The RNA-Binding and RNA-Melting Activities of the Multifunctional Protein Nucleobindin 1
Abstract
:1. Introduction
2. Results
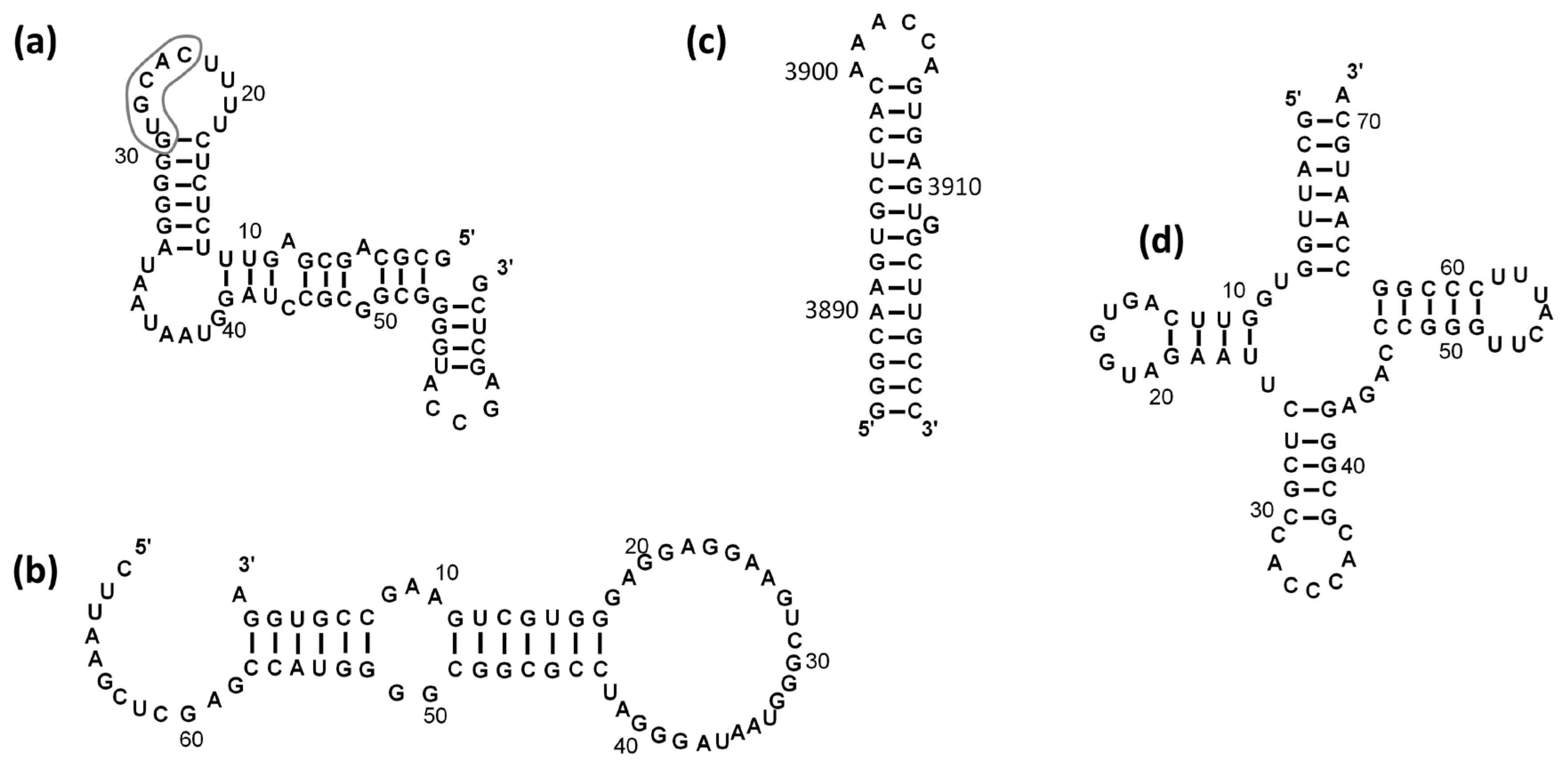
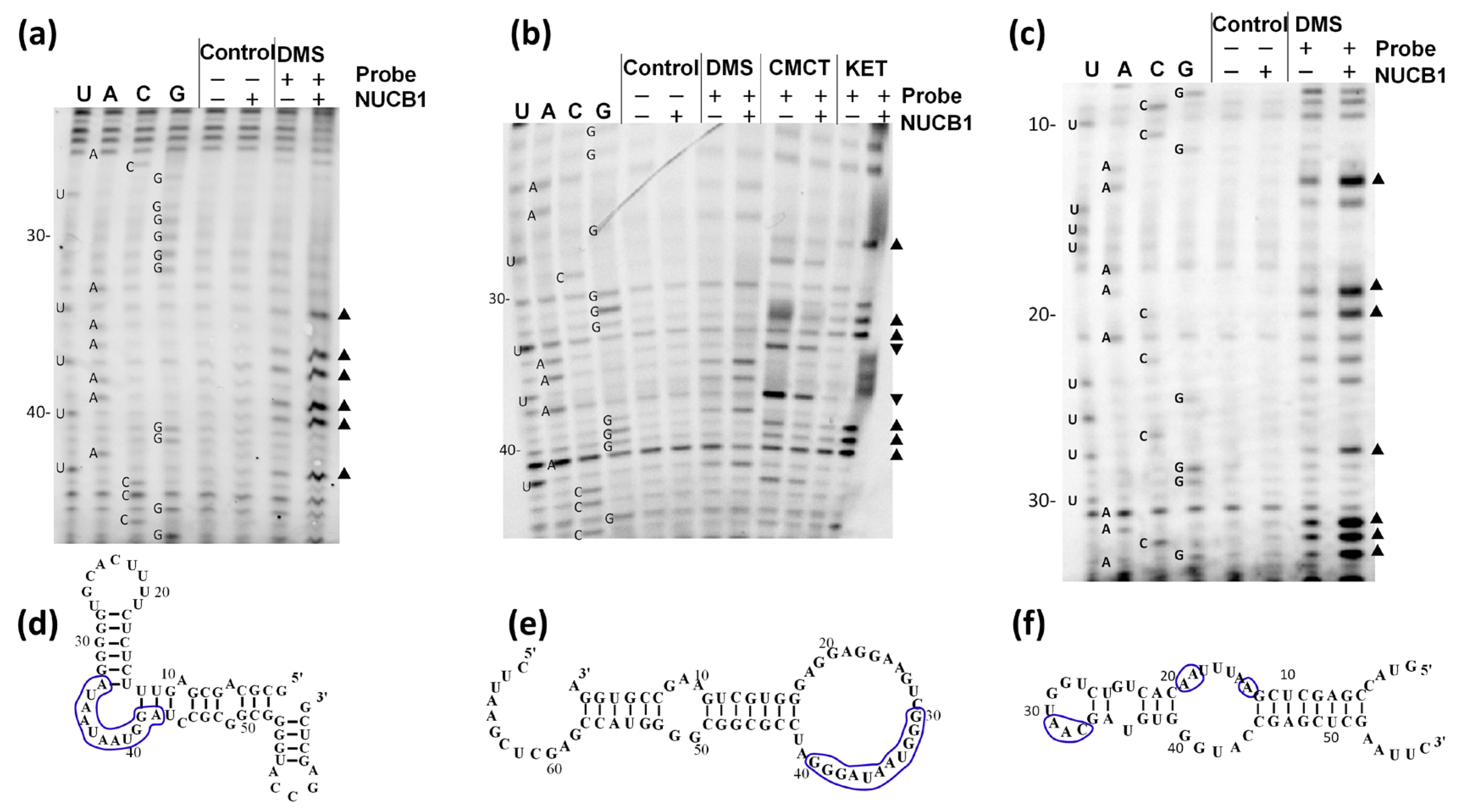
2.1. NUCB1 Is Able to Bind to RNA Fragments
2.2. NUCB1 Binds to the ssRNA GG Motif
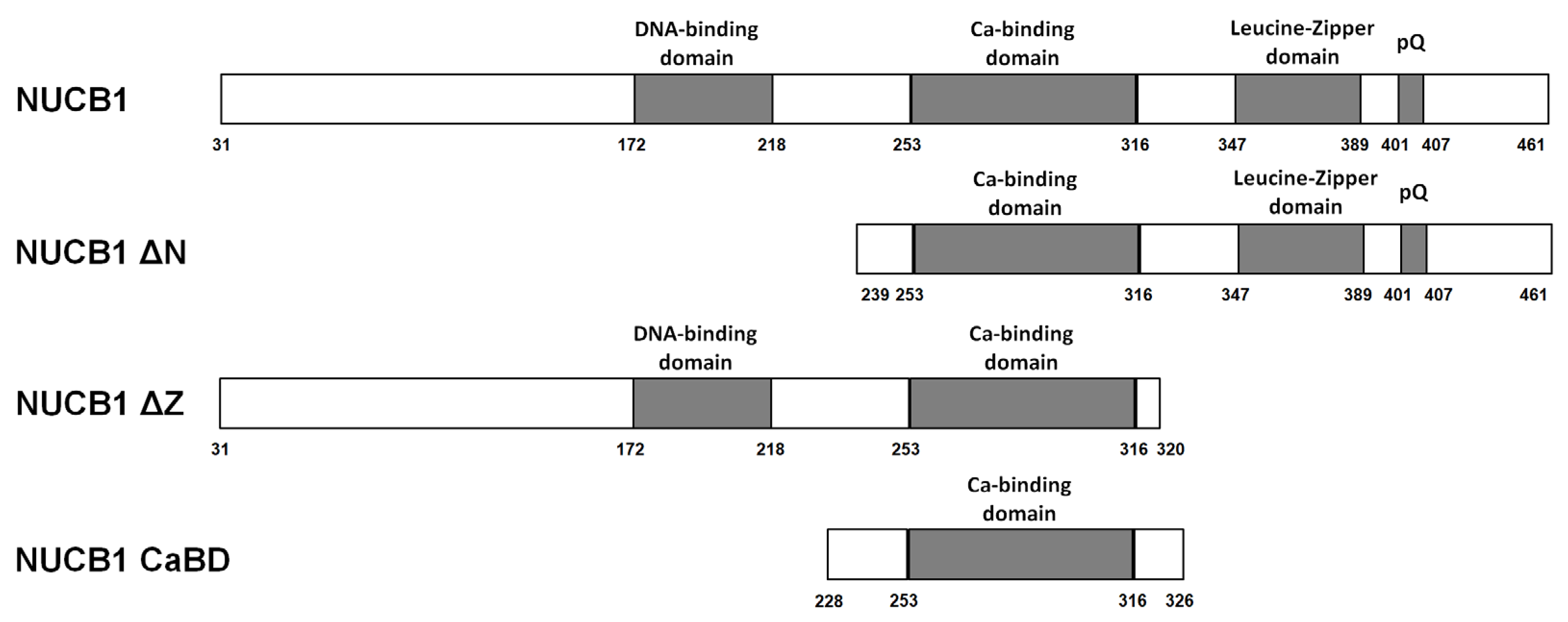
2.3. Truncated Forms of NUCB1 Bind to RNA Fragments
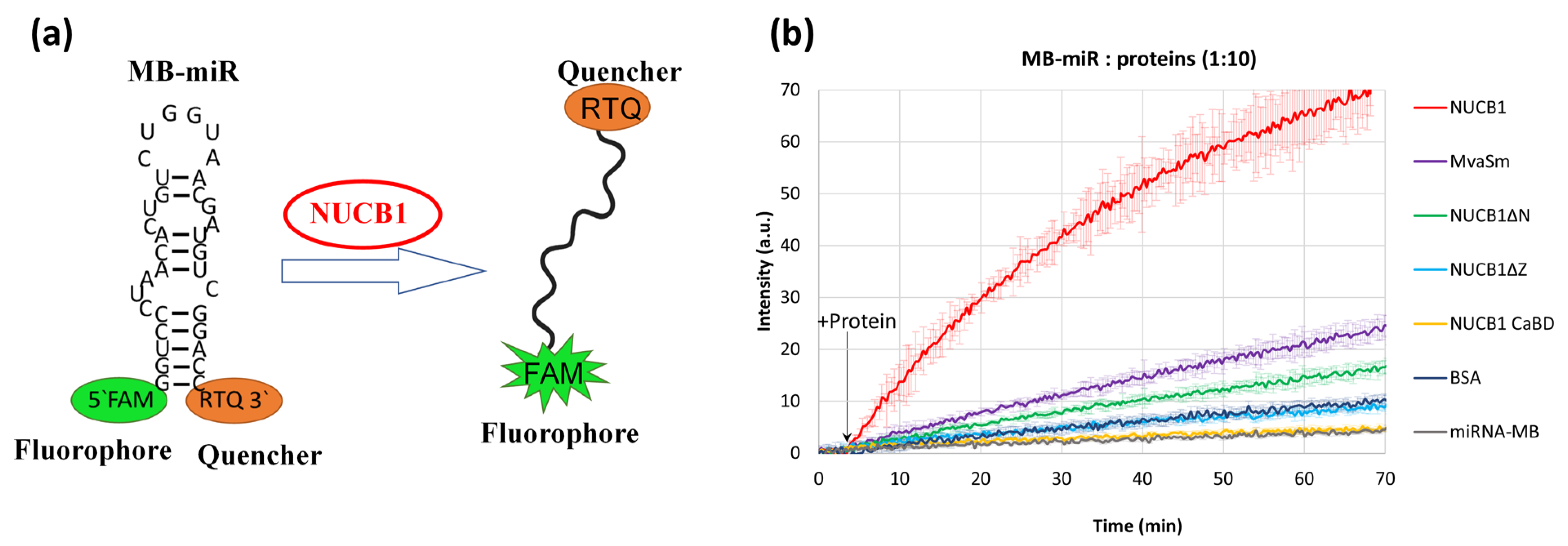
2.4. NUCB1 Can Melt RNA Hairpin Structures in RNAs
3. Discussion
3.1. Possible Physiological Role of the RNA-Binding Properties of NUCB1
3.2. NUCB1 Binds with High Specificity to a Single-Stranded GG RNA Sequences in a Ca2+-Independent Manner
3.3. Ca2+-Binding Domain of NUCB1 Is Sufficient for RNA Binding
3.4. NUCB1 Can Melt RNA Secondary Structures
4. Materials and Methods
4.1. Production and Purification of NUCB1 and Its Truncated Forms
4.2. Cloning and Purification of RNA Fragments
4.3. Analysis of NUCB1-RNA Interaction
- U-rich: 5′-GUGGUCAGUCGAGUGG-U18-3′
- A-rich: 5′-GUGGUCAGUCGAGUGG-A18-3′
- C-rich: 5′-GUGGUCAGUCGAGUGG-C6-3′
- C-rich(+AA): 5′-GUGGUCAGUCGAGUGG-AAC6-3′
- G-rich: 5′-GUGGUCAGUCGAGUGG-G6-3′
- miR-200a-3p: 5′-GUGGUCAGUCGAGUGGUAACACUGUCUGGUAACGAUGU-3′
- miR-203a-5p: 5′-GUGGUCAGUCGAGUGGAGUGGUUCUUAACAGUUCAACAGUU-3′
- miR-145-3p: 5′-GUGGUCAGUCGAGUGGGGAUUCCUGGAAAUACUGUUCU-3′
- miR-155-5p: 5′-GUGGUCAGUCGAGUGGUUAAUGCUAAUCGUGAUAGGGGUU-3′
- mRNA: 5′-AGGUGCCGAAGUCGUGGGAGGAGGAAGUCGGGUAAUAG-3′
- E-box RNA: 5′-GCGCAGCGAGUUUCUCUCUUUUCACGUGGGGGAUAAUAAU-3′
4.4. RNA Chemical Probing
4.4.1. Chemical Modifications of RNA
4.4.2. Fluorescent Primer Extension
4.5. Molecular Beacon Melting Assay
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Kanai, Y.; Tanuma, S. Purification of a novel B cell growth and differentiation factor associated with luous syndrome. Immunol. Lett. 1992, 21, 43–48. [Google Scholar] [CrossRef]
- Kanai, Y.; Takeda, O.; Kanai, Y.; Miura, K.; Kurosawa, Y. Novel autoimmune phenomena induced in vivo by a new DNA binding protein Nuc: A study on MRL/n mice. Immunol. Lett. 1993, 39, 83–89. [Google Scholar] [CrossRef]
- Leclerc, P.; Biarc, J.; St-Onge, M.; Gilbert, C.; Dussault, A.-A.; Laflamme, C.; Pouliot, M. Nucleobindin co-localizes and associates with cyclooxygenase (COX)-2 in human neutrophils. PLoS ONE 2008, 3, e2229. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kanuru, M.; Aradhyam, G.-K. Chaperone-like activity of calnuc prevents amyloid aggregation. Biochemistry 2017, 56, 149–159. [Google Scholar] [CrossRef] [PubMed]
