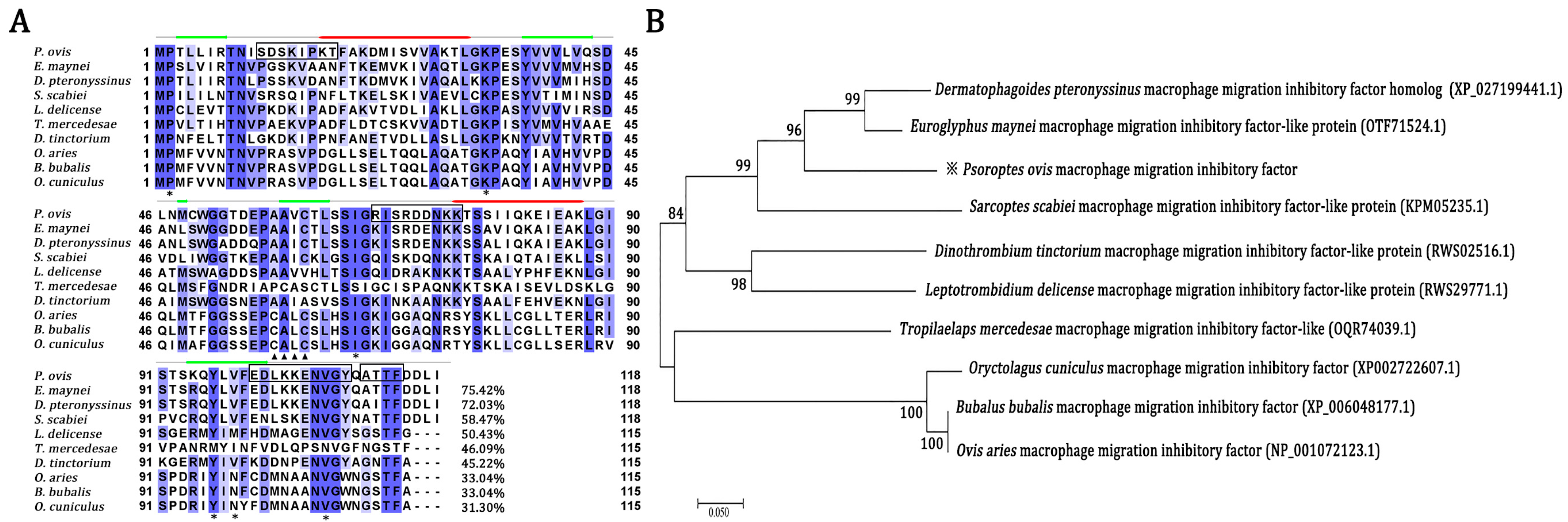
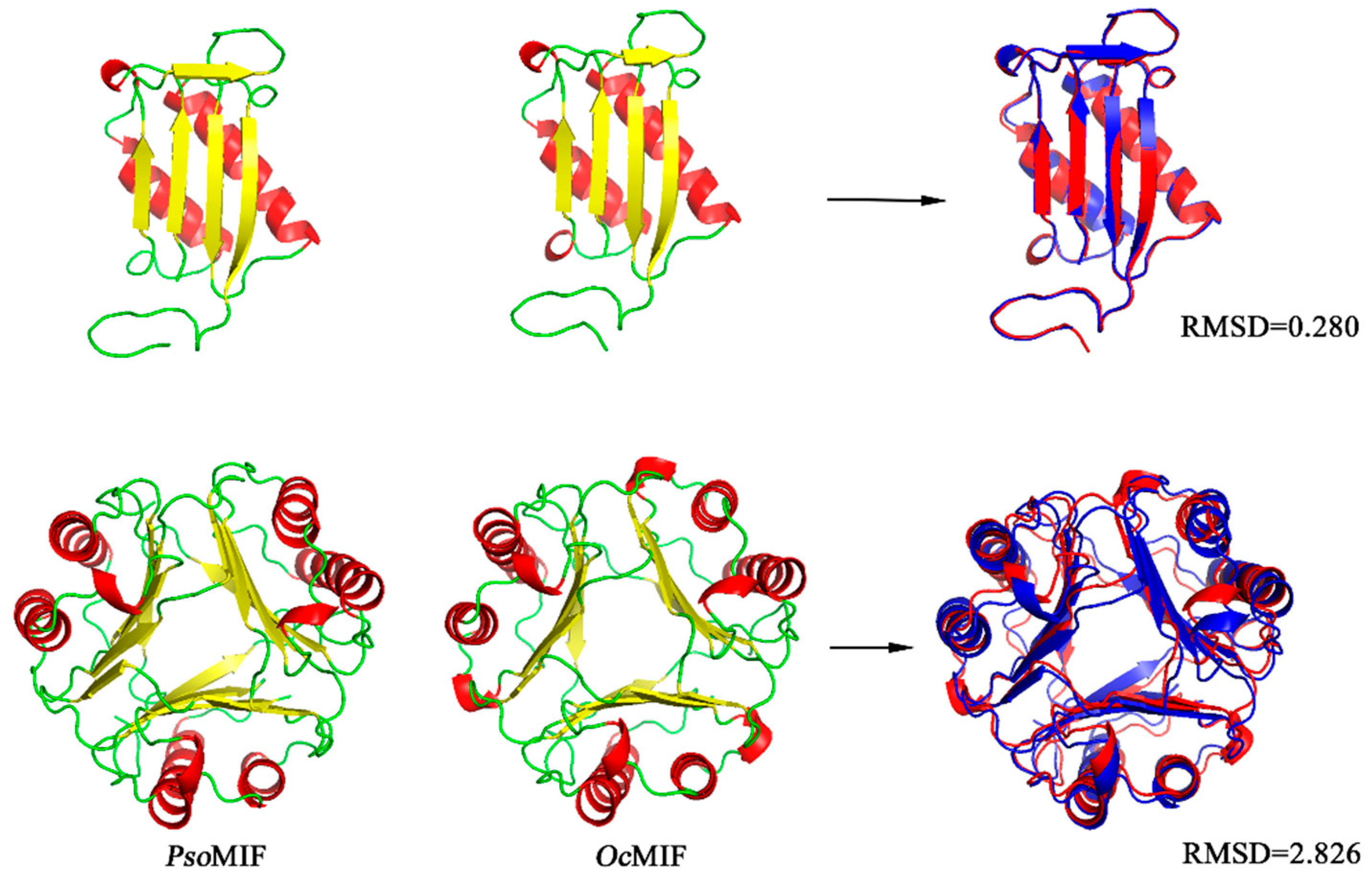
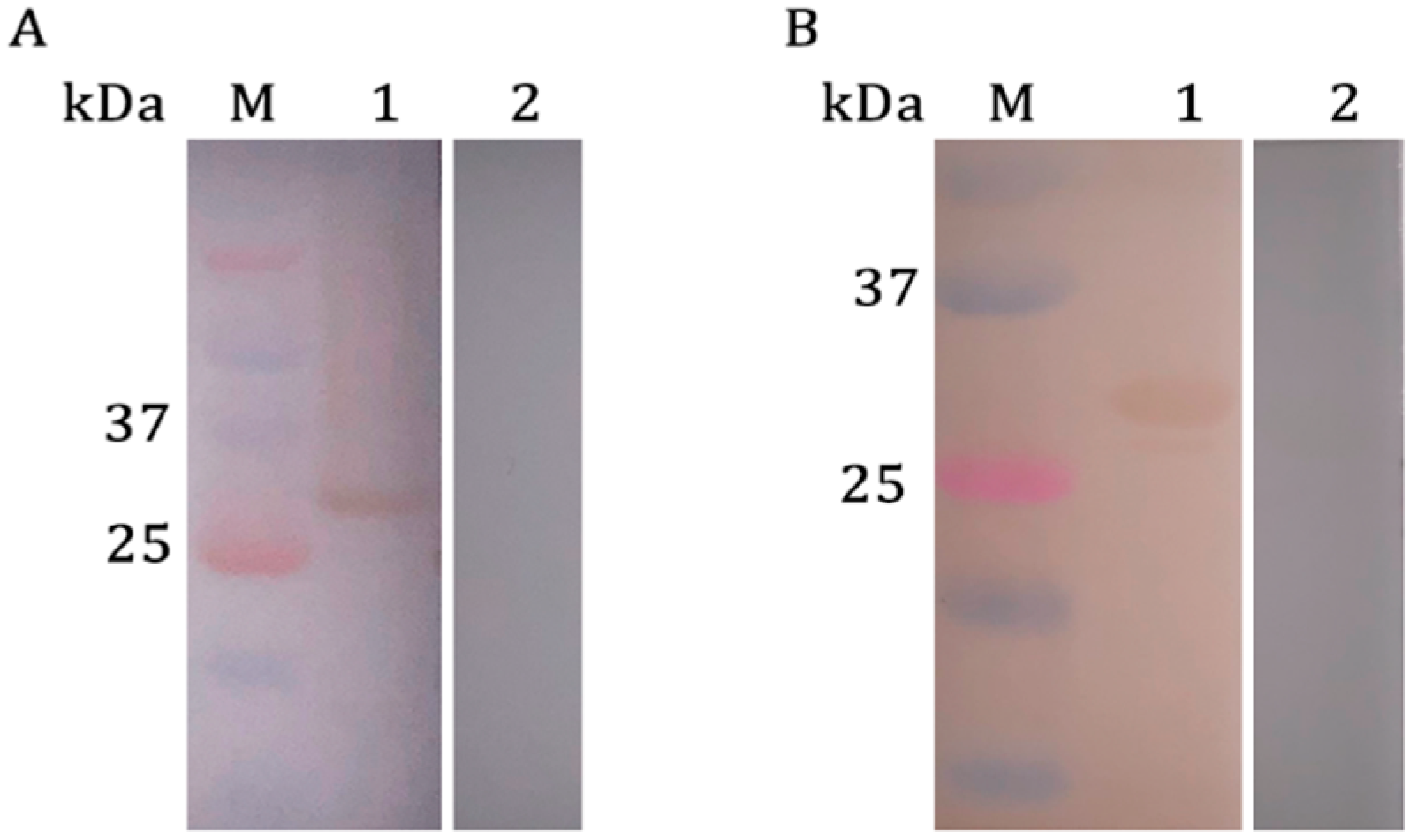
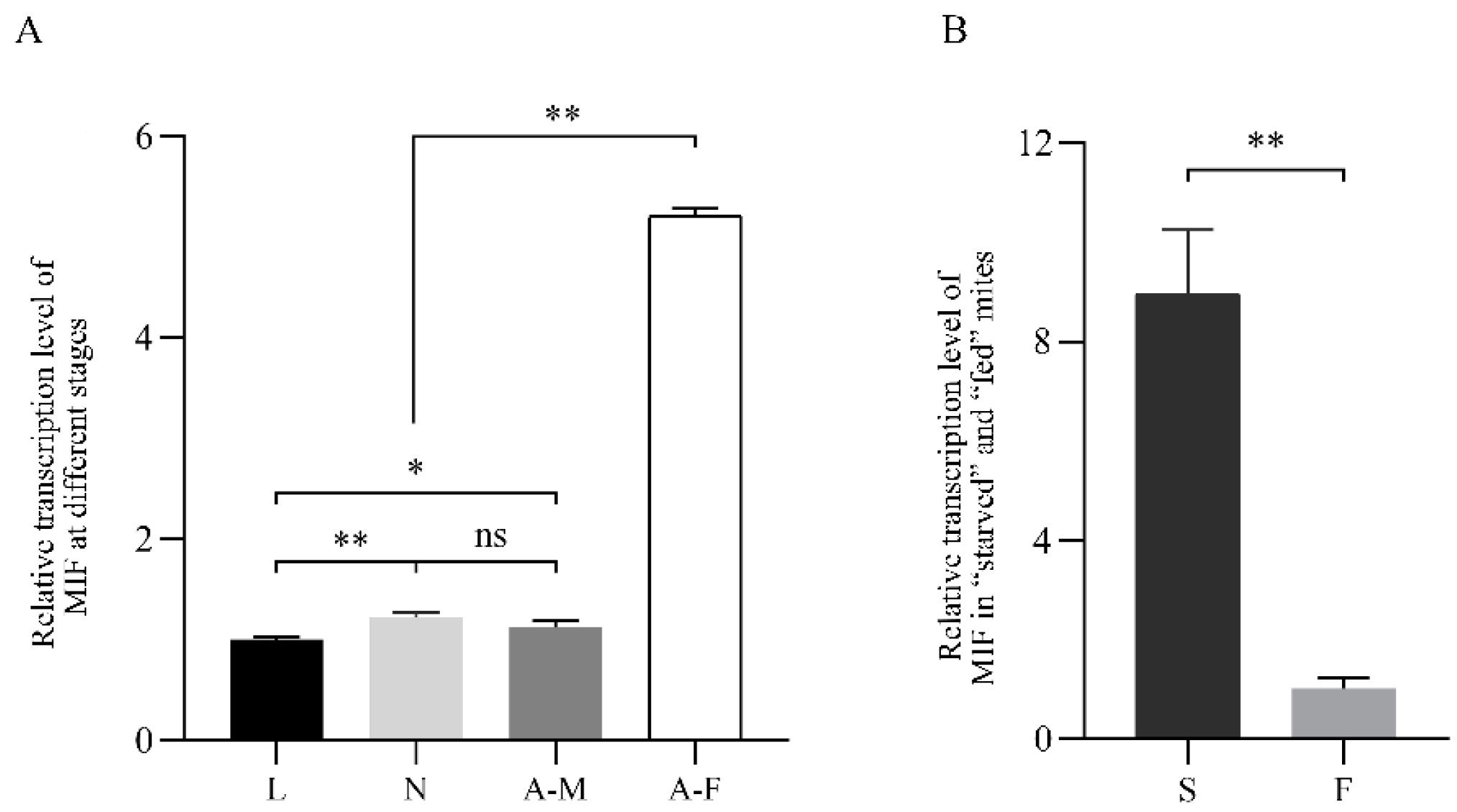

| ID | Tautomerase Active Site | Thiol-Protein Oxidoreductase Active Site | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Amino Acid Position | P2 | P33 | P65 | P96 | P98 | P107 | P57 | P58 | P59 | P60 |
| O. aries (sheep) | P | K | I | Y | N | V | C | A | L | C |
| B. bubalis (Water buffalo) | P | K | I | Y | N | V | C | A | L | C |
| O. cuniculus (rabbit) | P | K | I | Y | N | V | C | A | L | C |
| P. ovis | P | K | I | Y | V | V | A | A | V | C |
| E. maynei | P | K | I | Y | V | V | A | A | I | C |
