Single-Cell Sequencing-Based Validation of T Cell-Associated Diagnostic Model Genes and Drug Response in Crohn’s Disease
Abstract
1. Introduction
2. Results
2.1. Single-Cell Clustering and Cell Annotation
2.2. Cell Communication Analysis Related to T Cells in scRNA-Seq
2.3. T-Cell Enrichment Analysis in scRNA-Seq
2.4. Identification of T Cell-Associated Differentially Expressed Genes in CD
2.4.1. Differentially Expressed Gene Analysis of CD and Normal Samples
2.4.2. WGCNA Analysis between CD and Normal Samples
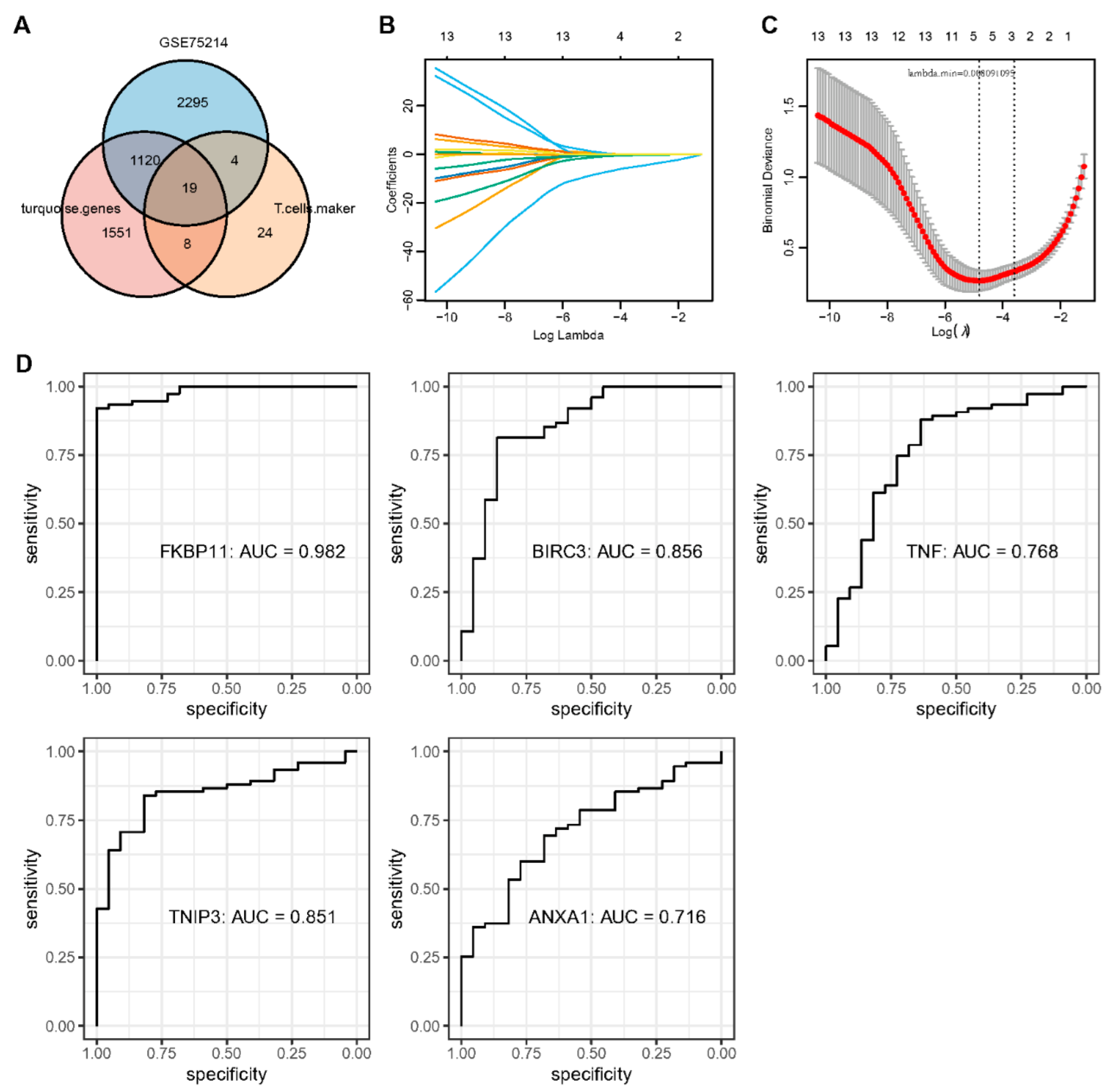
2.5. Identification of Key Genes and Analysis of Protein Interaction Networks
2.6. Correlation of Key Genes with Immune Cell Infiltration and Pathways
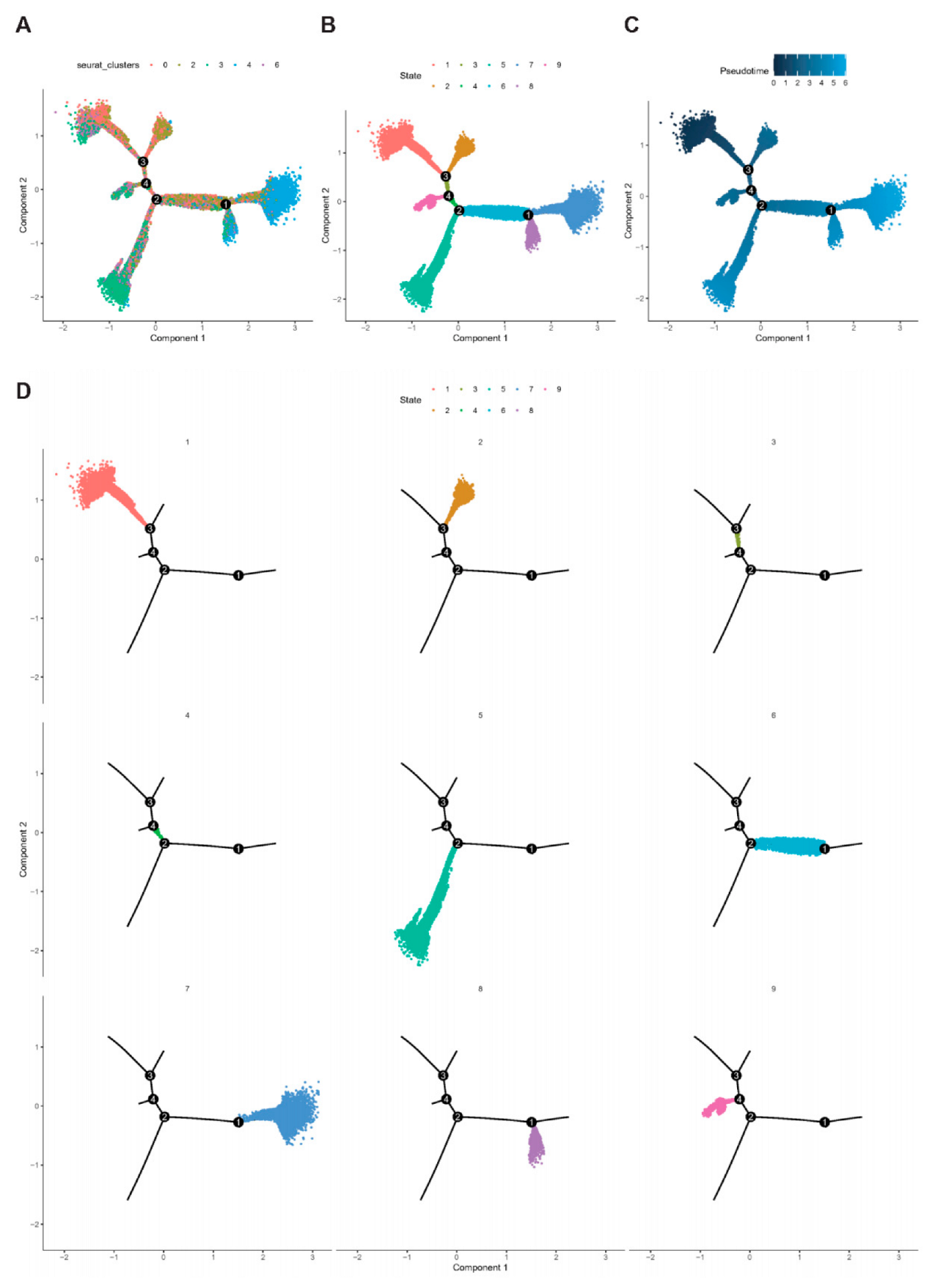
2.7. Trajectory of T Cell Maturation in CD
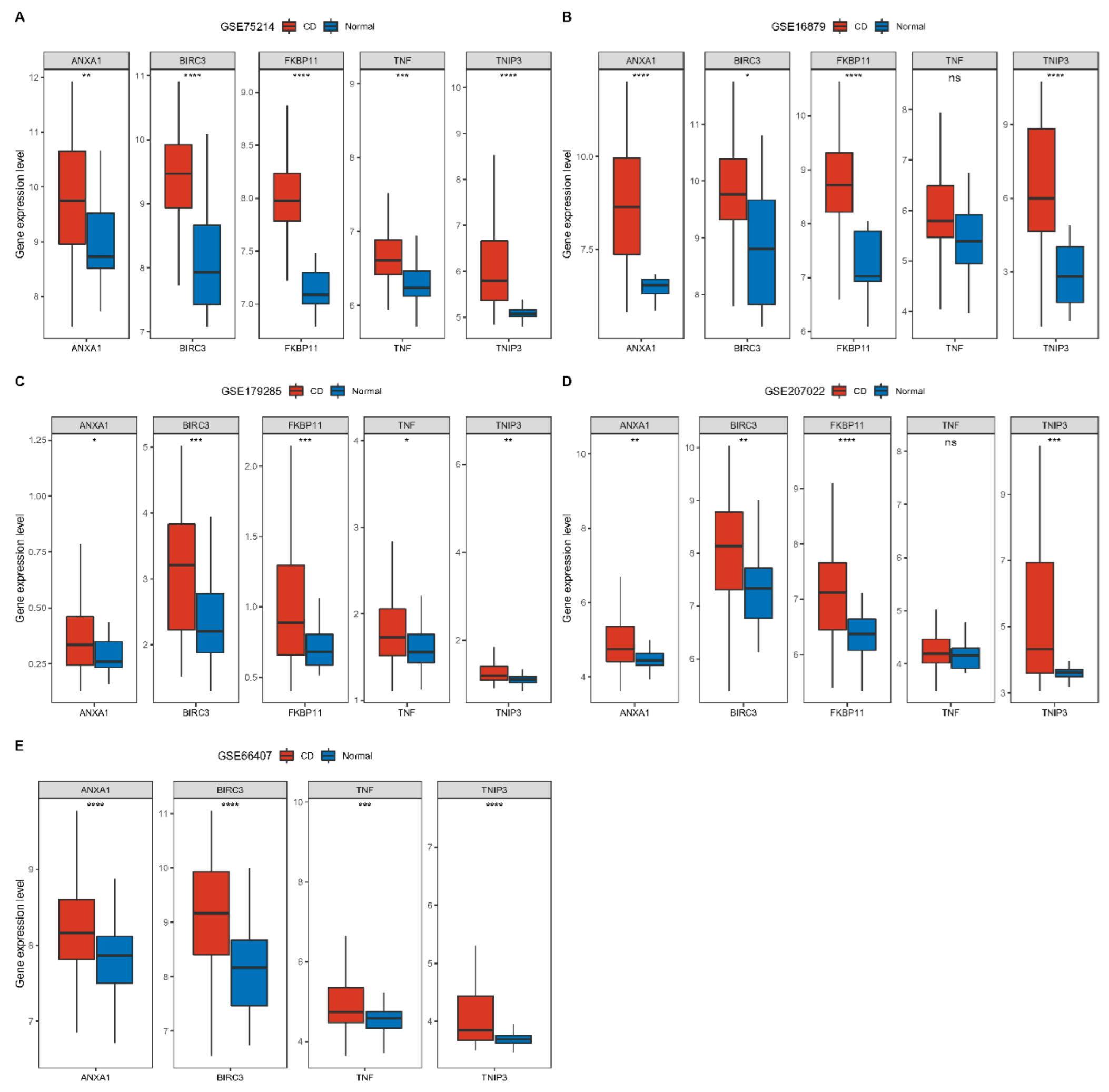
2.8. Construction and Validation of CD Diagnostic Models
2.9. Molecular Docking of Key Genes to Small Molecule Drugs
2.10. Gene Susceptibility Analysis to Drugs
2.11. Association of Key Genes with the Prognosis of Colon Cancer
3. Discussion
4. Materials and Methods
4.1. Data Sources and Preprocessing
4.1.1. Single-Cell Data Sources and Processing
4.1.2. Crohn’s Disease Expression Profile Data Sources and Processing
4.1.3. Colon Cancer Expression Profile Data Sources and Processing
4.2. Single-Cell Clustering and Cell Annotation Analysis
4.3. Cell Communication Analysis
4.4. Differential Gene Identification and Analysis
4.5. WGCNA and Functional Enrichment Analysis
4.6. Correlation of Key Genes with Immunity and Pathways
4.7. Establishment and Validation of Diagnostic Models
4.8. Real-Time Quantitative Polymerase Chain Reaction (RT-qPCR)
4.9. Molecular Docking
4.10. Association of Key Genes with the Prognosis of Colon Cancer
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Leong, R.W.; Lau, J.Y.; Sung, J.J. The epidemiology and phenotype of Crohn’s disease in the Chinese population. Inflamm. Bowel Dis. 2004, 10, 646–651. [Google Scholar] [CrossRef]
- Lichtenstein, G.R.; Shahabi, A.; Seabury, S.A.; Lakdawalla, D.N.; Espinosa, O.D.; Green, S.; Brauer, M.; Baldassano, R.N. Lifetime Economic Burden of Crohn’s Disease and Ulcerative Colitis by Age at Diagnosis. Clin. Gastroenterol. Hepatol. 2020, 18, 889–897.e810. [Google Scholar] [CrossRef] [PubMed]
- Meyer, A.; Dong, C.; Casagrande, C.; Chan, S.S.M.; Huybrechts, I.; Nicolas, G.; Rauber, F.; Levy, R.B.; Millett, C.; Oldenburg, B.; et al. Food Processing and Risk of Crohn’s Disease and Ulcerative Colitis: A European Prospective Cohort Study. Clin. Gastroenterol. Hepatol. 2022. [Google Scholar] [CrossRef] [PubMed]
