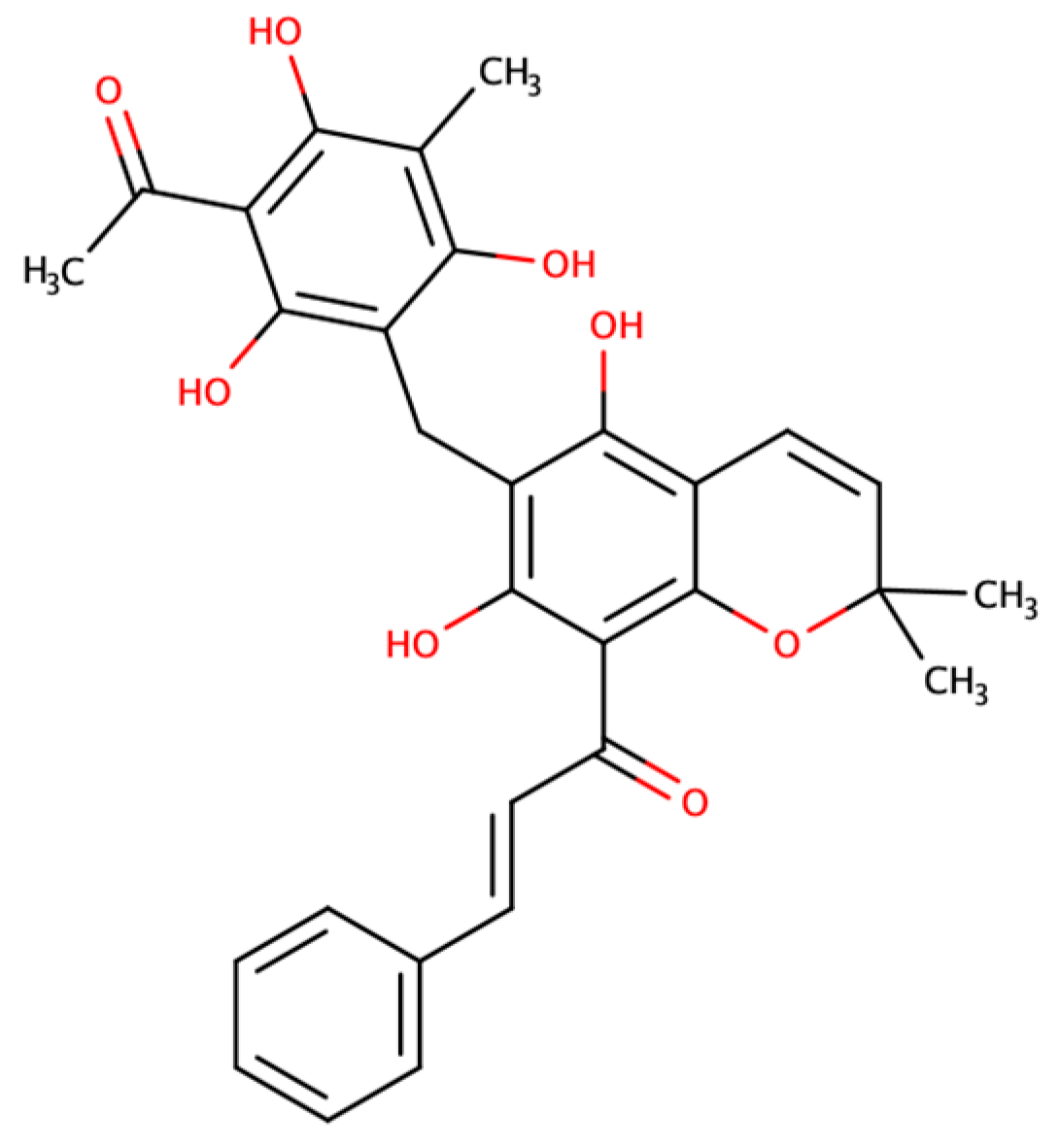
Unraveling the Aquaporin-3 Inhibitory Effect of Rottlerin by Experimental and Computational Approaches
Abstract
:1. Introduction
2. Results
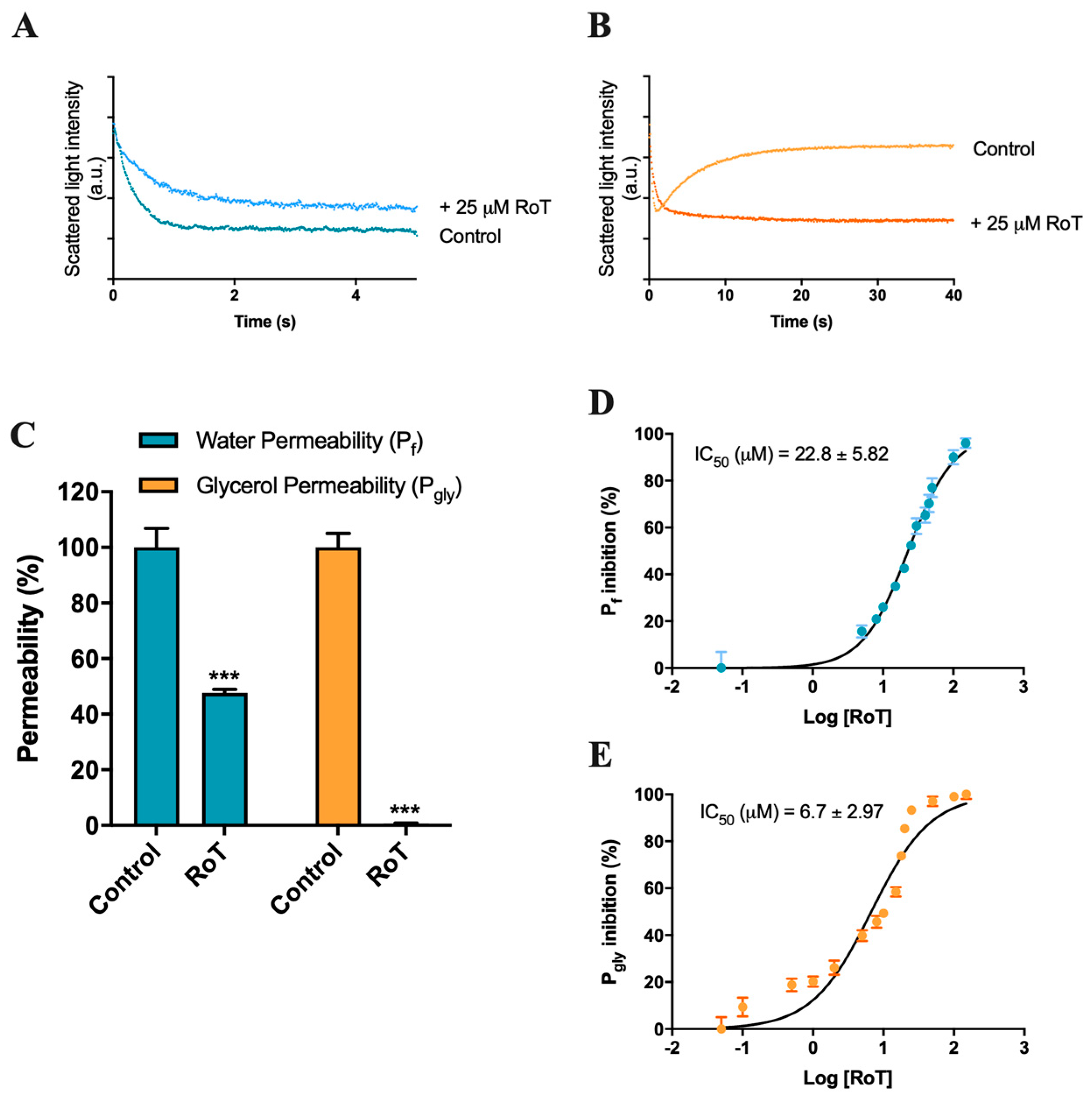
2.1. Effect of Rottlerin on AQP1 and AQP3 Activity
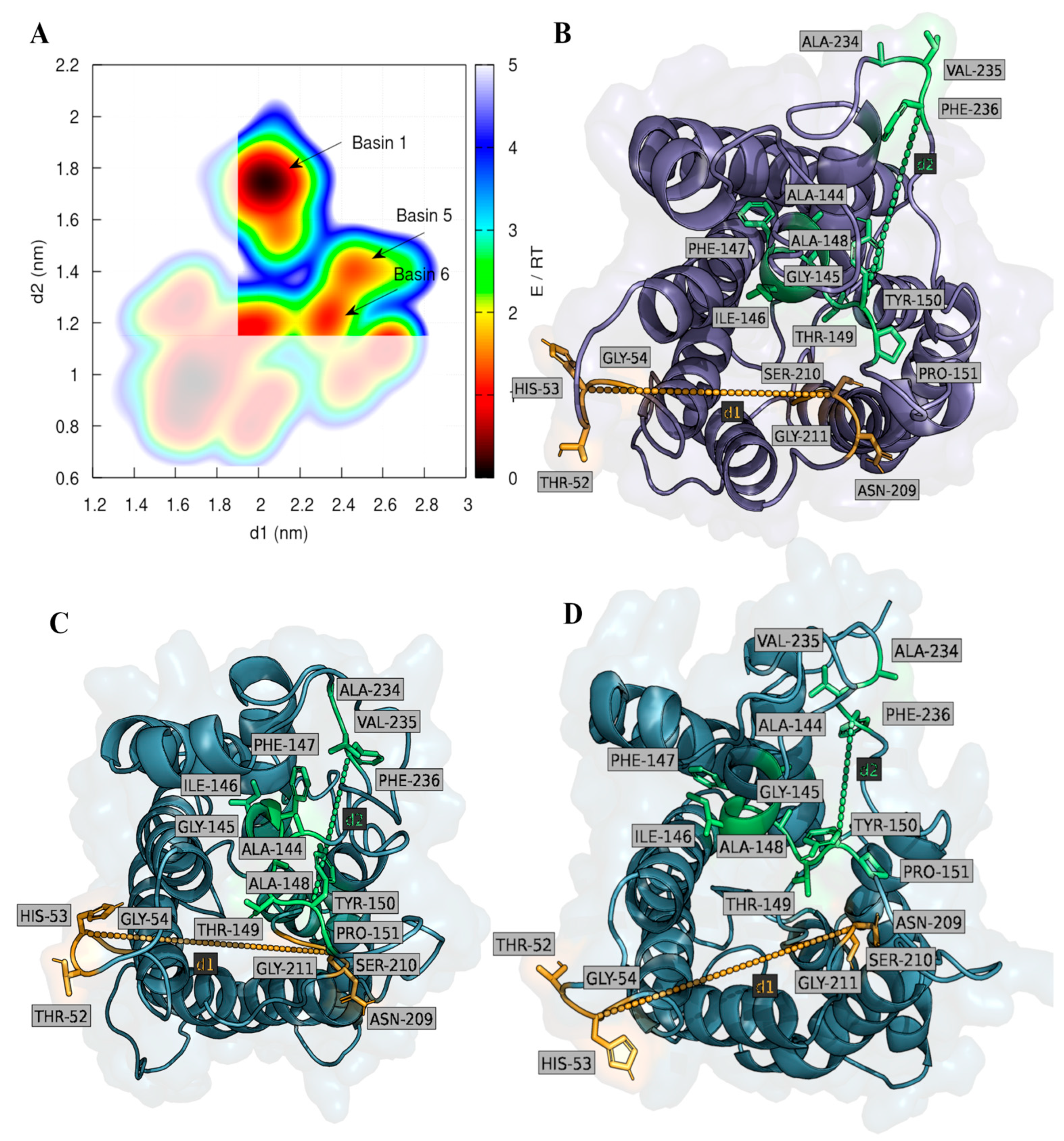
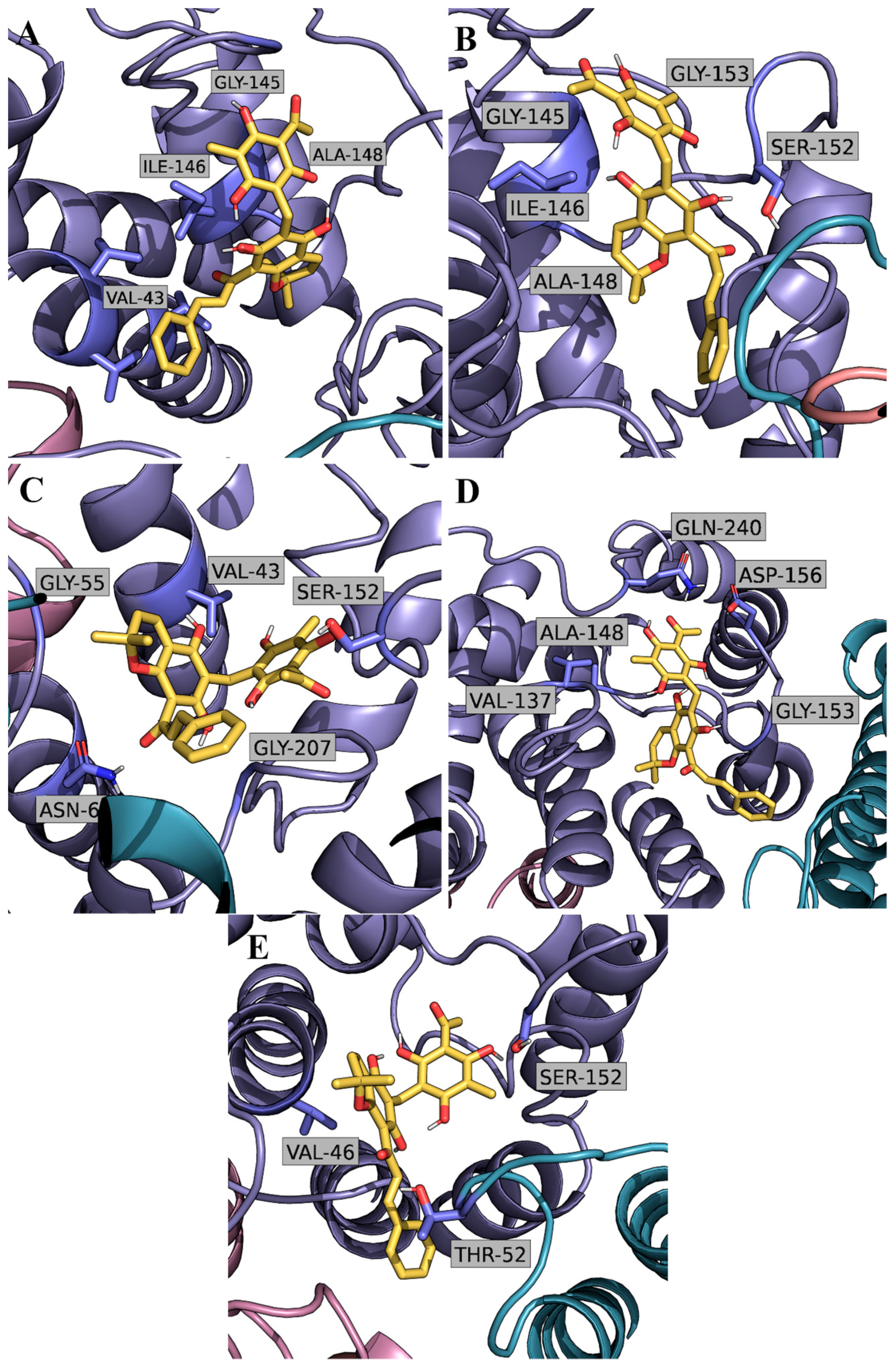
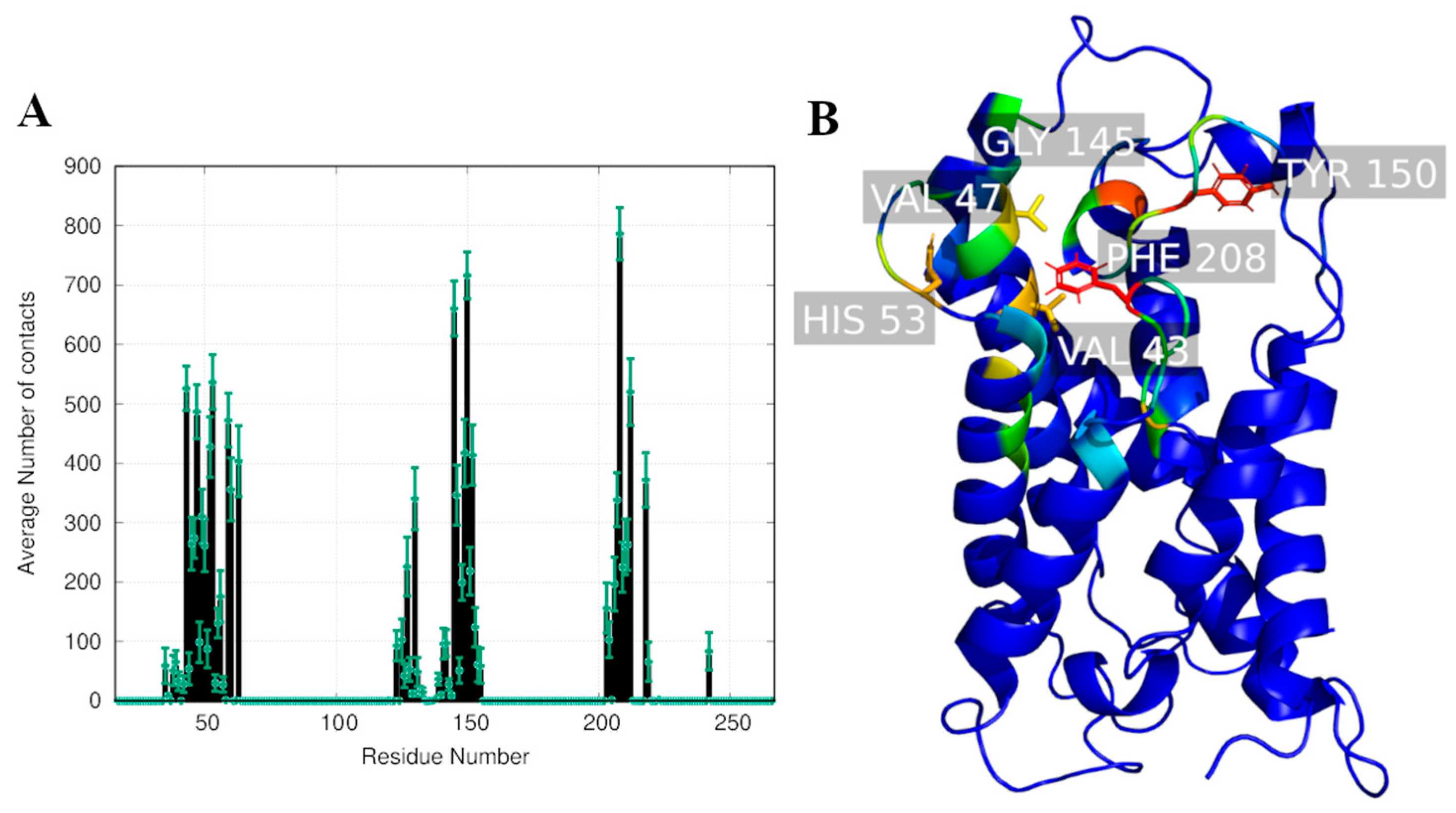
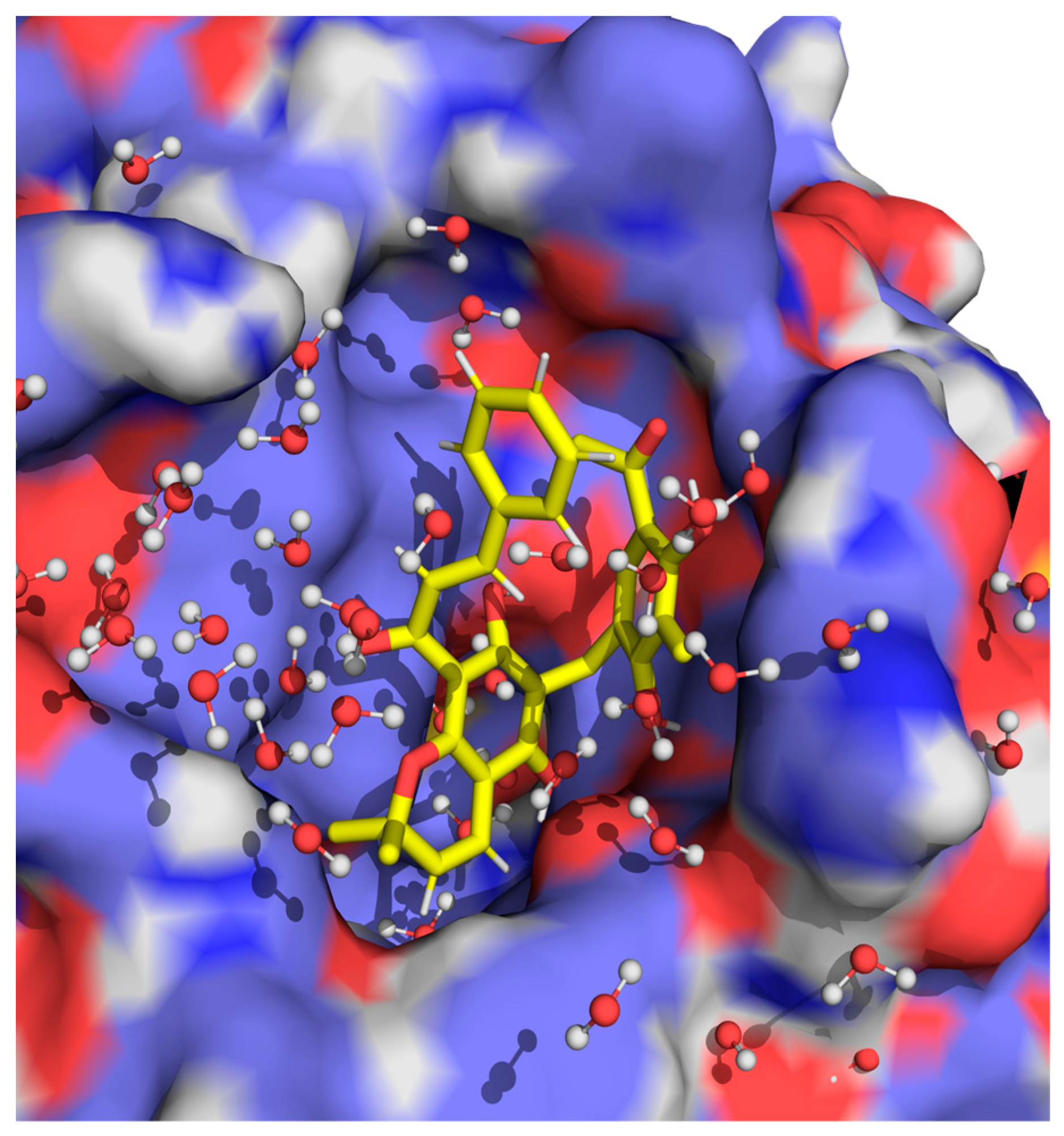
2.2. Computational Studies with AQP3
3. Discussion
4. Materials and Methods
4.1. Rottlerin
4.2. Ethics Statement
4.3. Red Blood Cells Sampling and Preparation
4.4. Yeast Strain and Growth Conditions
4.5. Permeability Assays
4.6. Statistical Analysis
4.7. Modeling the Tetrameric Structure of AQP3
4.8. Assemble the Simulation System and Sample the Conformational Space of AQP3
4.9. Molecular Docking
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Lounasmaa, M.; Widen, C.J.; Tuuf, C.M.; Huhtikangas, A. On the phloroglucinol derivatives of Mallotus philippinensis. Planta Med. 1975, 28, 16–31. [Google Scholar] [CrossRef] [PubMed]
- Ma, J.; Hou, Y.; Xia, J.; Zhu, X.; Wang, Z.P. Tumor suppressive role of rottlerin in cancer therapy. Am. J. Transl. Res. 2018, 10, 3345–3356. [Google Scholar] [PubMed]
- Gschwendt, M.; Muller, H.J.; Kielbassa, K.; Zang, R.; Kittstein, W.; Rincke, G.; Marks, F. Rottlerin, a novel protein kinase inhibitor. Biochem. Biophys. Res. Commun. 1994, 199, 93–98. [Google Scholar] [CrossRef]
- Maioli, E.; Daveri, E.; Maellaro, E.; Ietta, F.; Cresti, L.; Valacchi, G. Non-conventional rottlerin anticancer properties. Arch. Biochem. Biophys. 2018, 645, 50–53. [Google Scholar] [CrossRef]
- Torricelli, C.; Fortino, V.; Capurro, E.; Valacchi, G.; Pacini, A.; Muscettola, M.; Soucek, K.; Maioli, E. Rottlerin inhibits the nuclear factor kappaB/cyclin-D1 cascade in MCF-7 breast cancer cells. Life Sci. 2008, 82, 638–643. [Google Scholar] [CrossRef] [PubMed]
- Valacchi, G.; Pecorelli, A.; Sticozzi, C.; Torricelli, C.; Muscettola, M.; Aldinucci, C.; Maioli, E. Rottlerin exhibits antiangiogenic effects in vitro. Chem. Biol. Drug Des. 2011, 77, 460–470. [Google Scholar] [CrossRef] [PubMed]
- Ishii, R.; Horie, M.; Saito, K.; Arisawa, M.; Kitanaka, S. Prostaglandin E(2) production and induction of prostaglandin endoperoxide synthase-2 is inhibited in a murine macrophage-like cell line, RAW 264.7, by Mallotus japonicus phloroglucinol derivatives. Biochim. Biophys. Acta 2002, 1571, 115–123. [Google Scholar] [CrossRef] [PubMed]
