The Expression Pattern of tRNA-Derived Small RNAs in Adult Drosophila and the Function of tRF-Trp-CCA-014-H3C4 Network Analysis
Abstract
1. Introduction
2. Results
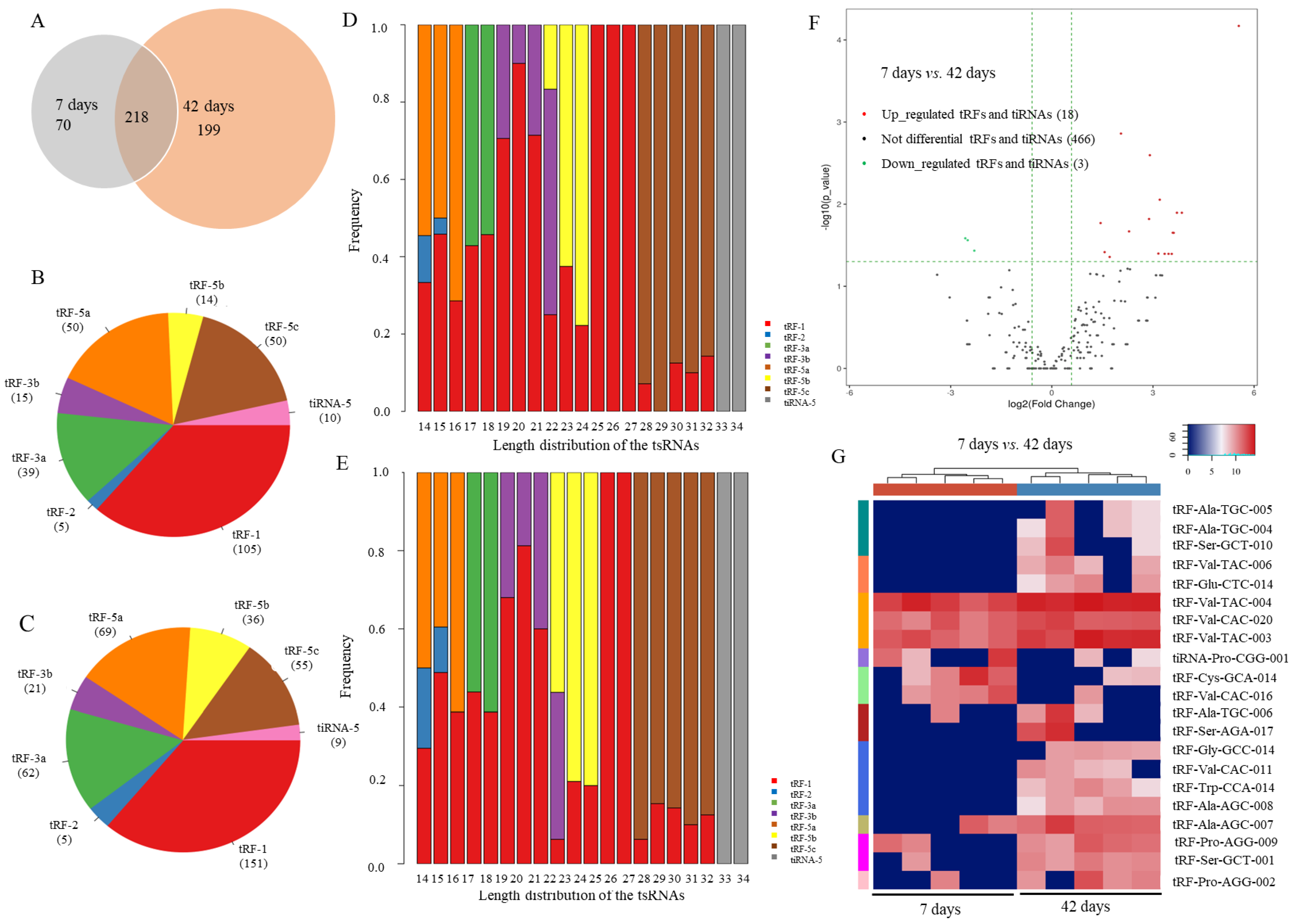
2.1. Expression Pattern of Drosophila tsRNAs
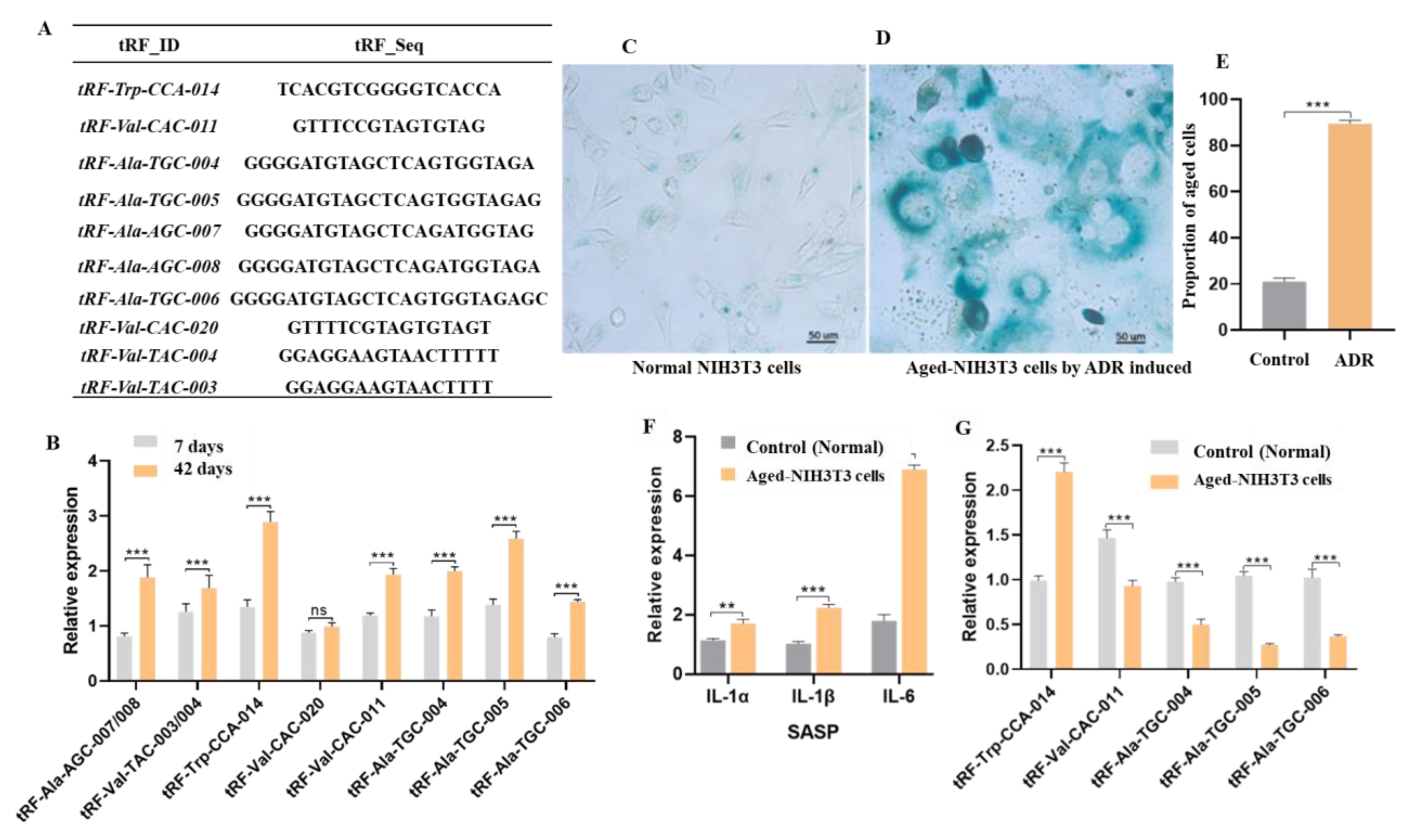
2.2. Verification of Differentially Expressed tsRNAs
2.3. Expression Features of tRF-Trp-CCA-014
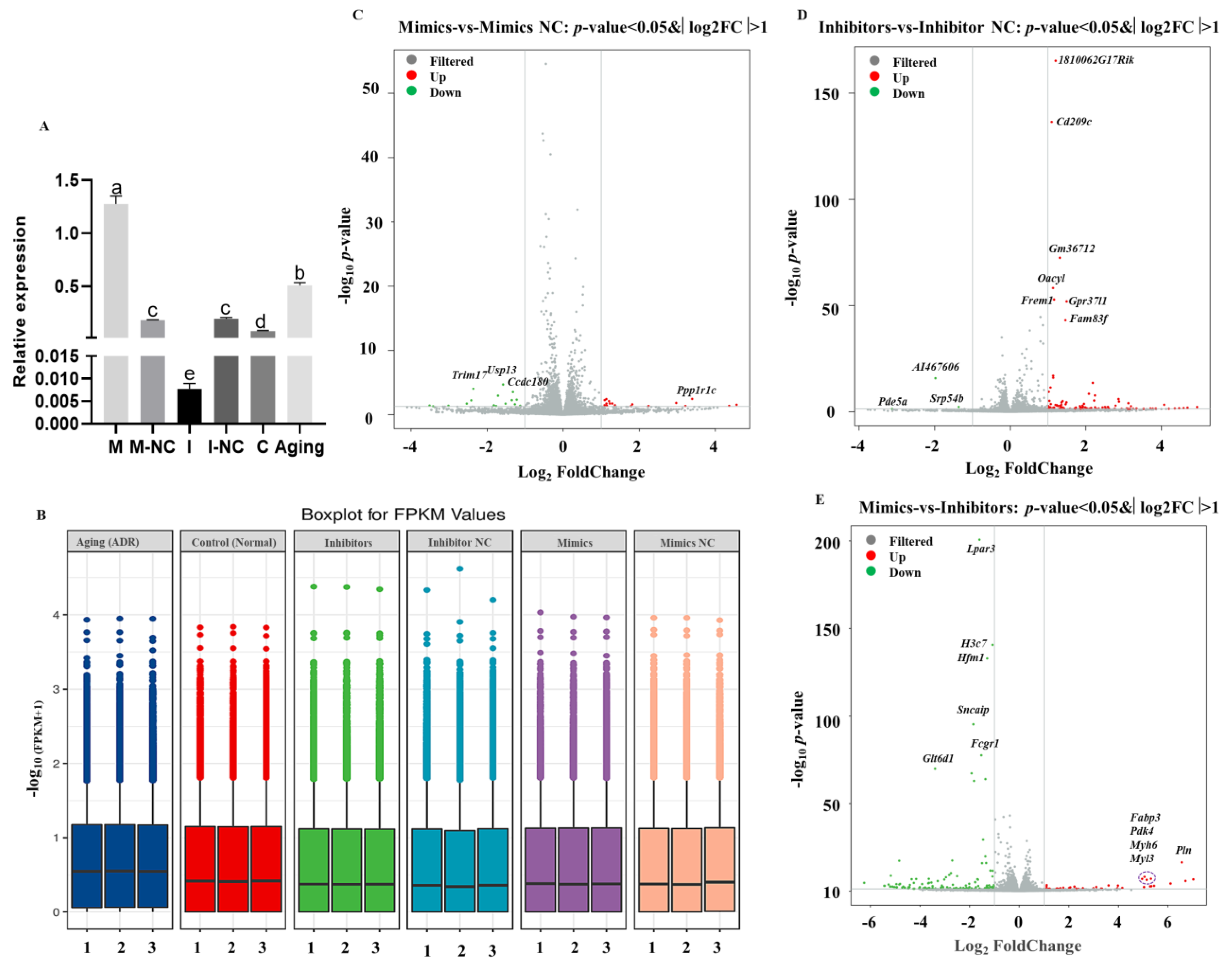
2.4. RNA-seq of Mouse NIH3T3 Cells
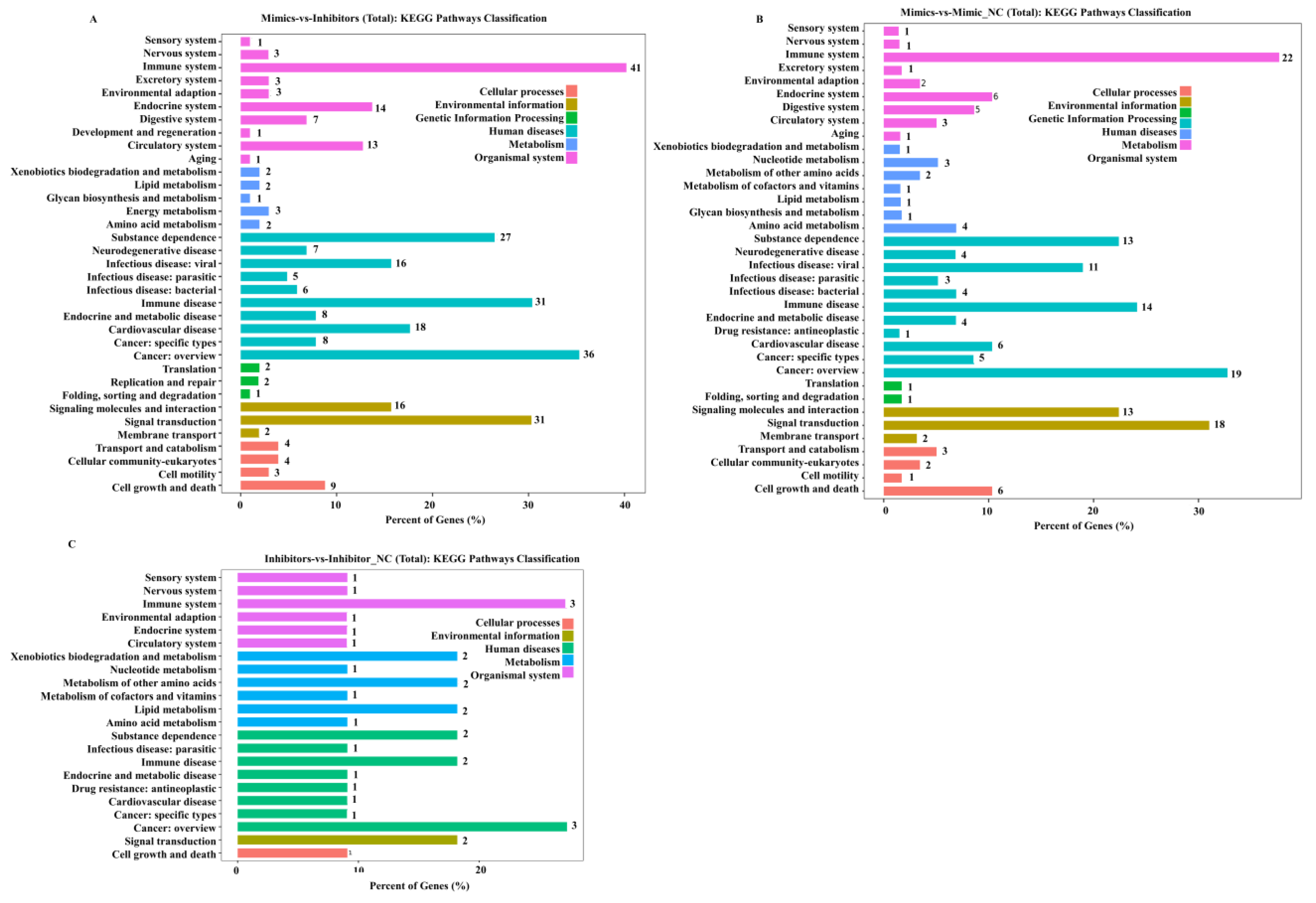
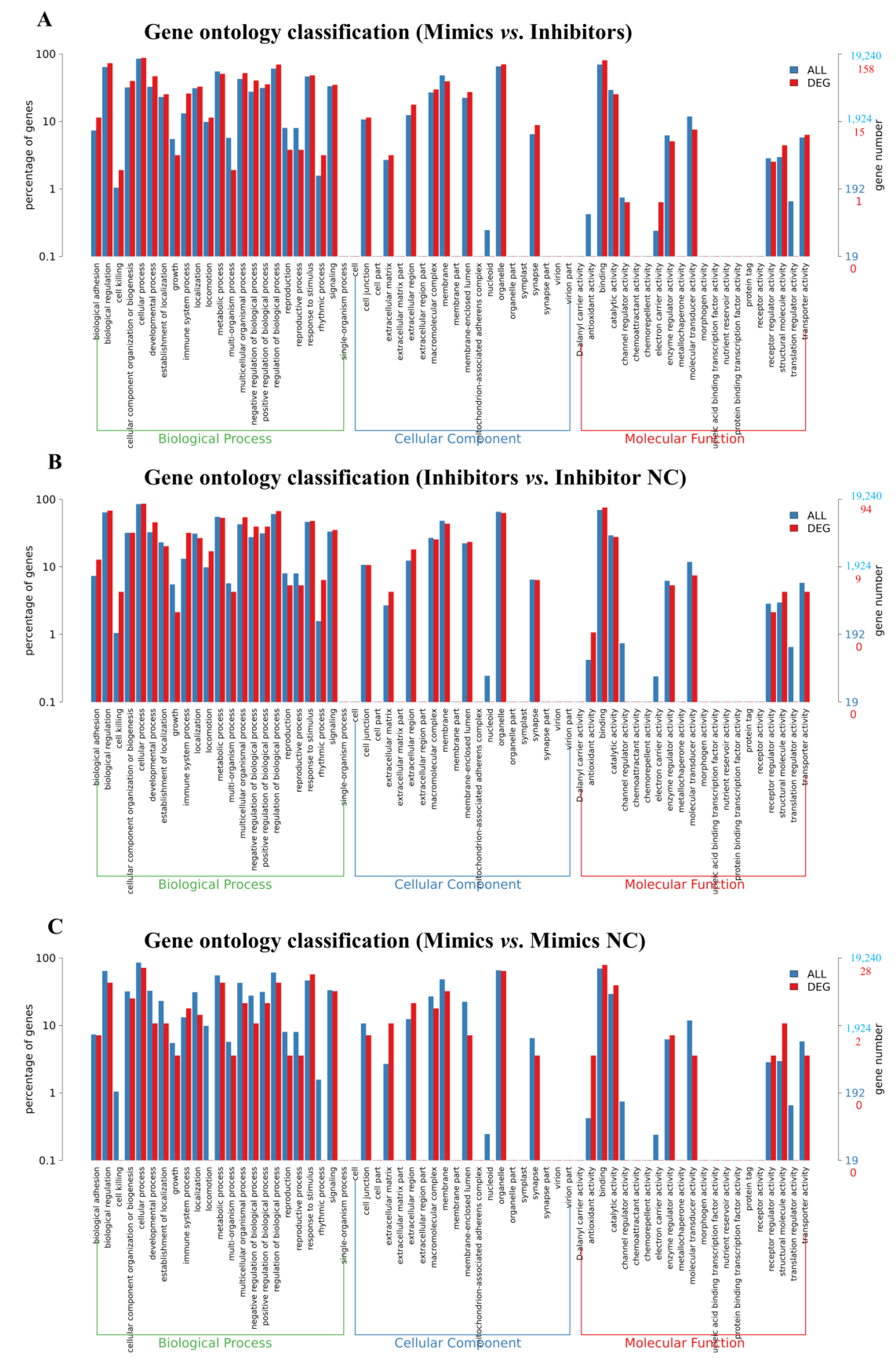
2.5. KEGG and GO Enrichment of DEGs
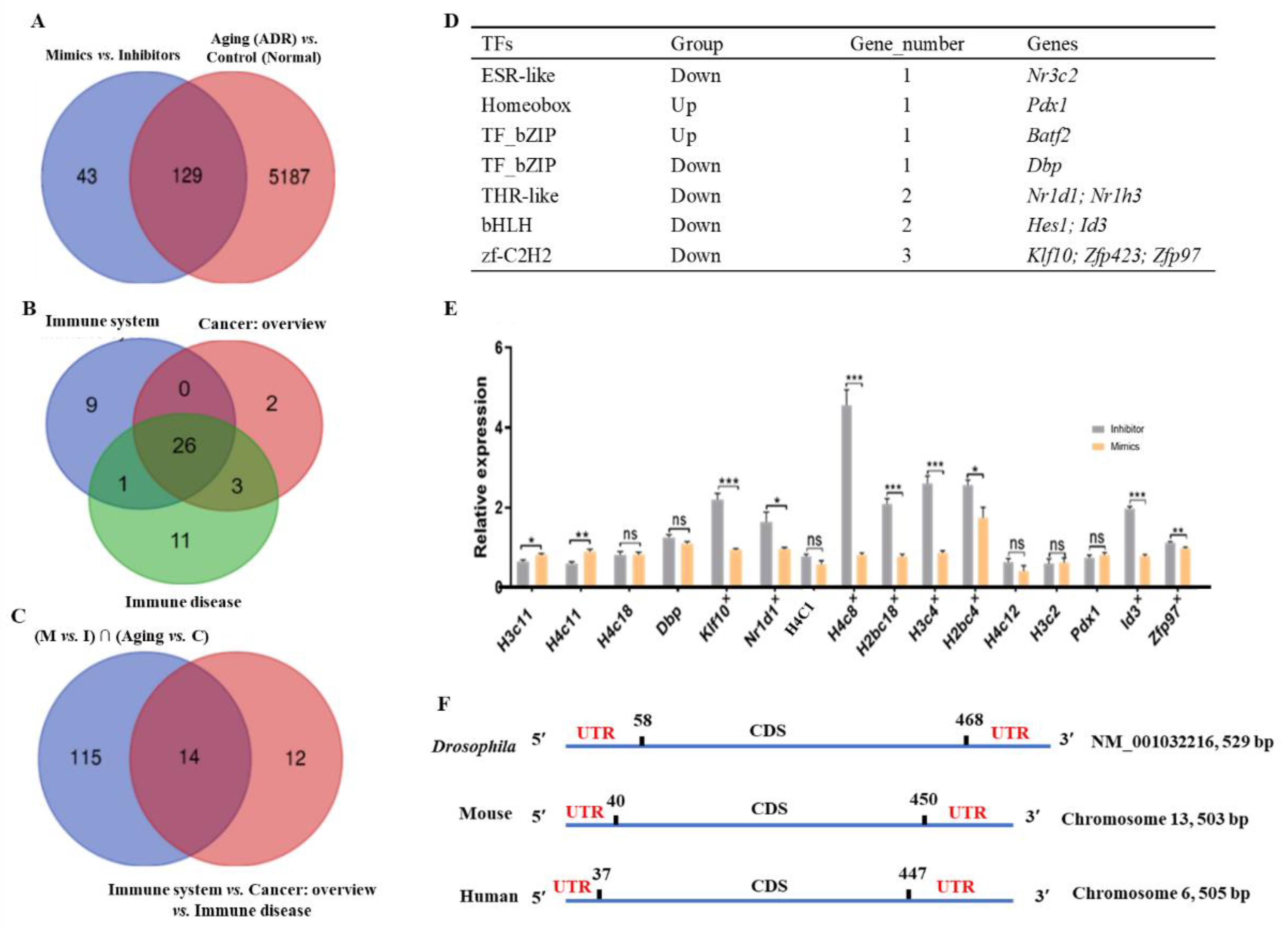
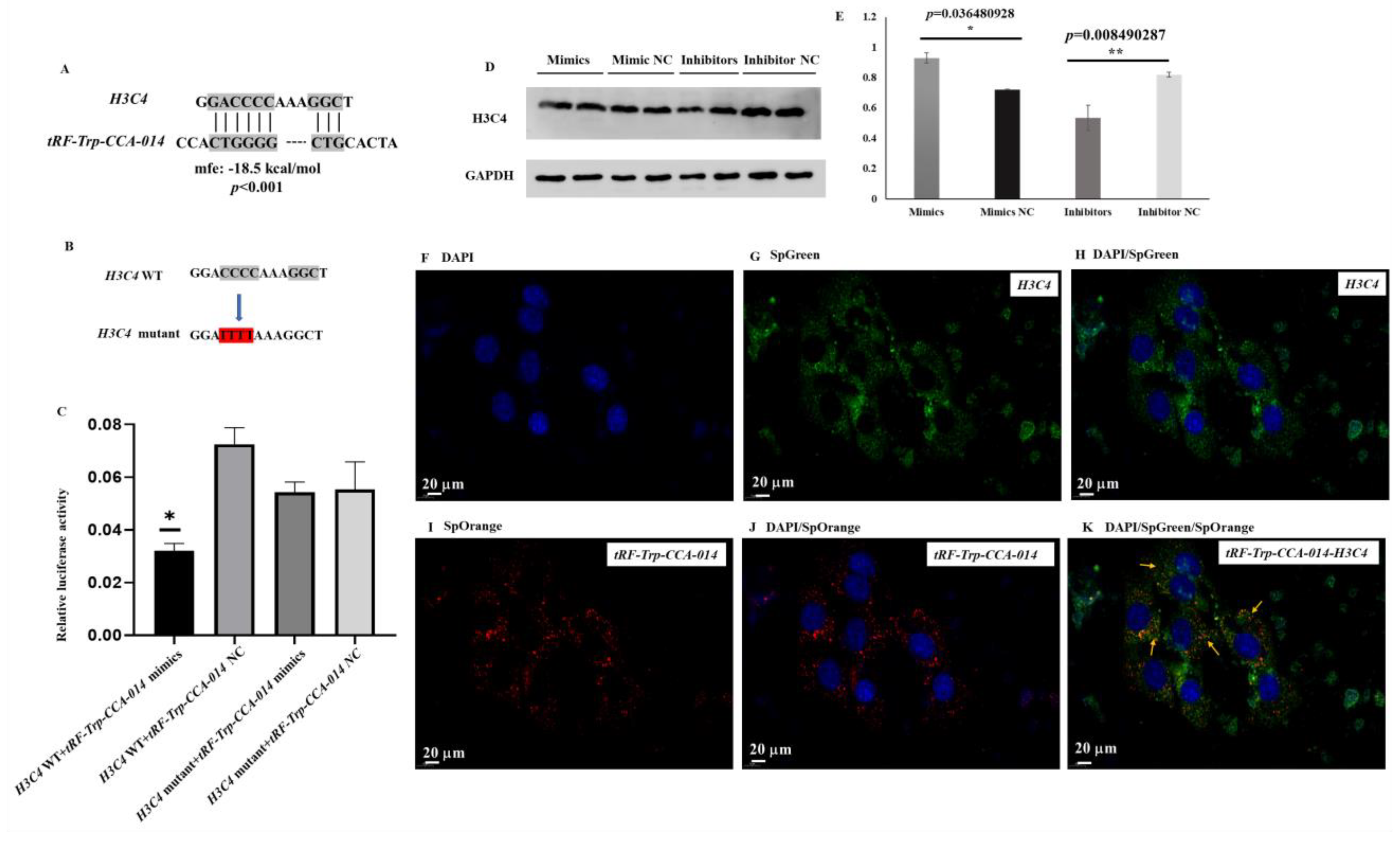
2.6. Analysis of tRF-Trp-CCA-014 Targets
2.7. Analysis of the tRF-Trp-CCA-014-H3C4 Network
3. Discussion
4. Materials and Methods
4.1. tsRNAs Analysis
4.2. GO and KEGG Enrichment
4.3. qPCR
4.4. Cell Culture and Cell Senescence Model
4.5. SA-β-gal Staining
4.6. Nuclear Cytoplasmic Separation
4.7. Tissue Expression Specificity of tRF-Trp-CCA-014
4.8. RNA-seq
4.9. Target Prediction of tRF-Trp-CCA-014
4.10. Dual-Luciferase Reporter Gene Assay
4.11. Western Blot
4.12. RNA in Situ Hybridization
4.13. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Weng, Q.; Wang, Y.; Xie, Y.; Yu, X.; Zhang, S.; Ge, J.; Li, Z.; Ye, G.; Guo, J. Extracellular vesicles-associated tRNA-derived fragments (tRFs): Biogenesis, biological functions, and their role as potential biomarkers in human diseases. J. Mol. Med. 2022, 100, 679–695. [Google Scholar] [CrossRef]
- Yu, X.; Xie, Y.; Zhang, S.; Song, X.; Xiao, B.; Yan, Z. tRNA-derived fragments: Mechanisms underlying their regulation of gene expression and potential applications as therapeutic targets in cancers and virus infections. Theranostics 2021, 11, 461–469. [Google Scholar] [CrossRef] [PubMed]
- Goodarzi, H.; Liu, X.; Nguyen, H.C.; Zhang, S.; Fish, L.; Tavazoie, S.F. Endogenous tRNA-Derived Fragments Suppress Breast Cancer Progression via YBX1 Displacement. Cell 2015, 161, 790–802. [Google Scholar] [CrossRef] [PubMed]
- Deng, H.; Wang, J.; Ye, D.; Chen, J.; Qiu, S.; Tang, M.; Zhou, C.; Shen, Y.; Fang, S.; Shen, Z.; et al. A 5′-tiRNA fragment that inhibits proliferation and migration of laryngeal squamous cell carcinoma by targeting PIK3CD. Genomics 2022, 114, 110392. [Google Scholar] [CrossRef] [PubMed]