- Bonito-Oliva, A.; Barbash, S.; Sakmar, T.P.; Graham, W.V. Nucleobindin 1 binds to multiple types of pre-fibrillar amyloid and inhibits fibrillization. Sci Rep. 2017, 7, 42880. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bonito-Oliva, A.; Schedin-Weiss, S.; Younesi, S.S.; Tiiman, A.; Adura, C.; Paknejad, N.; Brendel, M.; Romin, Y.; Parchem, R.J.; Graff, C.; et al. Conformation-specific antibodies against multiple amyloid protofibril species from a single amyloid immunogen. J. Cell Mol. Med. 2019, 23, 2103–2114. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kapoor, N.; Gupta, R.; Menon, S.T.; Folta-Stogniew, E.; Raleigh, D.R.; Sakmar, T.P. Nucleobindin 1 is a calcium-regulated guanine nucleotide dissociation inhibitor of Gαi1. J. Biol. Chem. 2010, 285, 31647–31660. [Google Scholar] [CrossRef] [Green Version]
- Xue, F.; Wu, Y.; Zhao, X.; Zhao, T.; Meng, Y.; Zhao, Z.; Guo, J.; Chen, W. CHIP mediates down-regulation of nucleobindin-1 in preosteoblast cell line models. Cell Signal. 2016, 28, 1058–1065. [Google Scholar] [CrossRef] [PubMed]
- Lin, P.; Li, F.; Zhang, Y.-W.; Huang, H.; Tong, G.; Farquhar, M.G.; Xu, H. Calnuc binds to Alzheimer’s beta-amyloid precursos protein and affects its biogenesis. J. Neurochem. 2007, 100, 1505–1514. [Google Scholar]
- Niphakis, M.J.; Lum, K.M.; Cognetta, A.B.; Correia, B.E.; Ichu, T.-A.; Olucha, J.; Brown, S.J.; Kundu, S.; Piscitelli, F.; Rosen, H.; et al. A global map of lipid-binding proteins and their ligandability in cells. Cell 2015, 161, 1668–1680. [Google Scholar] [CrossRef] [Green Version]
- Miura, K.; Titani, K.; Kurosawa, Y.; Kanai, Y. Molecular cloning of nucleobindin, a novel DNA-binding protein that contains both a signal peptide and a leucine zipper structure. Biochem. Biophys. Res. Commun. 1992, 187, 375–380. [Google Scholar] [CrossRef] [PubMed]
- Sinha, S.; Pattnaik, S.; Aradhyam, G.K. Molecular evolution guided functional analyses reveals Nuckeobindin-1 as a canonical E-box binding protein promoting Epithelial-to-Mesenchymal transition (EMT). Biochim. Biophys. Acta Proteins Proteom. 2019, 1867, 765–775. [Google Scholar] [CrossRef] [PubMed]