- Daikonya, A.; Katsuki, S.; Wu, J.B.; Kitanaka, S. Anti-allergic agents from natural sources (4): Anti-allergic activity of new phloroglucinol derivatives from Mallotus philippensis (Euphorbiaceae). Chem. Pharm. Bull. 2002, 50, 1566–1569. [Google Scholar] [CrossRef] [Green Version]
- Shivshankar, P.; Lei, L.; Wang, J.; Zhong, G. Rottlerin inhibits chlamydial intracellular growth and blocks chlamydial acquisition of sphingolipids from host cells. Appl. Environ. Microbiol. 2008, 74, 1243–1249. [Google Scholar] [CrossRef] [Green Version]
- Kulkarni, R.R.; Tupe, S.G.; Gample, S.P.; Chandgude, M.G.; Sarkar, D.; Deshpande, M.V.; Joshi, S.P. Antifungal dimeric chalcone derivative kamalachalcone E from Mallotus philippinensis. Nat. Prod. Res. 2014, 28, 245–250. [Google Scholar] [CrossRef]
- Ietta, F.; Maioli, E.; Daveri, E.; Gonzaga Oliveira, J.; da Silva, R.J.; Romagnoli, R.; Cresti, L.; Maria Avanzati, A.; Paulesu, L.; Barbosa, B.F.; et al. Rottlerin-mediated inhibition of Toxoplasma gondii growth in BeWo trophoblast-like cells. Sci. Rep. 2017, 7, 1279. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Maioli, E.; Greci, L.; Soucek, K.; Hyzdalova, M.; Pecorelli, A.; Fortino, V.; Valacchi, G. Rottlerin inhibits ROS formation and prevents NFkappaB activation in MCF-7 and HT-29 cells. J. Biomed. Biotechnol. 2009, 2009, 742936. [Google Scholar] [CrossRef] [Green Version]
- Zhang, D.; Anantharam, V.; Kanthasamy, A.; Kanthasamy, A.G. Neuroprotective effect of protein kinase C delta inhibitor rottlerin in cell culture and animal models of Parkinson’s disease. J. Pharmacol. Exp. Ther. 2007, 322, 913–922. [Google Scholar] [CrossRef]
- Ohno, I.; Eibl, G.; Odinokova, I.; Edderkaoui, M.; Damoiseaux, R.D.; Yazbec, M.; Abrol, R.; Goddard, W.A., 3rd; Yokosuka, O.; Pandol, S.J.; et al. Rottlerin stimulates apoptosis in pancreatic cancer cells through interactions with proteins of the Bcl-2 family. Am. J. Physiol. Gastrointest. Liver Physiol. 2010, 298, G63–G73. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Daveri, E.; Maellaro, E.; Valacchi, G.; Ietta, F.; Muscettola, M.; Maioli, E. Inhibitions of mTORC1 and 4EBP-1 are key events orchestrated by Rottlerin in SK-Mel-28 cell killing. Cancer Lett. 2016, 380, 106–113. [Google Scholar] [CrossRef]