- Hornschuh, M.; Wirthgen, E.; Wolfien, M.; Singh, K.P.; Wolkenhauer, O.; Dabritz, J. The role of epigenetic modifications for the pathogenesis of Crohn’s disease. Clin. Epigenetics 2021, 13, 108. [Google Scholar] [CrossRef] [PubMed]
- Suau, R.; Pardina, E.; Domenech, E.; Loren, V.; Manye, J. The Complex Relationship Between Microbiota, Immune Response and Creeping Fat in Crohn’s Disease. J. Crohns Colitis 2022, 16, 472–489. [Google Scholar] [CrossRef] [PubMed]
- Christophersen, A.; Zuhlke, S.; Lund, E.G.; Snir, O.; Dahal-Koirala, S.; Risnes, L.F.; Jahnsen, J.; Lundin, K.E.A.; Sollid, L.M. Pathogenic T Cells in Celiac Disease Change Phenotype on Gluten Challenge: Implications for T-Cell-Directed Therapies. Adv. Sci. 2022, 9, e2205912. [Google Scholar] [CrossRef]
- Rosati, E.; Rios Martini, G.; Pogorelyy, M.V.; Minervina, A.A.; Degenhardt, F.; Wendorff, M.; Sari, S.; Mayr, G.; Fazio, A.; Dowds, C.M.; et al. A novel unconventional T cell population enriched in Crohn’s disease. Gut 2022, 71, 2194–2204. [Google Scholar] [CrossRef]
- Jaeger, N.; Gamini, R.; Cella, M.; Schettini, J.L.; Bugatti, M.; Zhao, S.; Rosadini, C.V.; Esaulova, E.; Di Luccia, B.; Kinnett, B.; et al. Single-cell analyses of Crohn’s disease tissues reveal intestinal intraepithelial T cells heterogeneity and altered subset distributions. Nat. Commun. 2021, 12, 1921. [Google Scholar] [CrossRef]
- Mitsialis, V.; Wall, S.; Liu, P.; Ordovas-Montanes, J.; Parmet, T.; Vukovic, M.; Spencer, D.; Field, M.; McCourt, C.; Toothaker, J.; et al. Single-Cell Analyses of Colon and Blood Reveal Distinct Immune Cell Signatures of Ulcerative Colitis and Crohn’s Disease. Gastroenterology 2020, 159, 591–608.e510. [Google Scholar] [CrossRef]
- Becker, W.R.; Nevins, S.A.; Chen, D.C.; Chiu, R.; Horning, A.M.; Guha, T.K.; Laquindanum, R.; Mills, M.; Chaib, H.; Ladabaum, U.; et al. Single-cell analyses define a continuum of cell state and composition changes in the malignant transformation of polyps to colorectal cancer. Nat. Genet. 2022, 54, 985–995. [Google Scholar] [CrossRef]
- Wang, X.; Miao, J.; Wang, S.; Shen, R.; Zhang, S.; Tian, Y.; Li, M.; Zhu, D.; Yao, A.; Bao, W.; et al. Single-cell RNA-seq reveals the genesis and heterogeneity of tumor microenvironment in pancreatic undifferentiated carcinoma with osteoclast-like giant-cells. Mol. Cancer 2022, 21, 133. [Google Scholar] [CrossRef]
- Devlin, J.C.; Axelrad, J.; Hine, A.M.; Chang, S.; Sarkar, S.; Lin, J.D.; Ruggles, K.V.; Hudesman, D.; Cadwell, K.; Loke, P. Single-Cell Transcriptional Survey of Ileal-Anal Pouch Immune Cells From Ulcerative Colitis Patients. Gastroenterology 2021, 160, 1679–1693. [Google Scholar] [CrossRef] [PubMed]