- Mesquita-Ribeiro, R.; Fort, R.S.; Rathbone, A.; Farias, J.; Lucci, C.; James, V.; Sotelo-Silveira, J.; Duhagon, M.A.; Dajas-Bailador, F. Distinct small non-coding RNA landscape in the axons and released extracellular vesicles of developing primary cortical neurons and the axoplasm of adult nerves. RNA Biol. 2021, 18, 832–855. [Google Scholar] [CrossRef]
- Su, Z.; Monshaugen, I.; Wilson, B.; Wang, F.; Klungland, A.; Ougland, R.; Dutta, A. TRMT6/61A-dependent base methylation of tRNA-derived fragments regulates gene-silencing activity and the unfolded protein response in bladder cancer. Nat. Commun. 2022, 13, 2165. [Google Scholar] [CrossRef]
- Soares, A.R.; Fernandes, N.; Reverendo, M.; Araujo, H.R.; Oliveira, J.L.; Moura, G.M.; Santos, M.A. Conserved and highly expressed tRNA derived fragments in zebrafish. BMC Mol. Biol. 2015, 16, 22. [Google Scholar] [CrossRef]
- Shin, G.; Koo, H.J.; Seo, M.; Lee, S.V.; Nam, H.G.; Jung, G.Y. Transfer RNA-derived fragments in aging Caenorhabditis elegans originate from abundant homologous gene copies. Sci. Rep. 2021, 11, 12304. [Google Scholar] [CrossRef]
- Zhong, F.; Hu, Z.; Jiang, K.; Lei, B.; Wu, Z.; Yuan, G.; Luo, H.; Dong, C.; Tang, B.; Zheng, C.; et al. Complement C3 activation regulates the production of tRNA-derived fragments Gly-tRFs and promotes alcohol-induced liver injury and steatosis. Cell Res. 2019, 29, 548–561. [Google Scholar] [CrossRef]
- Mleczko, A.M.; Celichowski, P.; Bakowska-Zywicka, K. Transfer RNA-derived fragments target and regulate ribosome-associated aminoacyl-transfer RNA synthetases. Biochim. Biophys. Acta Gene Regul. Mech. 2018, 1861, 647–656. [Google Scholar] [CrossRef]
- Karaiskos, S.; Naqvi, A.S.; Swanson, K.E.; Grigoriev, A. Age-driven modulation of tRNA-derived fragments in Drosophila and their potential targets. Biol. Direct 2015, 10, 51. [Google Scholar] [CrossRef] [PubMed]
- Wang, B.G.; Yan, L.R.; Xu, Q.; Zhong, X.P. The role of Transfer RNA-Derived Small RNAs (tsRNAs) in Digestive System Tumors. J. Cancer 2020, 11, 7237–7245. [Google Scholar] [CrossRef] [PubMed]