- Manhas, D.; Gour, A.; Bhardwaj, N.; Sharma, D.K.; Sharma, K.; Vij, B.; Jain, S.K.; Singh, G.; Nandi, U. Pharmacokinetic Assessment of Rottlerin from Mallotus philippensis Using a Highly Sensitive Liquid Chromatography-Tandem Mass Spectrometry-Based Bioanalytical Method. ACS Omega 2021, 6, 32637–32646. [Google Scholar] [CrossRef] [PubMed]
- Papadopoulos, M.C.; Saadoun, S. Key roles of aquaporins in tumor biology. Biochim. Biophys. Acta 2015, 1848, 2576–2583. [Google Scholar] [CrossRef] [Green Version]
- Carbrey, J.M.; Agre, P. Discovery of the aquaporins and development of the field. In Handbook of Experimental Pharmacology; Springer: Berlin/Heidelberg, Germany, 2009; pp. 3–28. [Google Scholar] [CrossRef]
- Soveral, G.; Casini, A. Aquaporin modulators: A patent review (2010–2015). Expert Opin. Ther. Pat. 2017, 27, 49–62. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ishibashi, K. New members of mammalian aquaporins: AQP10-AQP12. In Handbook of Experimental Pharmacology; Springer: Berlin/Heidelberg, Germany, 2009; pp. 251–262. [Google Scholar] [CrossRef]
- Brown, D. The Discovery of Water Channels (Aquaporins). Ann. Nutr. Metab. 2017, 70 (Suppl. S1), 37–42. [Google Scholar] [CrossRef] [PubMed]
- Ishibashi, K.; Tanaka, Y.; Morishita, Y. The role of mammalian superaquaporins inside the cell. Biochim. Biophys. Acta 2014, 1840, 1507–1512. [Google Scholar] [CrossRef] [PubMed]
- Madeira, A.; Fernandez-Veledo, S.; Camps, M.; Zorzano, A.; Moura, T.F.; Ceperuelo-Mallafre, V.; Vendrell, J.; Soveral, G. Human aquaporin-11 is a water and glycerol channel and localizes in the vicinity of lipid droplets in human adipocytes. Obesity 2014, 22, 2010–2017. [Google Scholar] [CrossRef]
- Prata, C.; Hrelia, S.; Fiorentini, D. Peroxiporins in Cancer. Int. J. Mol. Sci. 2019, 20, 1371. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- De Ieso, M.L.; Yool, A.J. Mechanisms of Aquaporin-Facilitated Cancer Invasion and Metastasis. Front. Chem. 2018, 6, 135. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Smith, I.M.; Banerjee, S.; Moses, A.K.; Stroka, K.M. Aquaporin-mediated dysregulation of cell migration in disease states. Cell Mol. Life Sci. 2023, 80, 48. [Google Scholar] [CrossRef] [PubMed]
- Verkman, A.S.; Anderson, M.O.; Papadopoulos, M.C. Aquaporins: Important but elusive drug targets. Nat. Rev. Drug Discov. 2014, 13, 259–277. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tomita, Y.; Dorward, H.; Yool, A.J.; Smith, E.; Townsend, A.R.; Price, T.J.; Hardingham, J.E. Role of Aquaporin 1 Signalling in Cancer Development and Progression. Int. J. Mol. Sci. 2017, 18, 299. [Google Scholar] [CrossRef] [Green Version]