- Lavoie, C.; Meerloo, T.; Lin, P.; Farquhar, M.G. Calnuc, an EF-hand Ca(2+)-binding protein, is stored and processed in the Golgi and secreted by the constitutive-like pathway in AtT20 cells. Mol. Endocrinol. 2002, 16, 2462–2474. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Williams, P.; Tulke, S.; Ilegems, E.; Berggren, P.-O.; Broberger, C. Expression of nucleobindin 1 (NUCB1) in pancreatic islets and other endocrine tissues. Cell Tissue Res. 2014, 358, 331–342. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Larkin, H.; Costantino, S.; Seaman, M.N.J.; Lavoie, C. Calnuc function in endosomal sorting of lysosomal receptors. Traffic 2016, 17, 416–432. [Google Scholar] [CrossRef] [Green Version]
- Tulke, S.; Williams, P.; Hellysaz, A.; Ilegems, E.; Wendel, M.; Broberger, C. Nucleobindin 1 (NUCB1) is a Golgi-resident marker of neurons. Neuroscience 2016, 314, 179–188. [Google Scholar] [CrossRef] [PubMed]
- Lin, P.; Le-Niculescu, H.; Hofmeister, R.; McCaffery, J.M.; Jin, M.; Hennemann, H.; McQuistan, T.; de Vries, L.; Farquhar, M.G. The mammalian calcium-binding protein, nucleobindin (CALNUC), is a Golgi resident protein. J. Cell Biol. 1998, 141, 1515–1527. [Google Scholar] [CrossRef]
- Wang, S.N.; Miyauchi, M.; Koshikawa, N.; Maruyama, K.; Kubota, T.; Miura, K.; Kurosawa, Y.; Awaya, A.; Kanai, Y. Antigen expression associated with lymph node metastasis in gastric adenocarcinomas. Pathol. Int. 1994, 44, 844–849. [Google Scholar] [CrossRef]
- Somogyi, E.; Petersson, U.; Sugars, R.V.; Hultenby, K.; Wendel, M. Nucleobindin—A Ca2+-binding protein present in the cells and mineralized tissues of the tooth. Calcif. Tissue Int. 2004, 74, 366–376. [Google Scholar] [CrossRef]
- Tsukumo, Y.; Tsukahara, S.; Saito, S.; Tsuruo, T.; Tomida, A. A novel endoplasmic reticulum export signal: Proline at the +2-position from the signal peptide cleavage site. J. Biol. Chem. 2009, 284, 27500–27510. [Google Scholar] [CrossRef] [Green Version]
- Taniguchi, N.; Taniura, H.; Niinobe, M.; Takayama, C.; Tominaga-Yoshino, K.; Ogura, A.; Yoshikawa, K. The postmitotic growth suppressor necdin interacts with a calcium-binding protein (NEGA) in neuronal cytoplasm. J. Biol. Chem. 2000, 275, 31674–31681. [Google Scholar] [CrossRef] [Green Version]