- Khaliq, A.M.; Erdogan, C.; Kurt, Z.; Turgut, S.S.; Grunvald, M.W.; Rand, T.; Khare, S.; Borgia, J.A.; Hayden, D.M.; Pappas, S.G.; et al. Refining colorectal cancer classification and clinical stratification through a single-cell atlas. Genome Biol. 2022, 23, 113. [Google Scholar] [CrossRef]
- Fawkner-Corbett, D.; Antanaviciute, A.; Parikh, K.; Jagielowicz, M.; Gerós, A.S.; Gupta, T.; Ashley, N.; Khamis, D.; Fowler, D.; Morrissey, E.; et al. Spatiotemporal analysis of human intestinal development at single-cell resolution. Cell 2021, 184, 810–826.e823. [Google Scholar] [CrossRef] [PubMed]
- Razanskaite, V.; Kallis, C.; Young, B.; Williamson, P.R.; Bodger, K. Heterogeneity in outcome assessment for inflammatory bowel disease in routine clinical practice: A mixed-methods study in a sample of English hospitals. BMJ Open 2021, 11, e056413. [Google Scholar] [CrossRef]
- Cordero, R.Y.; Cordero, J.B.; Stiemke, A.B.; Datta, L.W.; Buyske, S.; Kugathasan, S.; McGovern, D.P.B.; Brant, S.R.; Simpson, C.L. Trans-ancestry, Bayesian Meta-analysis Discovers 20 Novel Risk Loci for Inflammatory Bowel Disease in an African American, East Asian, and European Cohort. Hum. Mol. Genet. 2022, 32, 873–882. [Google Scholar] [CrossRef]
- Anderson, C.A.; Massey, D.C.; Barrett, J.C.; Prescott, N.J.; Tremelling, M.; Fisher, S.A.; Gwilliam, R.; Jacob, J.; Nimmo, E.R.; Drummond, H.; et al. Investigation of Crohn’s disease risk loci in ulcerative colitis further defines their molecular relationship. Gastroenterology 2009, 136, 523–529.e523. [Google Scholar] [CrossRef]
- Andersen, V.; Pedersen, A.K.; Moller, S.; Green, A. Chronic Inflammatory Diseases—Diabetes Mellitus, Rheumatoid Arthritis, Coeliac Disease, Crohn’s Disease, and Ulcerative Colitis Among the Offspring of Affected Parents: A Danish Population-Based Registry Study. Clin. Epidemiol. 2021, 13, 13–20. [Google Scholar] [CrossRef]
- Askari, H.; Shojaei-Zarghani, S.; Raeis-Abdollahi, E.; Jahromi, H.K.; Abdullahi, P.R.; Daliri, K.; Tajbakhsh, A.; Rahmati, L.; Safarpour, A.R. The Role of Gut Microbiota in Inflammatory Bowel Disease-Current State of the Art. Mini Rev. Med. Chem. 2022. [Google Scholar] [CrossRef]
- Pedersen, T.K.; Brown, E.M.; Plichta, D.R.; Johansen, J.; Twardus, S.W.; Delorey, T.M.; Lau, H.; Vlamakis, H.; Moon, J.J.; Xavier, R.J.; et al. The CD4(+) T cell response to a commensal-derived epitope transitions from a tolerant to an inflammatory state in Crohn’s disease. Immunity 2022, 55, 1909–1923.e1906. [Google Scholar] [CrossRef] [PubMed]
- Duckworth, C.A. Understanding gene-environment interactions in a mouse model of Crohn’s disease. Dis. Model. Mech. 2011, 4, 7–8. [Google Scholar] [CrossRef] [PubMed]