- Marlar, S.; Jensen, H.H.; Login, F.H.; Nejsum, L.N. Aquaporin-3 in Cancer. Int. J. Mol. Sci. 2017, 18, 2106. [Google Scholar] [CrossRef] [Green Version]
- Rodrigues, C.; Pimpao, C.; Mosca, A.E.; Coxixo, A.S.; Lopes, D.; da Silva, I.V.; Pedersen, P.A.; Antunes, F.; Soveral, G. Human Aquaporin-5 Facilitates Hydrogen Peroxide Permeation Affecting Adaption to Oxidative Stress and Cancer Cell Migration. Cancers 2019, 11, 932. [Google Scholar] [CrossRef] [Green Version]
- Satooka, H.; Hara-Chikuma, M. Aquaporin-3 Controls Breast Cancer Cell Migration by Regulating Hydrogen Peroxide Transport and Its Downstream Cell Signaling. Mol. Cell Biol. 2016, 36, 1206–1218. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- de Almeida, A.; Soveral, G.; Casini, A. Gold compounds as aquaporin inhibitors: New opportunities for therapy and imaging. Med. Chem. Commun. 2014, 5, 1444–1453. [Google Scholar] [CrossRef]
- Pimpao, C.; da Silva, I.V.; Mosca, A.F.; Pinho, J.O.; Gaspar, M.M.; Gumerova, N.I.; Rompel, A.; Aureliano, M.; Soveral, G. The Aquaporin-3-Inhibiting Potential of Polyoxotungstates. Int. J. Mol. Sci. 2020, 21, 2467. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pimpao, C.; Wragg, D.; da Silva, I.V.; Casini, A.; Soveral, G. Aquaglyceroporin Modulators as Emergent Pharmacological Molecules for Human Diseases. Front. Mol. Biosci. 2022, 9, 845237. [Google Scholar] [CrossRef]
- Tesse, A.; Grossini, E.; Tamma, G.; Brenner, C.; Portincasa, P.; Marinelli, R.A.; Calamita, G. Aquaporins as Targets of Dietary Bioactive Phytocompounds. Front. Mol. Biosci. 2018, 5, 30. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Portincasa, P.; Calamita, G. Phytocompounds modulating Aquaporins: Clinical benefits are anticipated. Food Chem. 2019, 274, 642–650. [Google Scholar] [CrossRef]
- Preston, G.M.; Carroll, T.P.; Guggino, W.B.; Agre, P. Appearance of water channels in Xenopus oocytes expressing red cell CHIP28 protein. Science 1992, 256, 385–387. [Google Scholar] [CrossRef] [Green Version]
- Roudier, N.; Verbavatz, J.M.; Maurel, C.; Ripoche, P.; Tacnet, F. Evidence for the presence of aquaporin-3 in human red blood cells. J. Biol. Chem. 1998, 273, 8407–8412. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Madeira, A.; Moura, T.F.; Soveral, G. Detecting Aquaporin Function and Regulation. Front. Chem. 2016, 4, 3. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Campos, E.; Moura, T.F.; Oliva, A.; Leandro, P.; Soveral, G. Lack of Aquaporin 3 in bovine erythrocyte membranes correlates with low glycerol permeation. Biochem. Biophys. Res. Commun. 2011, 408, 477–481. [Google Scholar] [CrossRef] [Green Version]
