Wds-Mediated H3K4me3 Modification Regulates Lipid Synthesis and Transport in Drosophila
Abstract
:1. Introduction
2. Results
2.1. Disrupting Wds-Mezdiated H3K4me3 Modification in the Drosophila Fat Body Reduced Lipid Content
2.2. Fat Body-Specific Wds Knockdown Impaired the Transcription of Lipogenic Genes by Reducing H3K4me3 Deposition within Their Promoters
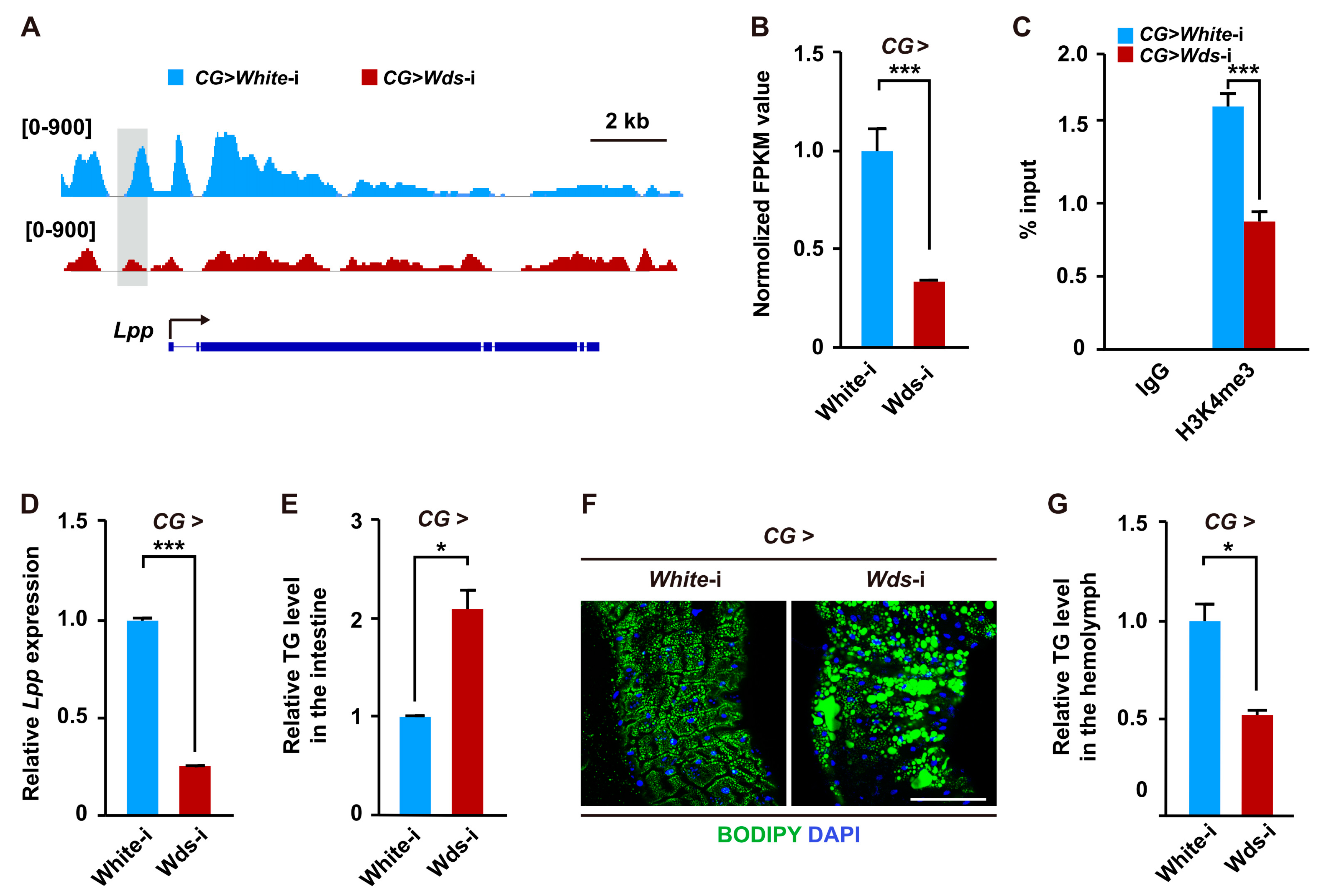
2.3. Wds-Mediated H3K4me3 Modification Regulates Lipid Transport from Intestine by Modulating the Expression of the Lpp Gene in the Fat Body
2.4. Transcription Factor Hsf Promotes Lipid Synthesis and Transport by Interacting with Wds to Modulate H3K4me3 Modification
2.5. HFD Condition Promotes Wds-Mediated H3K4me3 in the Drosophila Fat Body
3. Discussion
4. Materials and Methods
4.1. Drosophila Cultivation and Stocks
4.2. Immunostaining
4.3. RNA-seq
4.4. Cell Culture and Transfection
4.5. Co-Immunoprecipitation (Co-IP)
4.6. Western Blotting
4.7. Chromatin Immunoprecipitation Sequencing (ChIP-seq)
4.8. RNA Extraction and Real-Time Quantitative PCR (RT-qPCR)
4.9. TG Measurement
4.10. Databases
4.11. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Olzmann, J.A.; Carvalho, P. Dynamics and functions of lipid droplets. Nat. Rev. Mol. Cell. Biol. 2019, 20, 137–155. [Google Scholar] [CrossRef] [PubMed]
- Walther, T.C.; Farese, R.V., Jr. Lipid droplets and cellular lipid metabolism. Annu. Rev. Biochem. 2012, 81, 687–714. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Welte, M.A. As the fat flies: The dynamic lipid droplets of Drosophila embryos. Biochim. Biophys. Acta 2015, 1851, 1156–1185. [Google Scholar] [CrossRef] [Green Version]
- Nelliot, A.; Bond, N.; Hoshizaki, D.K. Fat-body remodeling in Drosophila melanogaster. Genesis 2006, 44, 396–400. [Google Scholar] [CrossRef] [PubMed]
- Li, S.; Yu, X.; Feng, Q. Fat body biology in the last decade. Annu. Rev. Entomol. 2019, 64, 315–333. [Google Scholar] [CrossRef]
- Moraes, K.C.M.; Montagne, J. Drosophila melanogaster: A powerful tiny animal model for the study of metabolic hepatic diseases. Front. Physiol. 2021, 12, 728407. [Google Scholar] [CrossRef]
- Ugrankar, R.; Liu, Y.; Provaznik, J.; Schmitt, S.; Lehmann, M. Lipin is a central regulator of adipose tissue development and function in Drosophila melanogaster. Mol. Cell. Biol. 2011, 31, 1646–1656. [Google Scholar] [CrossRef] [Green Version]
- Texada, M.J.; Koyama, T.; Rewitz, K. Regulation of body size and growth control. Genetics 2020, 216, 269–313. [Google Scholar] [CrossRef]
- Yongmei Xi, Y.Z. Fat body development and its function in energy storage and nutrient sensing in Drosophila melanogaster. J. Tissue Sci. Eng. 2015, 6, 1. [Google Scholar] [CrossRef] [Green Version]
