Genome-Wide Identification of the MADS-Box Gene Family during Male and Female Flower Development in Chayote (Sechium edule)
Abstract
1. Introduction
2. Results
2.1. Identification and Physicochemical Properties Analysis of the MADS-Box Gene Family of Sechium edule
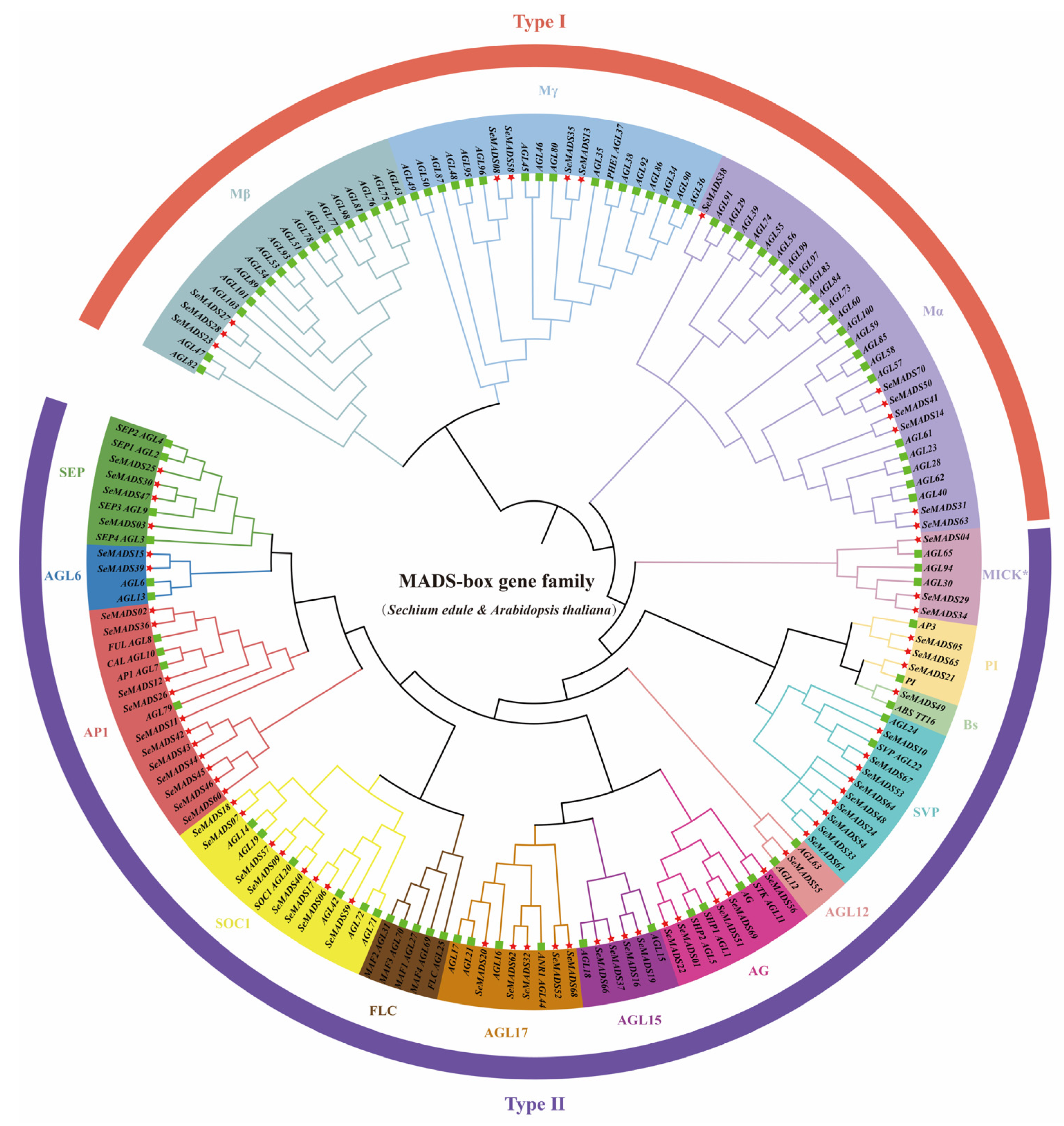
2.2. Phylogenetic Analysis of SeMADS-Box
2.3. Chromosomal Localization of SeMADS-BOX
2.4. Gene Structure and Conserved Motif Analysis of the SeMADS-Box
2.5. Promoter Region Analysis of SeMADS-Box
2.6. Collinearity Analyses of the SeMADS-Box within and between Species
2.7. GO and KEGG Analysis of SeMADS-Box
2.8. Protein-Protein Interaction Network Analysis of the SeMADS-Box
2.9. Expression Profiles of the SeMADS-Box
2.10. Expression Analysis of SeMADS-Box in Different Tissue and Stage
3. Discussion
4. Materials and Methods
4.1. Identification of MADS-Box Genes Family in Chayote
4.2. Construction of MADS-Box Phylogenetic Tree
4.3. MADS-Box Family Genes Chromosomal Localization
4.4. Gene Structure and Conserved Motif Analysis of MADS-Box
4.5. Cis-Element Analysis of MADS-Box
4.6. Collinearity Analyses of MADS-Box within and between Species
4.7. GO and KEGG Analysis of MADS-Box
4.8. MADS-Box Protein Interaction Network Analysis
4.9. MADS-Box Gene Expression Pattern
4.10. Plant Material, RNA Extraction and qRT-PCR Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Passmore, S.; Maine, G.T.; Elble, R.; Christ, C.; Tye, B.-K. Saccharomyces cerevisiae protein involved in plasmid maintenance is necessary for mating of MATα cells. J. Mol. Biol. 1988, 204, 593–606. [Google Scholar] [CrossRef]
- Yanofsky, M.F.; Ma, H.; Bowman, J.L.; Drews, G.N.; Feldmann, K.A.; Meyerowitz, E.M. The protein encoded by the Arabidopsis homeotic gene agamous resembles transcription factors. Nature 1990, 346, 35–39. [Google Scholar] [CrossRef]
- Sommer, H.; Beltrán, J.P.; Huijser, P.; Pape, H.; Lönnig, W.E.; Saedler, H.; Schwarz-Sommer, Z. Deficiens, a homeotic gene involved in the control of flower morphogenesis in Antirrhinum majus: The protein shows homology to transcription factors. EMBO J. 1990, 9, 605–613. [Google Scholar] [CrossRef]
- Norman, C.; Runswick, M.; Pollock, R.; Treisman, R. Isolation and properties of cDNA clones encoding SRF, a transcription factor that binds to the c-fos serum response element. Cell 1988, 55, 989–1003. [Google Scholar] [CrossRef]
- De Bodt, S.; Raes, J.; Van de Peer, Y.; Theißen, G. And then there were many: MADS goes genomic. Trends Plant Sci. 2003, 8, 475–483. [Google Scholar] [CrossRef]