- Kumar, P.; Kuscu, C.; Dutta, A. Biogenesis and Function of Transfer RNA-Related Fragments (tRFs). Trends Biochem. Sci. 2016, 41, 679–689. [Google Scholar] [CrossRef] [PubMed]
- Saikia, M.; Krokowski, D.; Guan, B.J.; Ivanov, P.; Parisien, M.; Hu, G.F.; Anderson, P.; Pan, T.; Hatzoglou, M. Genome-wide identification and quantitative analysis of cleaved tRNA fragments induced by cellular stress. J. Biol. Chem. 2012, 287, 42708–42725. [Google Scholar] [CrossRef]
- Ding, M.; Li, H.; Zheng, L. Drosophila exercise, an emerging model bridging the fields of exercise and aging in human. Front. Cell Dev. Biol. 2022, 10, 966531. [Google Scholar] [CrossRef] [PubMed]
- Zong, T.; Yang, Y.; Lin, X.; Jiang, S.; Zhao, H.; Liu, M.; Meng, Y.; Li, Y.; Zhao, L.; Tang, G.; et al. 5′-tiRNA-Cys-GCA regulates VSMC proliferation and phenotypic transition by targeting STAT4 in aortic dissection. Mol. Ther. Nucleic Acids 2021, 26, 295–306. [Google Scholar] [CrossRef] [PubMed]
- Guan, L.; Grigoriev, A. Age-Related Argonaute Loading of Ribosomal RNA Fragments. Microrna 2020, 9, 142–152. [Google Scholar] [CrossRef] [PubMed]
- Goktas, C.; Yigit, H.; Cosacak, M.I.; Akgul, B. (Differentially) Expressed tRNA-Derived Small RNAs Co-Sediment Primarily with Non-Polysomal Fractions in Drosophila. Genes 2017, 8, 333. [Google Scholar] [CrossRef] [PubMed]
- Cosacak, M.I.; Yigit, H.; Kizil, C.; Akgul, B. Re-Arrangements in the Cytoplasmic Distribution of Small RNAs Following the Maternal-to-Zygotic Transition in Drosophila Embryos. Genes 2018, 9, 82. [Google Scholar] [CrossRef]
- Sun, T.Y.; Zhang, L.X.; Feng, J.L.; Bao, L.Y.; Wang, J.Q.; Song, Z.Z.; Mao, Z.F.; Li, J.; Hu, Z. Characterization of cellular senescence in doxorubicin-induced aging mice. Exp. Gerontol. 2022, 163, 111800. [Google Scholar] [CrossRef]
- Wiley, C.D.; Velarde, M.C.; Lecot, P.; Liu, S.; Sarnoski, E.A.; Freund, A.; Shirakawa, K.; Lim, H.W.; Davis, S.S.; Ramanathan, A.; et al. Mitochondrial Dysfunction Induces Senescence with a Distinct Secretory Phenotype. Cell Metab. 2016, 23, 303–314. [Google Scholar] [CrossRef] [PubMed]