- Soveral, G.; Madeira, A.; Loureiro-Dias, M.C.; Moura, T.F. Water transport in intact yeast cells as assessed by fluorescence self-quenching. Appl. Environ. Microbiol. 2007, 73, 2341–2343. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Berman, H.M. The Protein Data Bank. Nucleic Acids Res. 2000, 28, 235–242. [Google Scholar] [CrossRef] [Green Version]
- Kandt, C.; Ash, W.L.; Peter Tieleman, D. Setting up and running molecular dynamics simulations of membrane proteins. Methods 2007, 41, 475–488. [Google Scholar] [CrossRef] [PubMed]
- Kučerka, N.; Tristram-Nagle, S.; Nagle, J.F. Structure of Fully Hydrated Fluid Phase Lipid Bilayers with Monounsaturated Chains. J. Membr. Biol. 2006, 208, 193–202. [Google Scholar] [CrossRef]
- Torricelli, C.; Salvadori, S.; Valacchi, G.; Soucek, K.; Slabakova, E.; Muscettola, M.; Volpi, N.; Maioli, E. Alternative Pathways of Cancer Cell Death by Rottlerin: Apoptosis versus Autophagy. Evid. Based Complement. Altern. Med. 2012, 2012, 980658. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Maioli, E.; Torricelli, C.; Valacchi, G. Rottlerin and cancer: Novel evidence and mechanisms. Sci. World J. 2012, 2012, 350826. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, J.; Wang, T.; Zhou, Y.C.; Gao, F.; Zhang, Z.H.; Xu, H.; Wang, S.L.; Shen, L.Z. Aquaporin 3 promotes epithelial-mesenchymal transition in gastric cancer. J. Exp. Clin. Cancer Res. 2014, 33, 38. [Google Scholar] [CrossRef] [Green Version]
- Silva, P.M.; da Silva, I.V.; Sarmento, M.J.; Silva, I.C.; Carvalho, F.A.; Soveral, G.; Santos, N.C. Aquaporin-3 and Aquaporin-5 Facilitate Migration and Cell-Cell Adhesion in Pancreatic Cancer by Modulating Cell Biomechanical Properties. Cells 2022, 11, 1308. [Google Scholar] [CrossRef]
- Soveral, G.; Nielsen, S.; Casini, A. Aquaporins in Health and Disease: New Molecular Targets for Drug Discovery; Soveral, G., Nielsen, S., Casini, A., Eds.; CRC Press, Taylor & Francis Group: Boca Raton, FL, USA, 2016. [Google Scholar]
- Madeira, A.; Camps, M.; Zorzano, A.; Moura, T.F.; Soveral, G. Biophysical assessment of human aquaporin-7 as a water and glycerol channel in 3T3-L1 adipocytes. PLoS ONE 2013, 8, e83442. [Google Scholar] [CrossRef] [PubMed]