- Alvarez-Buylla Elena, R.; Pelaz, S.; Liljegren Sarah, J.; Gold Scott, E.; Burgeff, C.; Ditta Gary, S.; Ribas de Pouplana, L.; Martínez-Castilla, L.; Yanofsky Martin, F. An ancestral MADS-box gene duplication occurred before the divergence of plants and animals. Proc. Natl. Acad. Sci. USA 2000, 97, 5328–5333. [Google Scholar] [CrossRef]
- Nam, J.; Kaufmann, K.; Theißen, G.N.; Nei, M. A simple method for predicting the functional differentiation of duplicate genes and its application to MIKC-type MADS-box genes. Nucleic Acids Res. 2005, 33, e12. [Google Scholar] [CrossRef]
- Henschel, K.; Kofuji, R.; Hasebe, M.; Saedler, H.; Münster, T.; Theißen, G. Two ancient classes of MIKC-type MADS-box genes are present in the Moss Physcomitrella patens. Mol. Biol. Evol. 2002, 19, 801–814. [Google Scholar] [CrossRef]
- Coen, E.S.; Meyerowitz, E.M. The war of the whorls: Genetic interactions controlling flower development. Nature 1991, 353, 31–37. [Google Scholar] [CrossRef]
- Zhang, L.; Chen, F.; Zhang, X.; Li, Z.; Zhao, Y.; Lohaus, R.; Chang, X.; Dong, W.; Ho, S.Y.W.; Liu, X.; et al. The water lily genome and the early evolution of flowering plants. Nature 2020, 577, 79–84. [Google Scholar] [CrossRef]
- Theißen, G. Development of floral organ identity: Stories from the MADS house. Curr. Opin. Plant Biol. 2001, 4, 75–85. [Google Scholar] [CrossRef]
- Theißen, G.; Saedler, H. Floral quartets. Nature 2001, 409, 469–471. [Google Scholar] [CrossRef] [PubMed]
- Huo, S.; Li, Y.; Li, R.; Chen, R.; Xing, H.; Wang, J.; Zhao, Y.; Song, X. Genome-wide analysis of the MADS-box gene family in Rhododendron hainanense Merr. and expression analysis under heat and waterlogging stresses. Ind. Crops Prod. 2021, 172, 114007. [Google Scholar] [CrossRef]
- Shao, Z.; He, M.; Zeng, Z.; Chen, Y.; Hanna, A.-D.; Zhu, H. Genome-wide identification and expression analysis of the MADS-Box gene family in sweet potato [Ipomoea batatas (L.) Lam]. Front. Genet. 2021, 12, 750137. [Google Scholar] [CrossRef] [PubMed]
- Song, G.; Han, X.; Ryner, J.T.; Thompson, A.; Wang, K. Utilizing MIKC-type MADS-box protein SOC1 for yield potential enhancement in maize. Plant Cell Rep. 2021, 40, 1679–1693. [Google Scholar] [CrossRef] [PubMed]
- Ning, X.; Yanli, C.; Hongli, C.; Xingxing, L.; Qi, W.; Liang, J.; Guixue, W.; Junli, H. Rice transcription factor OsMADS25 modulates root growth and confers salinity tolerance via the ABA-mediated regulatory pathway and ROS scavenging. PLoS Genet. 2018, 14, e1007662. [Google Scholar]