- Djouina, M.; Waxin, C.; Lepretre, F.; Tardivel, M.; Tillement, O.; Vasseur, F.; Figeac, M.; Bongiovanni, A.; Sebda, S.; Desreumaux, P.; et al. Gene/environment interaction in the susceptibility of Crohn’s disease patients to aluminum. Sci. Total. Environ. 2022, 850, 158017. [Google Scholar] [CrossRef]
- Vadstrup, K.; Galsgaard, E.D.; Jensen, H.; Lanier, L.L.; Ryan, J.C.; Chen, S.Y.; Nolan, G.P.; Vester-Andersen, M.K.; Pedersen, J.S.; Gerwien, J.; et al. NKG2D ligand expression in Crohn’s disease and NKG2D-dependent stimulation of CD8(+) T cell migration. Exp. Mol. Pathol. 2017, 103, 56–70. [Google Scholar] [CrossRef]
- Brand, S.; Beigel, F.; Olszak, T.; Zitzmann, K.; Eichhorst, S.T.; Otte, J.M.; Diepolder, H.; Marquardt, A.; Jagla, W.; Popp, A.; et al. IL-22 is increased in active Crohn’s disease and promotes proinflammatory gene expression and intestinal epithelial cell migration. Am. J. Physiol. Gastrointest Liver Physiol. 2006, 290, G827–G838. [Google Scholar] [CrossRef] [PubMed]
- Brown, C.Y.; Sadlon, T.; Hope, C.M.; Wong, Y.Y.; Wong, S.; Liu, N.; Withers, H.; Brown, K.; Bandara, V.; Gundsambuu, B.; et al. Molecular Insights Into Regulatory T-Cell Adaptation to Self, Environment, and Host Tissues: Plasticity or Loss of Function in Autoimmune Disease. Front. Immunol. 2020, 11, 1269. [Google Scholar] [CrossRef]
- Papamichael, K.; Dubinsky, M.C.; Cheifetz, A.S. Proactive Therapeutic Drug Monitoring of Adalimumab in Patients With Crohn’s Disease. Gastroenterology 2023, 164, 164–165. [Google Scholar] [CrossRef] [PubMed]
- Reinisch, W.; Gecse, K.; Halfvarson, J.; Irving, P.M.; Jahnsen, J.; Peyrin-Biroulet, L.; Rogler, G.; Schreiber, S.; Danese, S. Clinical Practice of Adalimumab and Infliximab Biosimilar Treatment in Adult Patients With Crohn’s Disease. Inflamm. Bowel Dis. 2021, 27, 106–122. [Google Scholar] [CrossRef]
- Suzuki, Y.; Matsui, T.; Ito, H.; Ashida, T.; Nakamura, S.; Motoya, S.; Matsumoto, T.; Sato, N.; Ozaki, K.; Watanabe, M.; et al. Circulating Interleukin 6 and Albumin, and Infliximab Levels Are Good Predictors of Recovering Efficacy After Dose Escalation Infliximab Therapy in Patients with Loss of Response to Treatment for Crohn’s Disease: A Prospective Clinical Trial. Inflamm. Bowel Dis. 2015, 21, 2114–2122. [Google Scholar] [CrossRef]
- Ma, C.; Huang, V.; Fedorak, D.K.; Kroeker, K.I.; Dieleman, L.A.; Halloran, B.P.; Fedorak, R.N. Crohn’s disease outpatients treated with adalimumab have an earlier secondary loss of response and requirement for dose escalation compared to infliximab: A real life cohort study. J. Crohns Colitis 2014, 8, 1454–1463. [Google Scholar] [CrossRef]
- Peyrin-Biroulet, L.; Laclotte, C.; Bigard, M.A. Adalimumab maintenance therapy for Crohn’s disease with intolerance or lost response to infliximab: An open-label study. Aliment. Pharmacol. Ther. 2007, 25, 675–680. [Google Scholar] [CrossRef]