- Pippa, S.; Mannironi, C.; Licursi, V.; Bombardi, L.; Colotti, G.; Cundari, E.; Mollica, A.; Coluccia, A.; Naccarato, V.; La Regina, G.; et al. Small molecule inhibitors of KDM5 histone demethylases increase the radiosensitivity of breast cancer cells overexpressing JARID1B. Molecules 2018, 24, 1739. [Google Scholar] [CrossRef] [Green Version]
- Petan, T.; Jarc, E.; Jusovic, M. Lipid droplets in cancer: Guardians of fat in a stressful world. Molecules 2018, 23, 1941. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jarc, E.; Petan, T. Lipid droplets and the management of cellular stress. Yale J. Biol. Med. 2019, 92, 435–452. [Google Scholar]
- Heier, C.; Kuhnlein, R.P. Triacylglycerol metabolism in Drosophila melanogaster. Genetics 2018, 210, 1163–1184. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Buszczak, M.; Lu, X.; Segraves, W.A.; Chang, T.Y.; Cooley, L. Mutations in the midway gene disrupt a Drosophila acyl coenzyme A: Diacylglycerol acyltransferase. Genetics 2002, 160, 1511–1518. [Google Scholar] [CrossRef] [PubMed]
- Parvy, J.P.; Napal, L.; Rubin, T.; Poidevin, M.; Perrin, L.; Wicker-Thomas, C.; Montagne, J. Drosophila melanogaster Acetyl-CoA-carboxylase sustains a fatty acid-dependent remote signal to waterproof the respiratory system. PLoS Genet. 2012, 8, e1002925. [Google Scholar] [CrossRef] [Green Version]
- Schmitt, S.; Ugrankar, R.; Greene, S.E.; Prajapati, M.; Lehmann, M. Drosophila Lipin interacts with insulin and TOR signaling pathways in the control of growth and lipid metabolism. J. Cell. Sci. 2015, 128, 4395–4406. [Google Scholar] [CrossRef] [Green Version]
- Canavoso, L.E.; Jouni, Z.E.; Karnas, K.J.; Pennington, J.E.; Wells, M.A. Fat metabolism in insects. Annu. Rev. Nutr. 2001, 21, 23–46. [Google Scholar] [CrossRef]
- Palm, W.; Sampaio, J.L.; Brankatschk, M.; Carvalho, M.; Mahmoud, A.; Shevchenko, A.; Eaton, S. Lipoproteins in Drosophila melanogaster—Assembly, function, and influence on tissue lipid composition. PLoS Genet. 2012, 8, e1002828. [Google Scholar] [CrossRef] [Green Version]
- Henne, W.M.; Reese, M.L.; Goodman, J.M. The assembly of lipid droplets and their roles in challenged cells. EMBO J. 2018, 37, e98947. [Google Scholar] [CrossRef]
- Fan, W.; Lam, S.M.; Xin, J.; Yang, X.; Liu, Z.; Liu, Y.; Wang, Y.; Shui, G.; Huang, X. Drosophila TRF2 and TAF9 regulate lipid droplet size and phospholipid fatty acid composition. PLoS Genet. 2017, 13, e1006664. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Song, W.; Veenstra, J.A.; Perrimon, N. Control of lipid metabolism by tachykinin in Drosophila. Cell. Rep. 2014, 9, 40–47. [Google Scholar] [CrossRef] [Green Version]