- Zhao, P.; Miao, Z.; Zhang, J.; Chen, S.; Liu, Q.; Xiang, C. Arabidopsis MADS-box factor AGL16 negatively regulates drought resistance via stomatal density and stomatal movement. J. Exp. Bot. 2020, 71, 6092–6106. [Google Scholar] [CrossRef]
- Fu, A.; Wang, Q.; Mu, J.; Ma, L.; Wen, C.; Zhao, X.; Gao, L.; Li, J.; Shi, K.; Wang, Y.; et al. Combined genomic, transcriptomic, and metabolomic analyses provide insights into chayote (Sechium edule) evolution and fruit development. Hortic. Res. 2021, 8, 35. [Google Scholar] [CrossRef]
- Pu, Y.; Luo, Q.; Wen, L.; Li, Y.; Meng, P.; Wang, X.; Tan, G. Origin, evolution, breeding, and omics of chayote, an important Cucurbitaceae vegetable crop. Front. Plant Sci. 2021, 12, 739091. [Google Scholar] [CrossRef] [PubMed]
- Vieira, E.F.; Pinho, O.; Ferreira, I.M.P.L.V.O.; Delerue-Matos, C. Chayote (Sechium edule): A review of nutritional composition, bioactivities and potential applications. Food Chem. 2019, 275, 557–568. [Google Scholar] [CrossRef]
- Díaz-de-Cerio, E.; Verardo, V.; Fernández-Gutiérrez, A.; Gómez-Caravaca, A.M. New insight into phenolic composition of chayote (Sechium edule (Jacq.) Sw.). Food Chem. 2019, 295, 514–519. [Google Scholar] [CrossRef] [PubMed]
- Su, L.; Cheng, S.; Liu, Y.; Xie, Y.; He, Z.; Jia, M.; Zhou, X.; Zhang, R.; Li, C. Transcriptome and metabolome analysis provide new insights into the process of tuberization of Sechium edule roots. Int. J. Mol. Sci. 2022, 23, 6390. [Google Scholar] [CrossRef] [PubMed]
- Parenicová, L.; de Folter, S.; Kieffer, M.; Horner, D.S.; Favalli, C.; Busscher, J.; Cook, H.E.; Ingram, R.M.; Kater, M.M.; Davies, B.; et al. Molecular and phylogenetic analyses of the complete MADS-Box transcription factor family in Arabidopsis: New openings to the MADS world. Plant Cell 2003, 15, 1538–1551. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Zhang, J.; Hu, Z.; Guo, X.; Tian, S.; Chen, G. Genome-wide analysis of the MADS-Box transcription factor family in Solanum lycopersicum. Int. J. Mol. Sci. 2019, 20, 2961. [Google Scholar] [CrossRef]
- Liu, J.; Zhang, J.; Zhang, J.; Miao, H.; Wang, J.; Gao, P.; Hu, W.; Jia, C.; Wang, Z.; Xu, B.; et al. Genome-wide analysis of banana MADS-box family closely related to fruit development and ripening. Sci. Rep. 2017, 7, 3467. [Google Scholar] [CrossRef]