- Guldener, U.; Heck, S.; Fielder, T.; Beinhauer, J.; Hegemann, J.H. A new efficient gene disruption cassette for repeated use in budding yeast. Nucleic Acids Res. 1996, 24, 2519–2524. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mosca, A.F.; de Almeida, A.; Wragg, D.; Martins, A.P.; Sabir, F.; Leoni, S.; Moura, T.F.; Prista, C.; Casini, A.; Soveral, G. Molecular Basis of Aquaporin-7 Permeability Regulation by pH. Cells 2018, 7, 207. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pronk, J.T. Auxotrophic yeast strains in fundamental and applied research. Appl. Environ. Microbiol. 2002, 68, 2095–2100. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Martins, A.P.; Marrone, A.; Ciancetta, A.; Galan Cobo, A.; Echevarria, M.; Moura, T.F.; Re, N.; Casini, A.; Soveral, G. Targeting aquaporin function: Potent inhibition of aquaglyceroporin-3 by a gold-based compound. PLoS ONE 2012, 7, e37435. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Altschul, S.F.; Gish, W.; Miller, W.; Myers, E.W.; Lipman, D.J. Basic local alignment search tool. J. Mol. Biol. 1990, 215, 403–410. [Google Scholar] [CrossRef] [PubMed]
- Gotfryd, K.; Mosca, A.F.; Missel, J.W.; Truelsen, S.F.; Wang, K.; Spulber, M.; Krabbe, S.; Helix-Nielsen, C.; Laforenza, U.; Soveral, G.; et al. Human adipose glycerol flux is regulated by a pH gate in AQP10. Nat. Commun. 2018, 9, 4749. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sali, A.; Blundell, T.L. Comparative protein modelling by satisfaction of spatial restraints. J. Mol. Biol. 1993, 234, 779–815. [Google Scholar] [CrossRef]
- Laskowski, R.A.; MacArthur, M.W.; Moss, D.S.; Thornton, J.M. PROCHECK: A program to check the stereochemical quality of protein structures. J. Appl. Crystallogr. 1993, 26, 283–291. [Google Scholar] [CrossRef]
- Schmidt, T.H.; Kandt, C. LAMBADA and InflateGRO2: Efficient membrane alignment and insertion of membrane proteins for molecular dynamics simulations. J. Chem. Inf. Model. 2012, 52, 2657–2669. [Google Scholar] [CrossRef]
- Magalhaes, P.R.; Reis, P.; Vila-Vicosa, D.; Machuqueiro, M.; Victor, B.L. Optimization of an in Silico Protocol Using Probe Permeabilities to Identify Membrane Pan-Assay Interference Compounds. J. Chem. Inf. Model. 2022, 62, 3034–3042. [Google Scholar] [CrossRef]