- Pacheko-fernandez, N.; Pakdel, M.; Blank, B.; Sanchez-Gonzaleam, I.; Weber, K.; Tran, M.L.; Hecht, T.K.-H.; Gautsch, R.; Beck, G.; Perez, F.; et al. Nucleobindin-1 regulates ECM degradation by promoting intra-Golgi trafficking of MMPs. J. Cell Biol. 2020, 219, e201907058. [Google Scholar] [CrossRef] [PubMed]
- De Alba, E.; Tjandra, N. Structural studies on the Ca2+-binding domain of human mucleobindin (calnuc). Biochemistry 2004, 43, 10039–10049. [Google Scholar] [CrossRef] [PubMed]
- Vignesh, R.; Aradhyam, G.K. A change in domain cooperativity drives the function of calnuc. Biochemistry 2020, 59, 2507–2517. [Google Scholar] [CrossRef] [PubMed]
- Kanai, Y.; Katagiri, T.; Mori, S.; Kubota, T. An established MRL/Mp-lpr/lpr cell line with null cell properties produces a B cell differentiation factor(s) that promotes anti-single-stranded DNA antibody production in MRL spleen cell culture. Int. Arch. Allergy Appl. Immunol. 1986, 81, 92–94. [Google Scholar] [CrossRef] [PubMed]
- Cassiday, L.A.; Maher, L.J. Having it both ways: Transcription factors that bind DNA and RNA. Nucleic Acids Res. 2002, 30, 4118–4126. [Google Scholar] [CrossRef] [Green Version]
- Hudson, W.H.; Ortlund, E.A. The structure, function and evolution of proteins that bind DNA and RNA. Nat. Rev. Mol. Cell Biol. 2014, 15, 749–760. [Google Scholar] [CrossRef] [Green Version]
- Luo, H.; Wang, L.; Bao, D.; Wang, L.; Zhao, H.; Lian, Y.; Yan, M.; Mohan, C.; Li, Q.-Z. Novel autoantibodies related to cell death and DNA repair pathways in systemic lupus erythematosus. Genom. Proteom. Bioinform. 2019, 17, 248–259. [Google Scholar] [CrossRef]
- Kanai, Y.; Takeda, O.; Miura, K.; Amagai, M.; Kaneko, T.; Kubota, T.; Kanai, Y.; Tanuma, S.; Kurosawa, Y. Induction of autoantibodies in normal mice by injection of nucleobindin and natural occurrence of antibodies against nucleobindin in autoimmune MRL/lpr/lpr mice. Immunol. Lett. 1995, 45, 35–42. [Google Scholar] [CrossRef]
- Carlsen, A.L.; Schetter, A.J.; Nielsen, C.T.; Lood, C.; Knudsen, S.; Voss, A.; Harris, C.C.; Hellmark, T.; Segelmark, M.; Jacobsen, S.; et al. Circulationg microRNA expression profiles associated with systemic lupus erythematosus. Arthritis Rheum. 2013, 65, 1324–1334. [Google Scholar] [CrossRef]
- Luo, B.; Zhou, K.; Liufu, Y.; Huang, X.; Zeng, H.; Zhang, Z. Novel insight into miRNA biology and its role in the pathogenesis of systemic lupus erythematosus. Front. Immunol. 2022, 13, 1059887. [Google Scholar] [CrossRef] [PubMed]