- Gagniere, C.; Beaugerie, L.; Pariente, B.; Seksik, P.; Amiot, A.; Abitbol, V.; Allez, M.; Cosnes, J.; Sokol, H. Benefit of infliximab reintroduction after successive failure of infliximab and adalimumab in Crohn’s disease. J. Crohns Colitis 2015, 9, 349–355. [Google Scholar] [CrossRef] [PubMed]
- R-Grau, M.D.C.; Chaparro, M.; Mesonero, F.; Acosta, M.B.-D.; Castro, L.; Castro, M.; Domènech, E.; Mancenido, N.; Pérez-Calle, J.L.; Taxonera, C.; et al. Effectiveness of anti-TNFα drugs in patients with Crohn’s disease who do not achieve remission with their first anti-TNFα agent. Dig. Liver Dis. 2016, 48, 613–619. [Google Scholar] [CrossRef]
- Singh, S.; Fumery, M.; Sandborn, W.J.; Murad, M.H. Systematic review and network meta-analysis: First- and second-line biologic therapies for moderate-severe Crohn’s disease. Aliment. Pharmacol. Ther. 2018, 48, 394–409. [Google Scholar] [CrossRef] [PubMed]
- Watanabe, K.; Matsumoto, T.; Hisamatsu, T.; Nakase, H.; Motoya, S.; Yoshimura, N.; Ishida, T.; Kato, S.; Nakagawa, T.; Esaki, M.; et al. Clinical and Pharmacokinetic Factors Associated With Adalimumab-Induced Mucosal Healing in Patients With Crohn’s Disease. Clin. Gastroenterol. Hepatol. 2018, 16, 542–549.e541. [Google Scholar] [CrossRef]
- Wiese, D.M.; Beaulieu, D.; Slaughter, J.C.; Horst, S.; Wagnon, J.; Duley, C.; Annis, K.; Nohl, A.; Herline, A.; Muldoon, R.; et al. Use of Endoscopic Ultrasound to Guide Adalimumab Treatment in Perianal Crohn’s Disease Results in Faster Fistula Healing. Inflamm. Bowel Dis. 2015, 21, 1594–1599. [Google Scholar] [CrossRef]
- Stournaras, E.; Qian, W.; Pappas, A.; Hong, Y.Y.; Shawky, R.; Investigators, U.I.B.; Raine, T.; Parkes, M.; Investigators, U.I.B. Thiopurine monotherapy is effective in ulcerative colitis but significantly less so in Crohn’s disease: Long-term outcomes for 11 928 patients in the UK inflammatory bowel disease bioresource. Gut 2021, 70, 677–686. [Google Scholar] [CrossRef]
- Lakatos, P.L.; Golovics, P.A.; David, G.; Pandur, T.; Erdelyi, Z.; Horvath, A.; Mester, G.; Balogh, M.; Szipocs, I.; Molnar, C.; et al. Has there been a change in the natural history of Crohn’s disease? Surgical rates and medical management in a population-based inception cohort from Western Hungary between 1977–2009. Am. J. Gastroenterol. 2012, 107, 579–588. [Google Scholar] [CrossRef]
- Coussens, L.M.; Werb, Z. Inflammation and cancer. Nature 2002, 420, 860–867. [Google Scholar] [CrossRef]
- Pflug, K.M.; Sitcheran, R. Targeting NF-kappaB-Inducing Kinase (NIK) in Immunity, Inflammation, and Cancer. Int. J. Mol. Sci. 2020, 21, 8470. [Google Scholar] [CrossRef] [PubMed]