- Ortiz-Montero, P.; Londoño-Vallejo, A.; Vernot, J.P. Senescence-associated IL-6 and IL-8 cytokines induce a self- and cross-reinforced senescence/inflammatory milieu strengthening tumorigenic capabilities in the MCF-7 breast cancer cell line. Cell Commun. Signal. 2017, 15, 17. [Google Scholar] [CrossRef] [PubMed]
- Martínez-Zamudio, R.I.; Dewald, H.K.; Vasilopoulos, T.; Gittens-Williams, L.; Fitzgerald-Bocarsly, P.; Herbig, U. Fitzgerald-Bocarsly, and U. Herbig. Senescence-associated β-galactosidase reveals the abundance of senescent CD8+ T cells in aging humans. Aging Cell 2021, 20, e13344. [Google Scholar] [PubMed]
- Li, W.S.; Li, Y.L.; Cao, R.; Ha, C.F.; Sun, S.; Yu, L.; Li, J. Differential Expression and Bioinformatics Analysis of tRF/tiRNA in Endometriosis Patients. Biomed. Res. Int. 2022, 2022, 9911472. [Google Scholar] [CrossRef]
- Molla-Herman, A.; Angelova, M.T.; Ginestet, M.; Carre, C.; Antoniewski, C.; Huynh, J.R. tRNA Fragments Populations Analysis in Mutants Affecting tRNAs Processing and tRNA Methylation. Front. Genet. 2020, 11, 518949. [Google Scholar] [CrossRef]
- Yin, L.; Sun, Y.; Wu, J.; Yan, S.; Deng, Z.; Wang, J.; Liao, S.; Yin, D.; Li, G. Discovering novel microRNAs and age-related nonlinear changes in rat brains using deep sequencing. Neurobiol. Aging 2015, 36, 1037–1044. [Google Scholar] [CrossRef]
- Karaiskos, S.; Grigoriev, A. Dynamics of tRNA fragments and their targets in aging mammalian brain. F1000Research 2016, 5, 2758. [Google Scholar] [CrossRef]
- Panowski, S.H.; Dillin, A. Signals of youth: Endocrine regulation of aging in Caenorhabditis elegans. Trends Endocrinol. Metab. 2009, 20, 259–264. [Google Scholar] [CrossRef]
- Chen, F.; Song, C.; Meng, F.; Zhu, Y.; Chen, X.; Fang, X.; Ma, D.; Wang, Y.; Zhang, C. 5′-tRF-GlyGCC promotes breast cancer metastasis by increasing fat mass and obesity-associated protein demethylase activity. Int. J. Biol. Macromol. 2023, 226, 397–409. [Google Scholar] [CrossRef]
- Green, D.; Fraser, W.D.; Dalmay, T. Transfer RNA-derived small RNAs in the cancer transcriptome. Pflugers Arch. 2016, 468, 1041–1047. [Google Scholar] [CrossRef]
- Yamasaki, S.; Ivanov, P.; Hu, G.F.; Anderson, P. Angiogenin cleaves tRNA and promotes stress-induced translational repression. J. Cell Biol. 2009, 185, 35–42. [Google Scholar] [CrossRef]