- Muralidhar, G.G.; Barbolina, M.V. The miR-200 family: Versatile players in epithelial ovarian cancer. Int. J. Mol. Sci. 2015, 16, 16833–16847. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pan, G.; Liu, Y.; Shang, L.; Zhou, F.; Yang, S. EMT-associated microRNAs and their roles in cancer stemness and drug resistance. Cancer Commun. 2021, 41, 199–217. [Google Scholar] [CrossRef]
- Zare, M.; Bastami, M.; Solali, S.; Alivand, M.R. Aberrant miRNA promoter methylation and EMT-involving miRNAs in breast cancer metastasis: Diagnosis and therapeutic implications. J. Cell Physiol. 2018, 233, 3729–3744. [Google Scholar] [CrossRef] [PubMed]
- Woodson, S.A.; Panja, S.; Santiago-Frangos, A. Proteins that chaperone RNA regulation. Microbiol. Spectr. 2018, 6. [Google Scholar] [CrossRef] [PubMed]
- Wassarman, K.M.; Repoila, F.; Rosenow, C.; Storz, G.; Gottesman, S. Identification of novel small RNAs using comparative genomics and microarrays. Genes Dev. 2001, 15, 1637–1651. [Google Scholar] [CrossRef] [Green Version]
- Lekontseva, N.; Mikhailina, A.; Fando, M.; Kravchenko, O.; Balobanov, V.; Tishchenko, S.; Nikulin, A. Crystal structures and RNA-binding properties of Lsm proteins from archaea Sulfolobus acidocaldarius and Methanococcus vannielii: Similarity and difference of the U-binding mode. Biochimie 2020, 175, 1–12. [Google Scholar] [CrossRef]
- Li, J.; Cao, Z.C.; Tang, Z.; Wang, K.; Tan, W. Molecular beacons for protein-DNA interaction studies. Methods Mol. Biol. 2008, 429, 209–224. [Google Scholar]
- Hu, Y.; Tang, H. MicroRNAs regulate the epithelial to mesenchymal transition (EMT) in cancer progression. Microrna 2014, 3, 108–117. [Google Scholar] [CrossRef]
- Singh, R.P.; Hahn, B.H.; Bischoff, D.S. Identification and contribution of inflammation-induced novel microRNA in the pathogenesis of systemic lupus erythematosus. Front. Immunol. 2022, 13, 848149. [Google Scholar] [CrossRef]
- Syeda, Z.A.; Langden, S.S.S.; Munkhzul, C.; Lee, M.; Song, S.J. Regulatory mechanism of microRNA expression in cancer. Int. J. Mol. Sci. 2020, 21, 1723. [Google Scholar] [CrossRef] [Green Version]
- O’Brien, J.; Hayder, H.; Zayed, Y.; Peng, C. Overview of microRNA biogenesis, mechanisms of actions, and circulation. Front. Endocrinol. 2018, 9, 402. [Google Scholar] [CrossRef] [Green Version]