- Molaei, M.; Vandehoef, C.; Karpac, J. NF-kappaB shapes metabolic adaptation by attenuating Foxo-mediated lipolysis in Drosophila. Dev. Cell. 2019, 49, 802–810.e6. [Google Scholar] [CrossRef]
- Sieber, M.H.; Thummel, C.S. The DHR96 nuclear receptor controls triacylglycerol homeostasis in Drosophila. Cell. Metab. 2009, 10, 481–490. [Google Scholar] [CrossRef] [Green Version]
- Birse, R.T.; Choi, J.; Reardon, K.; Rodriguez, J.; Graham, S.; Diop, S.; Ocorr, K.; Bodmer, R.; Oldham, S. High-fat-diet-induced obesity and heart dysfunction are regulated by the TOR pathway in Drosophila. Cell. Metab. 2010, 12, 533–544. [Google Scholar] [CrossRef] [Green Version]
- Praggastis, S.A.; Lam, G.; Horner, M.A.; Nam, H.J.; Thummel, C.S. The Drosophila E78 nuclear receptor regulates dietary triglyceride uptake and systemic lipid levels. Dev. Dyn. 2021, 250, 640–651. [Google Scholar] [CrossRef]
- Park, S.; Kim, G.W.; Kwon, S.H.; Lee, J.S. Broad domains of histone H3 lysine 4 trimethylation in transcriptional regulation and disease. FEBS J. 2020, 287, 2891–2902. [Google Scholar] [CrossRef] [Green Version]
- Bochynska, A.; Luscher-Firzlaff, J.; Luscher, B. Modes of interaction of KMT2 Histone H3 Lysine 4 methyltransferase/COMPASS complexes with chromatin. Cells 2018, 7, 17. [Google Scholar] [CrossRef] [Green Version]
- Kaser, S.; Castellano-Castillo, D.; Denechaud, P.-D.; Fajas, L.; Moreno-Indias, I.; Oliva-Olivera, W.; Tinahones, F.; Queipo-Ortuño, M.I.; Cardona, F. Human adipose tissue H3K4me3 histone mark in adipogenic, lipid metabolism and inflammatory genes is positively associated with BMI and HOMA-IR. PLoS ONE 2019, 14, e0215083. [Google Scholar]
- Lee, J.E.; Cho, Y.W.; Deng, C.X.; Ge, K. MLL3/MLL4-associated PAGR1 regulates adipogenesis by controlling induction of C/EBPbeta and C/EBPdelta. Mol. Cell. Biol. 2020, 40, e00209-20. [Google Scholar] [CrossRef]
- Sun, H.; Wang, Y.; Wang, Y.; Ji, F.; Wang, A.; Yang, M.; He, X.; Li, L. Bivalent regulation and related mechanisms of H3K4/27/9me3 in stem cells. Stem Cell. Rev. Rep. 2021, 18, 165–178. [Google Scholar] [CrossRef]
- Nanduri, R. Epigenetic regulators of white adipocyte browning. Epigenomes 2021, 5, 3. [Google Scholar] [CrossRef]
- Lee, J.; Saha, P.K.; Yang, Q.H.; Lee, S.; Park, J.Y.; Suh, Y.; Lee, S.K.; Chan, L.; Roeder, R.G.; Lee, J.W. Targeted inactivation of MLL3 histone H3-Lys-4 methyltransferase activity in the mouse reveals vital roles for MLL3 in adipogenesis. Proc. Natl. Acad. Sci. USA 2008, 105, 19229–19234. [Google Scholar] [CrossRef] [Green Version]
- Wan, Q.L.; Meng, X.; Wang, C.; Dai, W.; Luo, Z.; Yin, Z.; Ju, Z.; Fu, X.; Yang, J.; Ye, Q.; et al. Histone H3K4me3 modification is a transgenerational epigenetic signal for lipid metabolism in Caenorhabditis elegans. Nat. Commun. 2022, 13, 768. [Google Scholar] [CrossRef]