- Kumar, P.; Anaya, J.; Mudunuri, S.B.; Dutta, A. Meta-analysis of tRNA derived RNA fragments reveals that they are evolutionarily conserved and associate with AGO proteins to recognize specific RNA targets. BMC Biol. 2014, 12, 78. [Google Scholar] [CrossRef] [PubMed]
- Zhang, S.; Li, H.; Zheng, L.; Li, H.; Feng, C.; Zhang, W. Identification of functional tRNA-derived fragments in senescence-accelerated mouse prone 8 brain. Aging 2019, 11, 10485–10498. [Google Scholar] [CrossRef] [PubMed]
- Barwell, T.; DeVeale, B.; Poirier, L.; Zheng, J.; Seroude, F.; Seroude, L. Regulating the UAS/GAL4 system in adult Drosophila with Tet-off GAL80 transgenes. PeerJ 2017, 5, e4167. [Google Scholar] [CrossRef] [PubMed]
- Casella, G.; Munk, R.; Kim, K.M.; Piao, Y.L.; De, S.; Abdelmohsen, K.; Gorospe, M. Transcriptome signature of cellular senescence. Nucleic Acids Res. 2019, 47, 11476. [Google Scholar] [CrossRef] [PubMed]
- Ren, J.; Wu, X.; Shang, F.F.; Qi, Y.; Tang, Z.; Wen, C.; Cao, W.; Cheng, Q.; Tan, L.; Chen, H.; et al. The tRNA-Cys-GCA Derived tsRNAs Suppress Tumor Progression of Gliomas via Regulating VAV2. Dis. Mark. 2022, 2022, 8708312. [Google Scholar] [CrossRef] [PubMed]
- Tachiwana, H.; Osakabe, A.; Shiga, T.; Miya, Y.; Kimura, H.; Kagawa, W.; Kurumizaka, H. Structures of human nucleosomes containing major histone H3 variants. Acta Crystallogr. D Biol. Crystallogr. 2011, 67, 578–583. [Google Scholar] [CrossRef]
- Hake, S.B.; Garcia, B.A.; Duncan, E.M.; Kauer, M.; Dellaire, G.; Shabanowitz, J.; Bazett-Jones, D.P.; Allis, C.D.; Hunt, D.F. Expression patterns and post-translational modifications associated with mammalian histone H3 variants. J. Biol. Chem. 2006, 281, 559–568. [Google Scholar] [CrossRef]
- Tagami, H.; Ray-Gallet, D.; Almouzni, G.; Nakatani, Y. Histone H3.1 and H3.3 complexes mediate nucleosome assembly pathways dependent or independent of DNA synthesis. Cell 2004, 116, 51–61. [Google Scholar]
- Fingerman, I.M.; Li, H.C.; Briggs, S.D. A charge-based interaction between histone H4 and Dot1 is required for H3K79 methylation and telomere silencing: Identification of a new trans-histone pathway. Genes Dev. 2007, 21, 2018–2029. [Google Scholar] [CrossRef]