- Krylova, S.V.; Feng, D. The machinery of exosomes: Biogenesis, release, and uptake. Int. J. Mol. Sci. 2023, 31, 1337. [Google Scholar] [CrossRef] [PubMed]
- Xiao, D.; Ohlendorf, J.; Chen, Y.; Taylor, D.; Rai, S.; Waigel, S.; Zacharias, W.; Hao, H.; McMasters, K. Identifying mRNA, microRNA and protein profiles of melanoma exosomes. PLoS ONE 2012, 7, e46874. [Google Scholar] [CrossRef] [Green Version]
- Vignesh, R.; Sjolander, A.; Venkatraman, G.; Rayala, S.; Aradhyam, G. Aberrant environment and PS-binding to calnuc C-terminal tail drives exosomal packaging and its metastatic ability. Biochem. J. 2021, 478, 2265–2283. [Google Scholar] [CrossRef] [PubMed]
- Parsonnet, N.V.; Lammer, N.C.; Holmes, Z.E.; Batey, R.T.; Wuttke, D.S. Tha glucocorticoid receptor DNA-binding domain recognizes RNA hairpin structures with high affinity. Nucleic Acids Res. 2019, 47, 8180–8192. [Google Scholar] [CrossRef] [PubMed]
- Holmes, Z.E.; Hamilton, D.J.; Hwang, T.; Parsonnet, N.V.; Rinn, J.L.; Wuttke, D.S.; Batey, R.T. The Sox2 transcription factor binds RNA. Nat. Commun. 2020, 11, 1805. [Google Scholar] [CrossRef] [Green Version]
- Dickey, T.H.; Pyle, A.M. The SMAD3 transcription factor binds complex RNA structures with high affinity. Nucleic Acids Res. 2017, 45, 11980–11988. [Google Scholar] [CrossRef] [Green Version]
- Andrade, J.M.; Dos Santos, R.F.; Chelysheva, I.; Ignatova, Z.; Arraiano, C.M. The RNA-binding protein Hfq is important for ribosome biogenesis and affects translation fidelity. EMBO J. 2018, 37, e97631. [Google Scholar] [CrossRef]
- Mathisen, P.M.; Johnson, J.M.; Kawczak, J.A.; Tuohy, V.K. Visinin-like protein (VILIP) is a neuron-specific calcium-dependent double-stranded RNA-binding protein. J. Biol. Chem. 1999, 274, 31571–31576. [Google Scholar] [CrossRef] [Green Version]
- Patel, N.; Hoang, D.; Miller, N.; Ansaloni, S.; Huang, Q.; Rogers, J.T.; Lee, J.C.; Saunders, A.J. MicroRNAs can regulate human APP levels. Mol. Neurodegener. 2008, 3, 10. [Google Scholar] [CrossRef] [Green Version]
- Lobanov, M.; Sokolovskiy, I.; Galzitskaya, O. IsUnstruct: Prediction of the residue status to be ordered or disordered in the protein chain by a method based on the Ising model. J. Biomol. Struct. Dyn. 2013, 31, 1034–1043. [Google Scholar] [CrossRef] [PubMed]