- Schilling, S.; Kennedy, A.; Pan, S.; Jermiin, L.S.; Melzer, R. Genome-wide analysis of MIKC-type MADS-box genes in wheat: Pervasive duplications, functional conservation and putative neofunctionalization. New Phytol. 2020, 225, 511–529. [Google Scholar] [CrossRef] [PubMed]
- Wang, P.; Wang, S.; Chen, Y.; Xu, X.; Guang, X.; Zhang, Y. Genome-wide analysis of the MADS-Box gene family in Watermelon. Comput. Biol. Chem. 2019, 80, 341–350. [Google Scholar] [CrossRef]
- Hu, L.; Liu, S. Genome-wide analysis of the MADS-box gene family in cucumber. Genome 2012, 55, 245–256. [Google Scholar] [CrossRef]
- Hao, X.; Fu, Y.; Zhao, W.; Liu, L.; Bade, R.; Hasi, A.; Hao, J. Genome-wide identification and analysis of the MADS-box gene family in Melon. J. Am. Soc. Hortic. Sci. 2016, 141, 507–519. [Google Scholar] [CrossRef]
- Wang, B.; Hu, W.; Fang, Y.; Feng, X.; Fang, J.; Zou, T.; Zheng, S.; Ming, R.; Zhang, J. Comparative analysis of the MADS-Box genes revealed their potential functions for flower and fruit development in Longan (Dimocarpus longan). Front. Plant Sci. 2022, 12, 813798. [Google Scholar] [CrossRef]
- Chen, S.; Sun, W.; Xiong, Y.; Jiang, Y.; Liu, X.; Liao, X.; Zhang, D.; Jiang, S.; Li, Y.; Liu, B.; et al. The Phoebe genome sheds light on the evolution of magnoliids. Hortic. Res. 2020, 7, 146. [Google Scholar] [CrossRef] [PubMed]
- Qu, Y.; Kong, W.; Wang, Q.; Fu, X. Genome-wide identification MIKC-Type MADS-Box gene family and their roles during development of floral buds in wheel wingnut (Cyclocarya paliurus). Int. J. Mol. Sci. 2021, 22, 10128. [Google Scholar] [CrossRef]
- Liu, M.; Fu, Q.; Ma, Z.; Sun, W.; Huang, L.; Wu, Q.; Tang, Z.; Bu, T.; Li, C.; Chen, H. Genome-wide investigation of the MADS gene family and dehulling genes in tartary buckwheat (Fagopyrum tataricum). Planta 2019, 249, 1301–1318. [Google Scholar] [CrossRef]
- Flagel, L.E.; Wendel, J.F. Gene duplication and evolutionary novelty in plants. New Phytol. 2009, 183, 557–564. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Li, Y.; Zhang, X.; Cai, K.; Li, Y.; Wang, Q.; Qu, G.; Han, R.; Zhao, X. Genome-wide identification and expression analysis of the MADS-box gene family during female and male flower development in Juglans mandshurica. Front. Plant Sci. 2022, 13, 1020706. [Google Scholar] [CrossRef]
- Ning, K.; Han, Y.; Chen, Z.; Luo, C.; Wang, S.; Zhang, W.; Li, L.; Zhang, X.; Fan, S.; Wang, Q. Genome-wide analysis of MADS-box family genes during flower development in lettuce. Plant Cell Environ. 2019, 42, 1868–1881. [Google Scholar] [CrossRef]