- Levy, M.A.; Tian, J.; Gandelman, M.; Cheng, H.; Tsapekos, M.; Crego, S.R.; Maddela, R.; Sinnott, R. A Multivitamin Mixture Protects against Oxidative Stress-Mediated Telomere Shortening. J. Diet. Suppl. 2023, 1–18. [Google Scholar] [CrossRef] [PubMed]
- Sha, Z.; Hou, T.; Zhou, T.; Dai, Y.; Bao, Y.; Jin, Q.; Ye, J.; Lu, Y.M.; Wu, L.Q. Causal relationship between atrial fibrillation and leukocyte telomere length: A two sample, bidirectional Mendelian randomization study. Front. Cardiovasc. Med. 2023, 10, 1093255. [Google Scholar] [CrossRef] [PubMed]
- Yang, D.Y.; Xiao, F.; Li, J.M.; Wang, S.Q.; Fan, X.L.; Ni, Q.Y.; Li, Y.; Zhang, M.W.; Yan, T.M.; Yang, M.Y.; et al. Age-related ceRNA networks in adult Drosophila ageing. Front. Genet. 2023, 14, 1096902. [Google Scholar] [CrossRef] [PubMed]
- Martin, M. Cutadapt Removes Adapter Sequences From High-Throughput Sequencing Reads. EMBnet J. 2006, 17, 10–12. [Google Scholar] [CrossRef]
- Langmead, B.; Trapnell, C.; Pop, M.; Salzberg, S.L. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009, 10, R25. [Google Scholar] [CrossRef] [PubMed]
- McDermaid, A.; Monier, B.; Zhao, J.; Liu, B.; Ma, Q. Interpretation of differential gene expression results of RNA-seq data: Review and integration. Brief. Bioinform. 2019, 20, 2044–2054. [Google Scholar] [CrossRef] [PubMed]
- Young, M.D.; Wakefield, M.J.; Smyth, G.K.; Oshlack, A. Gene ontology analysis for RNA-seq: Accounting for selection bias. Genome Biol. 2010, 11, R14. [Google Scholar] [CrossRef]
- Yu, G.; Wang, L.G.; Han, Y.; He, Q.Y. ClusterProfiler: An R package for comparing biological themes among gene clusters. Omics J. Integr. Biol. 2012, 16, 284–287. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Namdar, P.; YekeFallah, L.; Jalalian, F.; Barikani, A.; Razaghpoor, A. Improving Door-to-Balloon Time for Patients With Acute ST-Elevation Myocardial Infarction: A Controlled Clinical Trial. Curr. Probl. Cardiol. 2021, 46, 100674. [Google Scholar] [CrossRef]
- McGarvey, P.; Huang, J.; McCoy, M.; Orvis, J.; Katsir, Y.; Lotringer, N.; Nesher, I.; Kavarana, M.; Sun, M.; Peet, R.; et al. De novo assembly and annotation of transcriptomes from two cultivars of Cannabis sativa with different cannabinoid profiles. Gene 2020, 762, 145026. [Google Scholar] [CrossRef] [PubMed]