- Marc, D.; Mercey, R.; Lantier, F. Scavenger, transducer, RNA chaperone? What ligands of the prion protein teach us about its function. Cell. Mol. Life Sci. 2007, 64, 815–829. [Google Scholar] [CrossRef]
- Bayfield, M.; Jyotsna Vinayak, J.; Kerkhofs, K.; Mansouri-Noori, F. La proteins couple use of sequence-specific and non-specific binding modes to engage RNA substrates. RNA Biol. 2021, 18, 168–177. [Google Scholar] [CrossRef] [PubMed]
- Naeeni, A.; Conte, M.; Bayfield, M. RNA chaperone activity of human La protein is mediated by variant RNA recognition motif. J. Biol. Chem. 2012, 287, 5472–5482. [Google Scholar] [CrossRef] [Green Version]
- Sommer, G.; Sendlmeier, C.; Heise, T. Salt-dependent modulation of the RNA chaperone activity of RNA-binding protein La. Methods Mol. Biol. 2020, 2106, 121–136. [Google Scholar]
- Mikhaylina, A.O.; Kostareva, O.S.; Sarskikh, A.V.; Fedorov, R.V.; Piendl, W.; Garber, M.B.; Tishchenko, S.V. Investigation of the regulatory function of archaeal ribosomal protein L4. Biochemistry 2014, 79, 69–76. [Google Scholar] [CrossRef]
- Katsamba, P.S.; Park, S.; Laird-Offringa, I.A. Kinetic studies of RNA-protein interactions using surface plasmon resonance. Methods 2002, 26, 95–104. [Google Scholar] [CrossRef] [PubMed]
- Nikonova, E.Y.; Mihaylina, A.O.; Nemchinova, M.S.; Garber, M.B.; Nikonov, O.S. Glycyl-tRNA synthetase as a potential universal regulator of translation initiation at IRES-I. Mol. Biol. 2018, 52, 10–18. [Google Scholar] [CrossRef]
- Tishchenko, S.V.; Kljashtorny, V.; Kostareva, O.; Nevskaya, N.; Nikulin, A.; Gulak, P.; Piendl, W.; Garber, M.; Nikonoov, S. Domain II of Thermus thermophilus ribosomal protein L1 hinders recognition of its mRNA. J. Mol. Biol. 2008, 383, 31–305. [Google Scholar] [CrossRef]
- Moazed, D.; Stern, S.; Noller, H.F. Rapid chemical probing of conformation in 16S ribosomal RNA and 30S ribosomal subunits using primer extension. J. Mol. Biol. 1986, 187, 399–416. [Google Scholar] [CrossRef] [PubMed]
- Stern, S.; Moazed, D.; Noller, H.F. Structural analysis of RNA using chemical and enzymatic probing monitored by primer extension. Methods Enzymol. 1988, 164, 481–489. [Google Scholar] [PubMed]
- Moazed, D.; Noller, H.F. Interaction of tRNA with 23S rRNA in the ribosomal A, P, and E sites. Cell 1989, 57, 585–597. [Google Scholar] [CrossRef] [PubMed]