- Pan, X.; Ouyang, Y.; Wei, Y.; Zhang, B.; Wang, J.; Zhang, H. Genome-wide analysis of MADS-box families and their expressions in flower organs development of pineapple (Ananas comosus (L.) Merr.). Front. Plant Sci. 2022, 13, 948587. [Google Scholar] [CrossRef]
- Ye, L.; Luo, M.; Wang, Z.; Bai, F.; Luo, X.; Gao, L.; Peng, J.; Chen, Q.; Zhang, L. Genome-wide analysis of MADS-box gene family in kiwifruit (Actinidia chinensis var. chinensis) and their potential role in floral sex differentiation. Front. Genet. 2022, 13, 1043178. [Google Scholar] [CrossRef]
- Yang, Z.; Nie, G.; Feng, G.; Xu, X.; Li, D.; Wang, X.; Huang, L.; Zhang, X. Genome-wide identification of MADS-box gene family in orchardgrass and the positive role of DgMADS114 and DgMADS115 under different abiotic stress. Int. J. Biol. Macromol. 2022, 223, 129–142. [Google Scholar] [CrossRef] [PubMed]
- Adamczyk, B.J.; Lehti-Shiu, M.D.; Fernandez, D.E. The MADS domain factors AGL15 and AGL18 act redundantly as repressors of the floral transition in Arabidopsis. Plant J. 2007, 50, 1007–1019. [Google Scholar] [CrossRef]
- Wang, D.; Hao, Z.; Long, X.; Wang, Z.; Zheng, X.; Ye, D.; Peng, Y.; Wu, W.; Hu, X.; Wang, G.; et al. The Transcriptome of Cunninghamia lanceolata male/female cone reveal the association between MIKC MADS-box genes and reproductive organs development. BMC Plant Biol. 2020, 20, 508. [Google Scholar] [CrossRef]
- Zhang, Q.; Hou, S.; Sun, Z.; Chen, J.; Meng, J.; Liang, D.; Wu, R.; Guo, Y. Genome-wide identification and analysis of the MADS-Box gene family in Theobroma cacao. Genes 2021, 12, 1799. [Google Scholar] [CrossRef]
- Lin, Z.; Cao, D.; Damaris, R.N.; Yang, P. Genome-wide identification of MADS-box gene family in sacred lotus (Nelumbo nucifera) identifies a SEPALLATA homolog gene involved in floral development. BMC Plant Biol. 2020, 20, 497. [Google Scholar] [CrossRef]
- Mi, Z.; Zhao, Q.; Lu, C.; Zhang, Q.; Li, L.; Liu, S.; Wang, S.; Wang, Z.; Niu, J. Genome-wide analysis and the expression pattern of the MADS-Box gene family in Bletilla striata. Plants 2021, 10, 2184. [Google Scholar] [CrossRef]
- Chen, C.; Chen, H.; Zhang, Y.; Thomas, H.R.; Frank, M.H.; He, Y.; Xia, R. TBtools: An integrative toolkit developed for interactive analyses of big biological data. Mol. Plant 2020, 13, 1194–1202. [Google Scholar] [CrossRef]
- Guan, H.; Wang, H.; Huang, J.; Liu, M.; Chen, T.; Shan, X.; Chen, H.; Shen, J. Genome-wide identification and expression analysis of MADS-Box family genes in Litchi (Litchi chinensis Sonn.) and their involvement in floral sex determination. Plants 2021, 10, 2142. [Google Scholar] [CrossRef]