- Kay, J.; Thadhani, E.; Samson, L.; Engelward, B. Inflammation-induced DNA damage, mutations and cancer. DNA Repair 2019, 83, 102673. [Google Scholar] [CrossRef]
- Padoan, A.; Plebani, M.; Basso, D. Inflammation and Pancreatic Cancer: Focus on Metabolism, Cytokines, and Immunity. Int. J. Mol. Sci. 2019, 20, 676. [Google Scholar] [CrossRef]
- Li, L.; Yu, R.; Cai, T.; Chen, Z.; Lan, M.; Zou, T.; Wang, B.; Wang, Q.; Zhao, Y.; Cai, Y. Effects of immune cells and cytokines on inflammation and immunosuppression in the tumor microenvironment. Int. Immunopharmacol. 2020, 88, 106939. [Google Scholar] [CrossRef]
- Xu, W.; Guo, Y.; Huang, Z.; Zhao, H.; Zhou, M.; Huang, Y.; Wen, D.; Song, J.; Zhu, Z.; Sun, M.; et al. Small heat shock protein CRYAB inhibits intestinal mucosal inflammatory responses and protects barrier integrity through suppressing IKKbeta activity. Mucosal Immunol. 2019, 12, 1291–1303. [Google Scholar] [CrossRef] [PubMed]
- Li, Q.; Wang, Y.; Lai, Y.; Xu, P.; Yang, Z. HspB5 correlates with poor prognosis in colorectal cancer and prompts epithelial-mesenchymal transition through ERK signaling. PLoS ONE 2017, 12, e0182588. [Google Scholar] [CrossRef]
- Martin, J.C.; Chang, C.; Boschetti, G.; Ungaro, R.; Giri, M.; Grout, J.A.; Gettler, K.; Chuang, L.S.; Nayar, S.; Greenstein, A.J.; et al. Single-Cell Analysis of Crohn’s Disease Lesions Identifies a Pathogenic Cellular Module Associated with Resistance to Anti-TNF Therapy. Cell 2019, 178, 1493–1508.e1420. [Google Scholar] [CrossRef]
- Hao, Y.; Hao, S.; Andersen-Nissen, E.; Mauck, W.M., 3rd; Zheng, S.; Butler, A.; Lee, M.J.; Wilk, A.J.; Darby, C.; Zager, M.; et al. Integrated analysis of multimodal single-cell data. Cell 2021, 184, 3573–3587.e3529. [Google Scholar] [CrossRef]
- Vancamelbeke, M.; Vanuytsel, T.; Farré, R.; Verstockt, S.; Ferrante, M.; Van Assche, G.; Rutgeerts, P.; Schuit, F.; Vermeire, S.; Arijs, I.; et al. Genetic and Transcriptomic Bases of Intestinal Epithelial Barrier Dysfunction in Inflammatory Bowel Disease. Inflamm. Bowel Dis. 2017, 23, 1718–1729. [Google Scholar] [CrossRef] [PubMed]
- Arijs, I.; De Hertogh, G.; Lemaire, K.; Quintens, R.; Van Lommel, L.; Van Steen, K.; Leemans, P.; Cleynen, I.; Van Assche, G.; Vermeire, S.; et al. Mucosal gene expression of antimicrobial peptides in inflammatory bowel disease before and after first infliximab treatment. PLoS ONE 2009, 4, e7984. [Google Scholar] [CrossRef] [PubMed]
- Keir, M.E.; Fuh, F.; Ichikawa, R.; Acres, M.; Hackney, J.A.; Hulme, G.; Carey, C.D.; Palmer, J.; Jones, C.J.; Long, A.K.; et al. Regulation and Role of αE Integrin and Gut Homing Integrins in Migration and Retention of Intestinal Lymphocytes during Inflammatory Bowel Disease. J. Immunol. 2021, 207, 2245–2254. [Google Scholar] [CrossRef] [PubMed]