- Wysocka, J.; Swigut, T.; Milne, T.A.; Dou, Y.; Zhang, X.; Burlingame, A.L.; Roeder, R.G.; Brivanlou, A.H.; Allis, C.D. WDR5 associates with histone H3 methylated at K4 and is essential for H3 K4 methylation and vertebrate development. Cell 2005, 121, 859–872. [Google Scholar] [CrossRef] [Green Version]
- Wysocka, J.; Swigut, T.; Xiao, H.; Milne, T.A.; Kwon, S.Y.; Landry, J.; Kauer, M.; Tackett, A.J.; Chait, B.T.; Badenhorst, P.; et al. A PHD finger of NURF couples histone H3 lysine 4 trimethylation with chromatin remodelling. Nature 2006, 442, 86–90. [Google Scholar] [CrossRef]
- Ang, Y.S.; Tsai, S.Y.; Lee, D.F.; Monk, J.; Su, J.; Ratnakumar, K.; Ding, J.; Ge, Y.; Darr, H.; Chang, B.; et al. Wdr5 mediates self-renewal and reprogramming via the embryonic stem cell core transcriptional network. Cell. 2011, 145, 183–197. [Google Scholar] [CrossRef] [Green Version]
- Song, Z.T.; Sun, L.; Lu, S.J.; Tian, Y.; Ding, Y.; Liu, J.X. Transcription factor interaction with COMPASS-like complex regulates histone H3K4 trimethylation for specific gene expression in plants. Proc. Natl. Acad. Sci. USA 2015, 112, 2900–2905. [Google Scholar] [CrossRef] [Green Version]
- Lee, J.Y.; Lee, J.H.; Cheon, C.K. Functional characterization of gomisin N in high-fat-induced Drosophila obesity models. Int. J. Mol. Sci. 2020, 21, 7209. [Google Scholar] [CrossRef]
- Chatterjee, N.; Perrimon, N. What fuels the fly:energy metabolism in Drosophila and its application to the study of obesity and diabetes. Sci. Adv. 2021, 7, eabg4336. [Google Scholar] [CrossRef]
- Heinrichsen, E.T.; Haddad, G.G. Role of high-fat diet in stress response of Drosophila. PLoS ONE 2012, 7, e42587. [Google Scholar] [CrossRef] [Green Version]
- Heier, C.; Klishch, S.; Stilbytska, O.; Semaniuk, U.; Lushchak, O. The Drosophila model to interrogate triacylglycerol biology. Biochim. Biophys. Acta Mol. Cell. Biol. Lipids 2021, 1866, 158924. [Google Scholar] [CrossRef]
- Yao, Y.; Ding, L.; Huang, X. Diverse functions of lipids and lipid metabolism in development. Small Methods 2019, 4, 1900564. [Google Scholar] [CrossRef]
- Hebbar, S.; Khandelwal, A.; Jayashree, R.; Hindle, S.J.; Chiang, Y.N.; Yew, J.Y.; Sweeney, S.T.; Schwudke, D. Lipid metabolic perturbation is an early-onset phenotype in adult spinster mutants: A Drosophila model for lysosomal storage disorders. Mol. Biol. Cell. 2017, 28, 3728–3740. [Google Scholar] [CrossRef]
- Majerowicz, D.; Gondim, K.C. Insect Lipid Metabolism: Insights into Gene Expression Regulation; Nova Science Publishers: Hauppauge, NY, USA, 2013; pp. 147–189. [Google Scholar]
- Cheng, Y.; Lu, T.; Guo, J.; Lin, Z.; Jin, Q.; Zhang, X.; Zou, Z. Helicoverpa armigera miR-2055 regulates lipid metabolism via fatty acid synthase expression. Open. Biol. 2022, 12, 210307. [Google Scholar] [CrossRef]