- Cai, X.; Zhang, L.; Xiao, L.; Wen, Z.; Hou, Q.; Yang, K. Genome-wide identification of GRF gene family and their contribution to abiotic stress response in pitaya (Hylocereus polyrhizus). Int. J. Biol. Macromol. 2022, 223, 618–635. [Google Scholar] [CrossRef]











| Gene Name | Gene ID | Length(aa) | MW(Da) | PI | Asp + Glu | Arg + Lys | GRAVY | SLP |
|---|---|---|---|---|---|---|---|---|
| SeMADS01 | Sed0027248 | 236 | 27,151.55 | 9.03 | 30 | 35 | −0.822 | Nucleus |
| SeMADS02 | Sed0005514 | 247 | 28,710.73 | 9.66 | 30 | 42 | −0.943 | Nucleus |
| SeMADS03 | Sed0021668 | 258 | 29,044.79 | 8.46 | 28 | 30 | −0.599 | Nucleus |
| SeMADS04 | Sed0017917 | 359 | 40,697.70 | 5.72 | 44 | 36 | −0.684 | Nucleus |
| SeMADS05 | Sed0023354 | 232 | 26,862.59 | 8.85 | 31 | 35 | −0.730 | Nucleus |
| SeMADS06 | Sed0023514 | 211 | 23,885.32 | 8.81 | 29 | 34 | −0.648 | Nucleus |
| SeMADS07 | Sed0028207 | 205 | 23,880.68 | 9.20 | 32 | 39 | −0.726 | Nucleus |
| SeMADS08 | Sed0013720 | 337 | 37,643.72 | 5.73 | 46 | 38 | −0.562 | Nucleus |
| SeMADS09 | Sed0009751 | 215 | 24,456.80 | 8.95 | 29 | 33 | −0.705 | Nucleus |
| SeMADS10 | Sed0012619 | 230 | 25,797.58 | 8.36 | 33 | 35 | −0.483 | Nucleus |
| SeMADS11 | Sed0018953 | 63 | 7133.35 | 9.92 | 7 | 13 | −0.297 | Nucleus |
| SeMADS12 | Sed0008766 | 223 | 26,259.86 | 9.13 | 29 | 34 | −0.850 | Nucleus |
| SeMADS13 | Sed0022320 | 223 | 25,657.81 | 9.40 | 26 | 34 | −0.613 | Nucleus |
| SeMADS14 | Sed0018775 | 217 | 23,771.38 | 9.36 | 27 | 33 | −0.275 | Nucleus |
| SeMADS15 | Sed0021213 | 245 | 28,232.24 | 8.61 | 30 | 34 | −0.580 | Nucleus |
| SeMADS16 | Sed0006178 | 232 | 26,458.42 | 7.07 | 35 | 35 | −0.559 | Nucleus |
| SeMADS17 | Sed0009958 | 210 | 23,801.38 | 9.28 | 29 | 37 | −0.612 | Nucleus |
| SeMADS18 | Sed0019121 | 213 | 24,861.65 | 9.20 | 34 | 41 | −0.800 | Nucleus |
| SeMADS19 | Sed0025694 | 235 | 26,337.13 | 5.94 | 35 | 30 | −0.419 | Nucleus |
| SeMADS20 | Sed0016651 | 239 | 27,546.54 | 9.32 | 33 | 39 | −0.699 | Nucleus |
| SeMADS21 | Sed0027083 | 211 | 24,902.45 | 8.74 | 33 | 36 | −0.919 | Nucleus |
| SeMADS22 | Sed0014035 | 240 | 27,810.45 | 9.47 | 28 | 39 | −0.870 | Nucleus |
| SeMADS23 | Sed0006669 | 356 | 41,143.66 | 7.14 | 41 | 41 | −0.741 | Nucleus |
| SeMADS24 | Sed0011763 | 220 | 24,888.25 | 5.93 | 36 | 33 | −0.677 | Nucleus |