- Kim, D.; Langmead, B.; Salzberg, S.L. HISAT: A fast spliced aligner with low memory requirements. Nat. Methods 2015, 12, 357–360. [Google Scholar] [CrossRef] [PubMed]
- Anders, S.; Pyl, P.T.; Huber, W. HTSeq—A Python framework to work with high-throughput sequencing data. Bioinformatics 2015, 31, 166–169. [Google Scholar] [CrossRef] [PubMed]
- Ghosh, S.; Chan, C.K. Analysis of RNA-Seq Data Using TopHat and Cufflinks. Methods Mol. Biol. 2016, 1374, 339–361. [Google Scholar]
- Li, Y.; Li, Z.; Liu, J.; Liu, Y.; Miao, G. miR-190-5p Alleviates Myocardial Ischemia-Reperfusion Injury by Targeting PHLPP1. Dis. Mark. 2021, 2021, 8709298. [Google Scholar] [CrossRef]
- Xu, W.; Zhou, B.; Wang, J.; Tang, L.; Hu, Q.; Wang, J.; Chen, H.; Zheng, J.; Yan, F.; Chen, H. tRNA-Derived Fragment tRF-Glu-TTC-027 Regulates the Progression of Gastric Carcinoma via MAPK Signaling Pathway. Front. Oncol. 2021, 11, 733763. [Google Scholar] [CrossRef]

| Gene Name | Forwards Primers | Reverse Primers |
|---|---|---|
| tRF-Trp-CCA-014 | GGCGGTCACGTCGGGG | AGTGCAGGGTCCGAGGTATT |
| tRF-Val-CAC-020 | GCGCGCGGTTTTCGTAG | AGTGCAGGGTCCGAGGTATT |
| tRF-Val-TAC-003 | GCGCGCGGAGGAAGTA | AGTGCAGGGTCCGAGGTATT |
| tRF-Val-TAC-004 | GCGCGCGGAGGAAGTAA | AGTGCAGGGTCCGAGGTATT |
| tRF-Ala-AGC-007 | GCGGGGGATGTAGCTCAGA | AGTGCAGGGTCCGAGGTATT |
| tRF-Ala-AGC-008 | GCGGGGGATGTAGCTCAGAT | AGTGCAGGGTCCGAGGTATT |
| tRF-Val-CAC-011 | GCGCGCGGTTTCCGTA | AGTGCAGGGTCCGAGGTATT |
| tRF-Ala-TGC-004 | GCGGGGGATGTAGCTCAGT | AGTGCAGGGTCCGAGGTATT |
| tRF-Ala-TGC-005 | CGGGGGATGTAGCTCAGTG | AGTGCAGGGTCCGAGGTATT |
| tRF-Ala-TGC-006 | CGGGGGATGTAGCTCAGTGG | AGTGCAGGGTCCGAGGTATT |
| U6 | TCTTGCTTCGGCAGAACATA | GATTTTGCGTGTCATCCTTG |
| RP49 | GCCCAAGGGTATCGACAACA | CTTGCGCTTCTTGGAGGAGA |
| GAPDH | TCTTCCAGGCGAACCACTTC | AAGACGTCCACGAAGCGAAT |
| H3C11 | GTGAAGAAGCCTCACCGCTA | TACGTGCCAGTGTAGAAGGC |
| H4C11 | TGCTCCATAGCCATGTCTGG | TTGGTGATGCCCTGGATGTT |
| H4C18 | ATCTCCGGCCTCATCTACGA | TAATTAGCCGCCGAATCCGT |
| Dbp | GCAGAGTCCTGTTCCTTGCT | ATTGTGTTGATGGAGGCGGT |
| Klf10 | CACCAGTGTCATCCGTCACA | GGCTCAGGCTTGGATCTGTT |