- Hermans, J.; Berendsen, H.J.C.; Vangunsteren, W.F.; Postma, J.P.M. A Consistent Empirical Potential for Water-Protein Interactions. Biopolymers 1984, 23, 1513–1518. [Google Scholar] [CrossRef]
- Reis, P.; Vila-Vicosa, D.; Rocchia, W.; Machuqueiro, M. PypKa: A Flexible Python Module for Poisson-Boltzmann-Based pK(a) Calculations. J. Chem. Inf. Model. 2020, 60, 4442–4448. [Google Scholar] [CrossRef]
- Pronk, S.; Pall, S.; Schulz, R.; Larsson, P.; Bjelkmar, P.; Apostolov, R.; Shirts, M.R.; Smith, J.C.; Kasson, P.M.; van der Spoel, D.; et al. GROMACS 4.5: A high-throughput and highly parallel open source molecular simulation toolkit. Bioinformatics 2013, 29, 845–854. [Google Scholar] [CrossRef] [Green Version]
- Scott, W.R.P.; Hunenberger, P.H.; Tironi, I.G.; Mark, A.E.; Billeter, S.R.; Fennen, J.; Torda, A.E.; Huber, T.; Kruger, P.; van Gunsteren, W.F. The GROMOS biomolecular simulation program package. J. Phys. Chem. A 1999, 103, 3596–3607. [Google Scholar] [CrossRef]
- Eichenberger, A.P.; Allison, J.R.; Dolenc, J.; Geerke, D.P.; Horta, B.A.C.; Meier, K.; Oostenbrink, C.; Schmid, N.; Steiner, D.; Wang, D.Q.; et al. GROMOS plus plus Software for the Analysis of Biomolecular Simulation Trajectories. J. Chem. Theory Comput. 2011, 7, 3379–3390. [Google Scholar] [CrossRef] [PubMed]
- Parrinello, M.; Rahman, A. Polymorphic transitions in single crystals: A new molecular dynamics method. J. Appl. Phys. 1981, 52, 7182–7190. [Google Scholar] [CrossRef]
- Nosé, S.; Klein, M.L. Constant pressure molecular dynamics for molecular systems. Mol. Phys. 2006, 50, 1055–1076. [Google Scholar] [CrossRef]
- Bussi, G.; Donadio, D.; Parrinello, M. Canonical sampling through velocity rescaling. J. Chem. Phys. 2007, 126, 14101. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kolafa, J.; Perram, J.W. Cutoff Errors in the Ewald Summation Formulae for Point Charge Systems. Mol. Simul. 1992, 9, 351–368. [Google Scholar] [CrossRef]
- Hess, B. P-LINCS: A Parallel Linear Constraint Solver for Molecular Simulation. J. Chem. Theory Comput. 2007, 4, 116–122. [Google Scholar] [CrossRef] [PubMed]
- Miyamoto, S.; Kollman, P.A. Settle: An analytical version of the SHAKE and RATTLE algorithm for rigid water models. J. Comput. Chem. 1992, 13, 952–962. [Google Scholar] [CrossRef]
- Wardi, Y. A Stochastic Steepest-Descent Algorithm. J. Optim. Theory App. 1988, 59, 307–323. [Google Scholar] [CrossRef]