| SeMADS25 | Sed0026823 | 246 | 28,172.01 | 8.97 | 29 | 33 | −0.638 | Nucleus |
| SeMADS26 | Sed0012551 | 247 | 28,499.47 | 8.67 | 35 | 38 | −0.754 | Nucleus |
| SeMADS27 | Sed0010395 | 290 | 33,536.67 | 5.33 | 50 | 35 | −0.711 | Nucleus |
| SeMADS28 | Sed0020082 | 353 | 41,225.84 | 5.52 | 54 | 41 | −0.471 | Nucleus |
| SeMADS29 | Sed0010468 | 341 | 38,775.45 | 6.27 | 40 | 36 | −0.716 | Nucleus |
| SeMADS30 | Sed0006335 | 243 | 28,008.95 | 8.59 | 29 | 32 | −0.645 | Nucleus |
| SeMADS31 | Sed0020653 | 219 | 24,716.41 | 9.59 | 20 | 29 | −0.453 | Nucleus |
| SeMADS32 | Sed0015557 | 236 | 27,208.20 | 9.50 | 25 | 35 | −0.695 | Nucleus |
| SeMADS33 | Sed0022681 | 215 | 24,209.69 | 9.34 | 28 | 34 | −0.670 | Nucleus |
| SeMADS34 | Sed0013054 | 346 | 39,213.01 | 5.52 | 43 | 34 | −0.696 | Nucleus |
| SeMADS35 | Sed0010970 | 218 | 24,726.63 | 8.86 | 27 | 31 | −0.343 | Nucleus |
| SeMADS36 | Sed0023078 | 246 | 28,402.28 | 8.33 | 33 | 35 | −0.782 | Nucleus |
| SeMADS37 | Sed0013411 | 200 | 22,993.30 | 9.95 | 25 | 37 | −0.742 | Nucleus |
| SeMADS38 | Sed0017436 | 254 | 28,871.34 | 9.26 | 37 | 43 | −0.518 | Nucleus |
| SeMADS39 | Sed0009092 | 246 | 28,305.01 | 8.93 | 29 | 34 | −0.712 | Nucleus |
| SeMADS40 | Sed0003074 | 214 | 24,685.14 | 6.36 | 39 | 37 | −0.797 | Nucleus |
| SeMADS41 | Sed0023327 | 208 | 23,157.95 | 9.44 | 24 | 32 | −0.326 | Nucleus |
| SeMADS42 | Sed0017192 | 91 | 10,494.20 | 9.38 | 8 | 14 | −0.305 | Nucleus |
| SeMADS43 | Sed0018117 | 212 | 24,447.88 | 9.38 | 31 | 39 | −0.822 | Nucleus |
| SeMADS44 | Sed0024122 | 191 | 21,856.08 | 9.87 | 20 | 34 | −0.665 | Nucleus |
| SeMADS45 | Sed0002596 | 239 | 28,379.56 | 8.50 | 31 | 34 | −0.490 | Nucleus |
| SeMADS46 | Sed0012087 | 185 | 21,618.52 | 5.02 | 34 | 27 | −0.864 | Nucleus |
| SeMADS47 | Sed0023127 | 243 | 27,905.73 | 8.60 | 29 | 32 | −0.671 | Nucleus |
| SeMADS48 | Sed0004913 | 108 | 12,916.34 | 9.99 | 8 | 20 | 0.069 | Nucleus |
| SeMADS49 | Sed0000693 | 354 | 40,628.93 | 5.81 | 47 | 39 | −0.671 | Nucleus |
| SeMADS50 | Sed0007340 | 250 | 28,333.50 | 6.31 | 32 | 28 | −0.882 | Nucleus |
| SeMADS51 | Sed0021520 | 248 | 28,501.53 | 9.50 | 29 | 39 | −0.751 | Nucleus |
| SeMADS52 | Sed0004053 | 234 | 26,720.89 | 9.22 | 30 | 36 | −0.500 | Nucleus |
| SeMADS53 | Sed0027590 | 229 | 25,984.65 | 8.95 | 34 | 38 | −0.650 | Nucleus |
| SeMADS54 | Sed0016270 | 218 | 24,221.75 | 9.21 | 30 | 35 | −0.569 | Nucleus |
| SeMADS55 | Sed0005726 | 205 | 23,752.59 | 6.14 | 29 | 27 | −0.412 | Nucleus |
| SeMADS56 | Sed0005214 | 224 | 25,715.39 | 9.35 | 27 | 35 | −0.600 | Nucleus |