| RNA | ka (M−1·s−1) | kd (s−1) | KD (nM) |
|---|---|---|---|
| IRES | Not detected | ||
| tRNAGly | Not detected | ||
| C-rich | Not detected | ||
| U-rich | 2.9 × 105 | 3.2 × 10−4 | 1.1 ± 0.1 |
| 1.9 × 105 * | 3.6 × 10−4 * | 1.9 ± 0.3 * | |
| A-rich | 2.9 × 105 | 3.4 × 10−4 | 1.2 ± 0.1 |
| 2.1 × 105 * | 2.9 × 10−4 * | 1.4 ± 0.1 * | |
| G-rich | 3.4 × 105 | 5.1 × 10−4 | 1.5 ± 0.2 |
| 3.8 × 105 * | 4.5 × 10−4 * | 1.2 ± 0.3 * | |
| SRP19 | 1.3 × 105 | 1.7 × 10−3 | 13.2 ± 0.6 |
| 2.5 × 105 * | 4.3 × 10−3 * | 17.2 ± 0.5 * | |
| E-box | 5.8 × 105 | 6.7 × 10−3 | 11.4 ± 0.8 |
| 8.2 × 105 * | 8.4 × 10−3 * | 10.2 ± 0.7 * |
| microRNA | Sequence |
|---|---|
| miR-200a-3p | 5′-UAACACUGUCUGGUAACGAUGU-3′ |
| miR-203a-5p | 5′-AGUGGUUCUUAACAGUUCAACAGUU-3′ |
| miR-145-3p | 5′-GGAUUCCUGGAAAUACUGUUCU-3′ |
| miR-155-5p | 5′-UUAAUGCUAAUCGUGAUAGGGGUU-3′ |
| RNA | ka (M−1·s−1) | kd (s−1) | KD (nM) |
|---|---|---|---|
| miR-145-3p | 5.4 × 103 | 2.6·10−5 | 4.9 ± 0.2 |
| miR-155-5p | 2.2 × 103 | 3.6·10−6 | 1.7 ± 0.1 |
| miR-200a-3p | 1.9 × 104 | 7.4·10−5 | 3.8 ± 0.1 |
| miR-203a-5p | 3.5 × 104 | 3.2·10−4 | 9.3 ± 0.4 |
| RNA | Truncated NUCB1 Variants | ||
|---|---|---|---|
| ∆N | ∆Z | CaBD | |
| miR-200a-3p | 0.6 ± 0.09 | 0.3 ± 0.01 | 5.2 ± 0.2 |
| C-rich | Not detected | Not detected | Not detected |
| A-rich | 1.4 ± 0.1 | 0.3 ± 0.02 | 5.7 ± 0.5 |
| U-rich | 1.7 ± 0.3 | 0.4 ± 0.03 | 0.2 ± 0.04 |
| G-rich | 5.3 ± 0.6 | 3.1 ± 0.2 | 18.2 ± 3 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mikhaylina, A.; Svoeglazova, A.; Stolboushkina, E.; Tishchenko, S.; Kostareva, O. The RNA-Binding and RNA-Melting Activities of the Multifunctional Protein Nucleobindin 1. Int. J. Mol. Sci. 2023, 24, 6193. https://doi.org/10.3390/ijms24076193
Mikhaylina A, Svoeglazova A, Stolboushkina E, Tishchenko S, Kostareva O. The RNA-Binding and RNA-Melting Activities of the Multifunctional Protein Nucleobindin 1. International Journal of Molecular Sciences. 2023; 24(7):6193. https://doi.org/10.3390/ijms24076193
Chicago/Turabian StyleMikhaylina, Alisa, Arina Svoeglazova, Elena Stolboushkina, Svetlana Tishchenko, and Olga Kostareva. 2023. "The RNA-Binding and RNA-Melting Activities of the Multifunctional Protein Nucleobindin 1" International Journal of Molecular Sciences 24, no. 7: 6193. https://doi.org/10.3390/ijms24076193