- Malde, A.K.; Zuo, L.; Breeze, M.; Stroet, M.; Poger, D.; Nair, P.C.; Oostenbrink, C.; Mark, A.E. An Automated Force Field Topology Builder (ATB) and Repository: Version 1.0. J. Chem. Theory Comput. 2011, 7, 4026–4037. [Google Scholar] [CrossRef] [PubMed]
- Koziara, K.B.; Stroet, M.; Malde, A.K.; Mark, A.E. Testing and validation of the Automated Topology Builder (ATB) version 2.0: Prediction of hydration free enthalpies. J. Comput. Aid. Mol. Des. 2014, 28, 221–233. [Google Scholar] [CrossRef] [PubMed]
- Gaspar, H.; Bronze, S.; Oliveira, C.; Victor, B.L.; Machuqueiro, M.; Pacheco, R.; Caldeira, M.J.; Santos, S. Proactive response to tackle the threat of emerging drugs: Synthesis and toxicity evaluation of new cathinones. Forensic. Sci. Int. 2018, 290, 146–156. [Google Scholar] [CrossRef] [PubMed]
- Fetrow, J.S.; Ravindranath, P.A.; Forli, S.; Goodsell, D.S.; Olson, A.J.; Sanner, M.F. AutoDockFR: Advances in Protein-Ligand Docking with Explicitly Specified Binding Site Flexibility. PLOS Comput. Biol. 2015, 11, e1004586. [Google Scholar] [CrossRef] [Green Version]
- Morris, G.M.; Huey, R.; Lindstrom, W.; Sanner, M.F.; Belew, R.K.; Goodsell, D.S.; Olson, A.J. AutoDock4 and AutoDockTools4: Automated docking with selective receptor flexibility. J. Comput. Chem. 2009, 30, 2785–2791. [Google Scholar] [CrossRef] [PubMed] [Green Version]


Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Paccetti-Alves, I.; Batista, M.S.P.; Pimpão, C.; Victor, B.L.; Soveral, G. Unraveling the Aquaporin-3 Inhibitory Effect of Rottlerin by Experimental and Computational Approaches. Int. J. Mol. Sci. 2023, 24, 6004. https://doi.org/10.3390/ijms24066004
Paccetti-Alves I, Batista MSP, Pimpão C, Victor BL, Soveral G. Unraveling the Aquaporin-3 Inhibitory Effect of Rottlerin by Experimental and Computational Approaches. International Journal of Molecular Sciences. 2023; 24(6):6004. https://doi.org/10.3390/ijms24066004
Chicago/Turabian StylePaccetti-Alves, Inês, Marta S. P. Batista, Catarina Pimpão, Bruno L. Victor, and Graça Soveral. 2023. "Unraveling the Aquaporin-3 Inhibitory Effect of Rottlerin by Experimental and Computational Approaches" International Journal of Molecular Sciences 24, no. 6: 6004. https://doi.org/10.3390/ijms24066004
APA StylePaccetti-Alves, I., Batista, M. S. P., Pimpão, C., Victor, B. L., & Soveral, G. (2023). Unraveling the Aquaporin-3 Inhibitory Effect of Rottlerin by Experimental and Computational Approaches. International Journal of Molecular Sciences, 24(6), 6004. https://doi.org/10.3390/ijms24066004