| SeMADS57 | Sed0019435 | 216 | 24,636.04 | 7.15 | 32 | 32 | −0.664 | Nucleus |
| SeMADS58 | Sed0024714 | 370 | 42,194.47 | 8.49 | 42 | 45 | −0.460 | Nucleus |
| SeMADS59 | Sed0024599 | 162 | 18,937.95 | 9.62 | 22 | 32 | −0.665 | Nucleus |
| SeMADS60 | Sed0014292 | 219 | 25,201.49 | 5.78 | 35 | 32 | −0.769 | Nucleus |
| SeMADS61 | Sed0025303 | 213 | 24,128.59 | 8.50 | 32 | 34 | −0.645 | Nucleus |
| SeMADS62 | Sed0012885 | 236 | 27,235.31 | 9.28 | 30 | 37 | −0.698 | Nucleus |
| SeMADS63 | Sed0009196 | 219 | 24,805.32 | 9.62 | 21 | 29 | −0.623 | Nucleus |
| SeMADS64 | Sed0009551 | 343 | 38,508.87 | 4.94 | 41 | 28 | −0.640 | Nucleus |
| SeMADS65 | Sed0007855 | 223 | 26,016.73 | 8.66 | 32 | 35 | −0.674 | Nucleus |
| SeMADS66 | Sed0024257 | 283 | 32,502.48 | 5.35 | 48 | 38 | −0.648 | Nucleus |
| SeMADS67 | Sed0017033 | 220 | 24,858.38 | 6.24 | 33 | 32 | −0.585 | Nucleus |
| SeMADS68 | Sed0000564 | 226 | 25,924.68 | 8.34 | 30 | 32 | −0.554 | Nucleus |
| SeMADS69 | Sed0004665 | 674 | 77,846.80 | 9.74 | 74 | 106 | −0.466 | Nucleus |
| SeMADS70 | Sed0027724 | 220 | 24,635.77 | 7.78 | 28 | 29 | −0.766 | Nucleus |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Cheng, S.; Jia, M.; Su, L.; Liu, X.; Chu, Q.; He, Z.; Zhou, X.; Lu, W.; Jiang, C. Genome-Wide Identification of the MADS-Box Gene Family during Male and Female Flower Development in Chayote (Sechium edule). Int. J. Mol. Sci. 2023, 24, 6114. https://doi.org/10.3390/ijms24076114
Cheng S, Jia M, Su L, Liu X, Chu Q, He Z, Zhou X, Lu W, Jiang C. Genome-Wide Identification of the MADS-Box Gene Family during Male and Female Flower Development in Chayote (Sechium edule). International Journal of Molecular Sciences. 2023; 24(7):6114. https://doi.org/10.3390/ijms24076114
Chicago/Turabian StyleCheng, Shaobo, Mingyue Jia, Lihong Su, Xuanxuan Liu, Qianwen Chu, Zhongqun He, Xiaoting Zhou, Wei Lu, and Chengyao Jiang. 2023. "Genome-Wide Identification of the MADS-Box Gene Family during Male and Female Flower Development in Chayote (Sechium edule)" International Journal of Molecular Sciences 24, no. 7: 6114. https://doi.org/10.3390/ijms24076114
APA StyleCheng, S., Jia, M., Su, L., Liu, X., Chu, Q., He, Z., Zhou, X., Lu, W., & Jiang, C. (2023). Genome-Wide Identification of the MADS-Box Gene Family during Male and Female Flower Development in Chayote (Sechium edule). International Journal of Molecular Sciences, 24(7), 6114. https://doi.org/10.3390/ijms24076114