- Sha, Q.Q.; Zhang, J.; Fan, H.Y. Function and Regulation of Histone H3 Lysine-4 Methylation During Oocyte Meiosis and Maternal-to-Zygotic Transition. Front. Cell. Dev. Biol. 2020, 8, 597498. [Google Scholar] [CrossRef]
- Shilatifard, A. The COMPASS family of histone H3K4 methylases: Mechanisms of regulation in development and disease pathogenesis. Annu. Rev. Biochem. 2012, 81, 65–95. [Google Scholar] [CrossRef] [Green Version]
- Juarez-Carreno, S.; Vallejo, D.M.; Carranza-Valencia, J.; Palomino-Schatzlein, M.; Ramon-Canellas, P.; Santoro, R.; de Hartog, E.; Ferres-Marco, D.; Romero, A.; Peterson, H.P.; et al. Body-fat sensor triggers ribosome maturation in the steroidogenic gland to initiate sexual maturation in Drosophila. Cell. Rep. 2021, 37, 109830. [Google Scholar] [CrossRef]
- MacDonald, M.E.; Schlegel, A.; Stainier, D.Y.R. Lessons from “Lower” organisms: What worms, flies, and zebrafish can teach us about human energy metabolism. PLoS Genet. 2007, 3, e199. [Google Scholar]
- Pirkkala, L.; Nykanen, P.; Sistonen, L. Roles of the heat shock transcription factors in regulation of the heat shock response and beyond. FASEB J. 2001, 15, 1118–1131. [Google Scholar] [CrossRef] [Green Version]
- Gonsalves, S.E.; Moses, A.M.; Razak, Z.; Robert, F.; Westwood, J.T. Whole-genome analysis reveals that active heat shock factor binding sites are mostly associated with non-heat shock genes in Drosophila melanogaster. PLoS ONE 2011, 6, e15934. [Google Scholar] [CrossRef] [Green Version]
- Gomez-Pastor, R.; Burchfiel, E.T.; Thiele, D.J. Regulation of heat shock transcription factors and their roles in physiology and disease. Nat. Rev. Mol. Cell. Biol. 2018, 19, 4–19. [Google Scholar] [CrossRef]
- Ma, X.; Xu, L.; Alberobello, A.T.; Gavrilova, O.; Bagattin, A.; Skarulis, M.; Liu, J.; Finkel, T.; Mueller, E. Celastrol protects against obesity and metabolic dysfunction through activation of a HSF1-PGC1alpha transcriptional axis. Cell. Metab. 2015, 22, 695–708. [Google Scholar] [CrossRef] [Green Version]
- Li, Y.; Wang, D.; Ping, X.; Zhang, Y.; Zhang, T.; Wang, L.; Jin, L.; Zhao, W.; Guo, M.; Shen, F.; et al. Local hyperthermia therapy induces browning of white fat and treats obesity. Cell. 2022, 185, 949–966 e19. [Google Scholar] [CrossRef]
- Hayashida, N. Set1/MLL complex is indispensable for the transcriptional ability of heat shock transcription factor 2. Biochem. Biophys. Res. Commun. 2015, 467, 805–812. [Google Scholar] [CrossRef]
- Heinrichsen, E.T.; Zhang, H.; Robinson, J.E.; Ngo, J.; Diop, S.; Bodmer, R.; Joiner, W.J.; Metallo, C.M.; Haddad, G.G. Metabolic and transcriptional response to a high-fat diet in Drosophila melanogaster. Mol. Metab. 2014, 3, 42–54. [Google Scholar] [CrossRef]