- Smith, J.J.; Deane, N.G.; Wu, F.; Merchant, N.B.; Zhang, B.; Jiang, A.; Lu, P.; Johnson, J.C.; Schmidt, C.; Bailey, C.E.; et al. Experimentally derived metastasis gene expression profile predicts recurrence and death in patients with colon cancer. Gastroenterology 2010, 138, 958–968. [Google Scholar] [CrossRef]
- Marisa, L.; de Reyniès, A.; Duval, A.; Selves, J.; Gaub, M.P.; Vescovo, L.; Etienne-Grimaldi, M.C.; Schiappa, R.; Guenot, D.; Ayadi, M.; et al. Gene expression classification of colon cancer into molecular subtypes: Characterization, validation, and prognostic value. PLoS Med. 2013, 10, e1001453. [Google Scholar] [CrossRef] [PubMed]
- Gautier, L.; Cope, L.; Bolstad, B.M.; Irizarry, R.A. affy—Analysis of Affymetrix GeneChip data at the probe level. Bioinformatics 2004, 20, 307–315. [Google Scholar] [CrossRef]
- Jin, S.; Guerrero-Juarez, C.F.; Zhang, L.; Chang, I.; Ramos, R.; Kuan, C.H.; Myung, P.; Plikus, M.V.; Nie, Q. Inference and analysis of cell-cell communication using CellChat. Nat. Commun. 2021, 12, 1088. [Google Scholar] [CrossRef]
- Langfelder, P.; Horvath, S. WGCNA: An R package for weighted correlation network analysis. BMC Bioinform. 2008, 9, 559. [Google Scholar] [CrossRef]
- Wu, T.; Hu, E.; Xu, S.; Chen, M.; Guo, P.; Dai, Z.; Feng, T.; Zhou, L.; Tang, W.; Zhan, L.; et al. clusterProfiler 4.0: A universal enrichment tool for interpreting omics data. Innovation 2021, 2, 100141. [Google Scholar] [CrossRef] [PubMed]
- Yoshihara, K.; Shahmoradgoli, M.; Martínez, E.; Vegesna, R.; Kim, H.; Torres-Garcia, W.; Treviño, V.; Shen, H.; Laird, P.W.; Levine, D.A.; et al. Inferring tumour purity and stromal and immune cell admixture from expression data. Nat. Commun. 2013, 4, 2612. [Google Scholar] [CrossRef]
- Becht, E.; Giraldo, N.A.; Lacroix, L.; Buttard, B.; Elarouci, N.; Petitprez, F.; Selves, J.; Laurent-Puig, P.; Sautès-Fridman, C.; Fridman, W.H.; et al. Estimating the population abundance of tissue-infiltrating immune and stromal cell populations using gene expression. Genome Biol. 2016, 17, 218. [Google Scholar] [CrossRef]
- Hänzelmann, S.; Castelo, R.; Guinney, J. GSVA: Gene set variation analysis for microarray and RNA-seq data. BMC Bioinform. 2013, 14, 7. [Google Scholar] [CrossRef] [PubMed]
- Charoentong, P.; Finotello, F.; Angelova, M.; Mayer, C.; Efremova, M.; Rieder, D.; Hackl, H.; Trajanoski, Z. Pan-cancer Immunogenomic Analyses Reveal Genotype-Immunophenotype Relationships and Predictors of Response to Checkpoint Blockade. Cell Rep. 2017, 18, 248–262. [Google Scholar] [CrossRef] [PubMed]