| D. pteronyssinus | P | K | I | Y | V | V | A | A | I | C |
| S. scabiei | P | K | I | Y | V | V | A | A | I | C |
| L. delicense | P | K | I | Y | M | V | A | A | V | V |
| T. mercedesae | P | K | S | M | I | N | P | C | A | S |
| D. tinctorium | P | K | I | Y | V | V | A | A | I | A |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gu, X.; Ge, Y.; Wang, Y.; Huang, C.; Yang, G.; Xie, Y.; Xu, J.; He, R.; Zhong, Z.; Yang, D.; et al. Macrophage Migration Inhibitory Factor in Psoroptes ovis: Molecular Characterization and Potential Role in Eosinophil Accumulation of Skin in Rabbit and Its Implication in the Host–Parasite Interaction. Int. J. Mol. Sci. 2023, 24, 5985. https://doi.org/10.3390/ijms24065985
Gu X, Ge Y, Wang Y, Huang C, Yang G, Xie Y, Xu J, He R, Zhong Z, Yang D, et al. Macrophage Migration Inhibitory Factor in Psoroptes ovis: Molecular Characterization and Potential Role in Eosinophil Accumulation of Skin in Rabbit and Its Implication in the Host–Parasite Interaction. International Journal of Molecular Sciences. 2023; 24(6):5985. https://doi.org/10.3390/ijms24065985
Chicago/Turabian StyleGu, Xiaobin, You Ge, Ya Wang, Cuirui Huang, Guangyou Yang, Yue Xie, Jing Xu, Ran He, Zhijun Zhong, Deying Yang, and et al. 2023. "Macrophage Migration Inhibitory Factor in Psoroptes ovis: Molecular Characterization and Potential Role in Eosinophil Accumulation of Skin in Rabbit and Its Implication in the Host–Parasite Interaction" International Journal of Molecular Sciences 24, no. 6: 5985. https://doi.org/10.3390/ijms24065985
APA StyleGu, X., Ge, Y., Wang, Y., Huang, C., Yang, G., Xie, Y., Xu, J., He, R., Zhong, Z., Yang, D., He, Z., & Peng, X. (2023). Macrophage Migration Inhibitory Factor in Psoroptes ovis: Molecular Characterization and Potential Role in Eosinophil Accumulation of Skin in Rabbit and Its Implication in the Host–Parasite Interaction. International Journal of Molecular Sciences, 24(6), 5985. https://doi.org/10.3390/ijms24065985