- Song, W.; Cheng, D.; Hong, S.; Sappe, B.; Hu, Y.; Wei, N.; Zhu, C.; O'Connor, M.B.; Pissios, P.; Perrimon, N. Midgut-derived activin regulates glucagon-like action in the fat body and glycemic control. Cell. Metab. 2017, 25, 386–399. [Google Scholar] [CrossRef] [Green Version]
- Komori, H.; Xiao, Q.; Janssens, D.H.; Dou, Y.; Lee, C.Y. Trithorax maintains the functional heterogeneity of neural stem cells through the transcription factor buttonhead. eLife 2014, 3, e03502. [Google Scholar] [CrossRef]
- Achary, B.G.; Campbell, K.M.; Co, I.S.; Gilmour, D.S. RNAi screen in Drosophila larvae identifies histone deacetylase 3 as a positive regulator of the hsp70 heat shock gene expression during heat shock. Biochim. Biophys. Acta 2014, 1839, 355–363. [Google Scholar] [CrossRef] [Green Version]
- Li, Z.; Qian, W.; Song, W.; Zhao, T.; Yang, Y.; Wang, W.; Wei, L.; Zhao, D.; Li, Y.; Perrimon, N.; et al. A salivary gland-secreted peptide regulates insect systemic growth. Cell. Rep. 2022, 38, 110397. [Google Scholar] [CrossRef]
- Fu, Z.W.; Li, J.H.; Feng, Y.R.; Yuan, X.; Lu, Y.T. The metabolite methylglyoxal-mediated gene expression is associated with histone methylglyoxalation. Nucleic Acids Res. 2021, 49, 1886–1899. [Google Scholar] [CrossRef] [PubMed]
- Mortazavi, A.; Williams, B.A.; McCue, K.; Schaeffer, L.; Wold, B. Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat. Methods 2008, 5, 621–628. [Google Scholar] [CrossRef] [PubMed]
- Hong, S.; Song, W.; Zushin, P.H.; Liu, B.; Jedrychowski, M.P.; Mina, A.I.; Deng, Z.; Cabarkapa, D.; Hall, J.A.; Palmer, C.J.; et al. Phosphorylation of Beta-3 adrenergic receptor at serine 247 by ERK MAP kinase drives lipolysis in obese adipocytes. Mol. Metab. 2018, 12, 25–38. [Google Scholar] [CrossRef] [PubMed]




Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhao, T.; Wang, M.; Li, Z.; Li, H.; Yuan, D.; Zhang, X.; Guo, M.; Qian, W.; Cheng, D. Wds-Mediated H3K4me3 Modification Regulates Lipid Synthesis and Transport in Drosophila. Int. J. Mol. Sci. 2023, 24, 6125. https://doi.org/10.3390/ijms24076125
Zhao T, Wang M, Li Z, Li H, Yuan D, Zhang X, Guo M, Qian W, Cheng D. Wds-Mediated H3K4me3 Modification Regulates Lipid Synthesis and Transport in Drosophila. International Journal of Molecular Sciences. 2023; 24(7):6125. https://doi.org/10.3390/ijms24076125
Chicago/Turabian StyleZhao, Tujing, Min Wang, Zheng Li, Hao Li, Dongqin Yuan, Xing Zhang, Mengge Guo, Wenliang Qian, and Daojun Cheng. 2023. "Wds-Mediated H3K4me3 Modification Regulates Lipid Synthesis and Transport in Drosophila" International Journal of Molecular Sciences 24, no. 7: 6125. https://doi.org/10.3390/ijms24076125