| Nr1d1 | GAAGTGTCTCTCCGTTGGCA | GAAGTGTCTCTCCGTTGGCA |
| H4C1 | ACTGGCTAGTGAGCTTCCTT | TCGTAGATGAGGCCGGAGAT |
| H4C8 | ATCTCCGGCCTCATCTACGA | AAGGGCCTTTTGTAGCAGAAAT |
| H2bC18 | ATCACTTCCCGGGAGATCCA | AGCCTTTTGGGTAAAGCCGA |
| H3C4 | GCCTACCTTGTGGGTCTGTT | AAGAGCCTTTGGTTAATTCCGT |
| H2bC4 | TACAACAAGCGCTCGACCAT | GGAATTCGCTACGGAGGCTT |
| H4c12 | GTGAACGACATCTTCGAGCG | GGTGCTAGACGTCAACCCT |
| Pdx1 | AGCGTTCCAATACGGACCAG | TGCTCAGCCGTTCTGTTTCT |
| H3c2 | GTTGCTTGTTTCTACCATGCCC | TCGAGCGCTTGTTGTAATGC |
| Id3 | CTGAAGAGCTAGCACACGCT | CTCTCGACACCCCATTCTCG |
| Zfp97 | TCCGGAATCCTTTTCGCTGG | TGCCTGAGCTTCCTTCACAG |
| IL-1α | TCTCAGATTCACAACTGTTCGTG | AGAAAATGAGGTCGGTCTCACTA |
| IL-1β | GAAATGCCACCTTTTGACAGTG | TGGATGCTCTCATCAGGACAG |
| IL-6 | ATCCAGTTGCCTTCTTGGGACTGA | TAAGCCTCCGACTTGTGAAGTGGT |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yang, D.; Xiao, F.; Yuan, Y.; Li, J.; Wang, S.; Fan, X.; Ni, Q.; Li, Y.; Zhang, M.; Gu, X.; et al. The Expression Pattern of tRNA-Derived Small RNAs in Adult Drosophila and the Function of tRF-Trp-CCA-014-H3C4 Network Analysis. Int. J. Mol. Sci. 2023, 24, 6169. https://doi.org/10.3390/ijms24076169
Yang D, Xiao F, Yuan Y, Li J, Wang S, Fan X, Ni Q, Li Y, Zhang M, Gu X, et al. The Expression Pattern of tRNA-Derived Small RNAs in Adult Drosophila and the Function of tRF-Trp-CCA-014-H3C4 Network Analysis. International Journal of Molecular Sciences. 2023; 24(7):6169. https://doi.org/10.3390/ijms24076169
Chicago/Turabian StyleYang, Deying, Feng Xiao, Ya Yuan, Jiamei Li, Siqi Wang, Xiaolan Fan, Qingyong Ni, Yan Li, Mingwang Zhang, Xiaobin Gu, and et al. 2023. "The Expression Pattern of tRNA-Derived Small RNAs in Adult Drosophila and the Function of tRF-Trp-CCA-014-H3C4 Network Analysis" International Journal of Molecular Sciences 24, no. 7: 6169. https://doi.org/10.3390/ijms24076169
APA StyleYang, D., Xiao, F., Yuan, Y., Li, J., Wang, S., Fan, X., Ni, Q., Li, Y., Zhang, M., Gu, X., Yan, T., Yang, M., & He, Z. (2023). The Expression Pattern of tRNA-Derived Small RNAs in Adult Drosophila and the Function of tRF-Trp-CCA-014-H3C4 Network Analysis. International Journal of Molecular Sciences, 24(7), 6169. https://doi.org/10.3390/ijms24076169