- Rody, A.; Holtrich, U.; Pusztai, L.; Liedtke, C.; Gaetje, R.; Ruckhaeberle, E.; Solbach, C.; Hanker, L.; Ahr, A.; Metzler, D.; et al. T-cell metagene predicts a favorable prognosis in estrogen receptor-negative and HER2-positive breast cancers. Breast Cancer Res. BCR 2009, 11, R15. [Google Scholar] [CrossRef] [PubMed]
- Danilova, L.; Ho, W.J.; Zhu, Q.; Vithayathil, T.; De Jesus-Acosta, A.; Azad, N.S.; Laheru, D.A.; Fertig, E.J.; Anders, R.; Jaffee, E.M.; et al. Programmed Cell Death Ligand-1 (PD-L1) and CD8 Expression Profiling Identify an Immunologic Subtype of Pancreatic Ductal Adenocarcinomas with Favorable Survival. Cancer Immunol. Res. 2019, 7, 886–895. [Google Scholar] [CrossRef]
- Sui, X.; Lei, L.; Chen, L.; Xie, T.; Li, X. Inflammatory microenvironment in the initiation and progression of bladder cancer. Oncotarget 2017, 8, 93279–93294. [Google Scholar] [CrossRef] [PubMed]
- Xiao, Y.; Ma, D.; Zhao, S.; Suo, C.; Shi, J.; Xue, M.Z.; Ruan, M.; Wang, H.; Zhao, J.; Li, Q.; et al. Multi-Omics Profiling Reveals Distinct Microenvironment Characterization and Suggests Immune Escape Mechanisms of Triple-Negative Breast Cancer. Clin. Cancer Res. Off. J. Am. Assoc. Cancer Res. 2019, 25, 5002–5014. [Google Scholar] [CrossRef] [PubMed]
- Chen, T.; Guestrin, C. Xgboost: A scalable tree boosting system. In Proceedings of the 22nd Acm Sigkdd International Conference on Knowledge Discovery and Data Mining, San Francisco, CA, USA, 13–17 August 2016; pp. 785–794. [Google Scholar]
- Chen, T.; He, T.; Benesty, M.; Khotilovich, V.; Tang, Y.; Cho, H.; Chen, K. Xgboost: Extreme gradient boosting. R Package 2015, 1, 1–4. [Google Scholar]







Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Dai, Z.; Zhang, J.; Xu, W.; Du, P.; Wang, Z.; Liu, Y. Single-Cell Sequencing-Based Validation of T Cell-Associated Diagnostic Model Genes and Drug Response in Crohn’s Disease. Int. J. Mol. Sci. 2023, 24, 6054. https://doi.org/10.3390/ijms24076054
Dai Z, Zhang J, Xu W, Du P, Wang Z, Liu Y. Single-Cell Sequencing-Based Validation of T Cell-Associated Diagnostic Model Genes and Drug Response in Crohn’s Disease. International Journal of Molecular Sciences. 2023; 24(7):6054. https://doi.org/10.3390/ijms24076054
Chicago/Turabian StyleDai, Zhujiang, Jie Zhang, Weimin Xu, Peng Du, Zhongchuan Wang, and Yun Liu. 2023. "Single-Cell Sequencing-Based Validation of T Cell-Associated Diagnostic Model Genes and Drug Response in Crohn’s Disease" International Journal of Molecular Sciences 24, no. 7: 6054. https://doi.org/10.3390/ijms24076054
APA StyleDai, Z., Zhang, J., Xu, W., Du, P., Wang, Z., & Liu, Y. (2023). Single-Cell Sequencing-Based Validation of T Cell-Associated Diagnostic Model Genes and Drug Response in Crohn’s Disease. International Journal of Molecular Sciences, 24(7), 6054. https://doi.org/10.3390/ijms24076054





