Potential Roles of m6A and FTO in Synaptic Connectivity and Major Depressive Disorder
Abstract
:1. Introduction
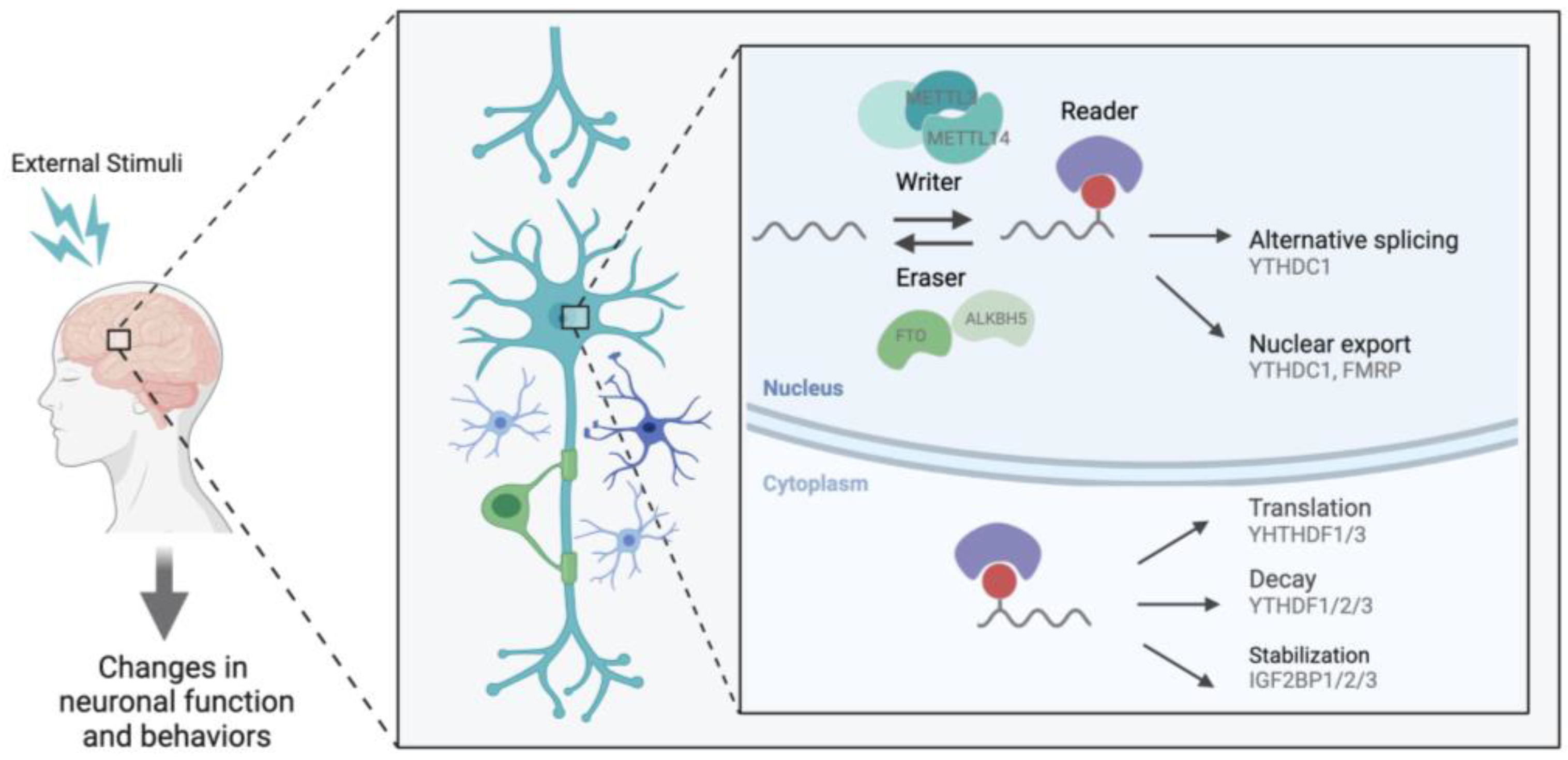
2. m6A in the Brain
3. Activity-Dependent Role of m6A
3.1. m6A Localizes Transcripts to the Synapse
3.2. m6A and Synaptic Plasticity from Learning and Memory Studies
3.3. m6A Regulates Pathways Implicated in Psychiatric Disorders
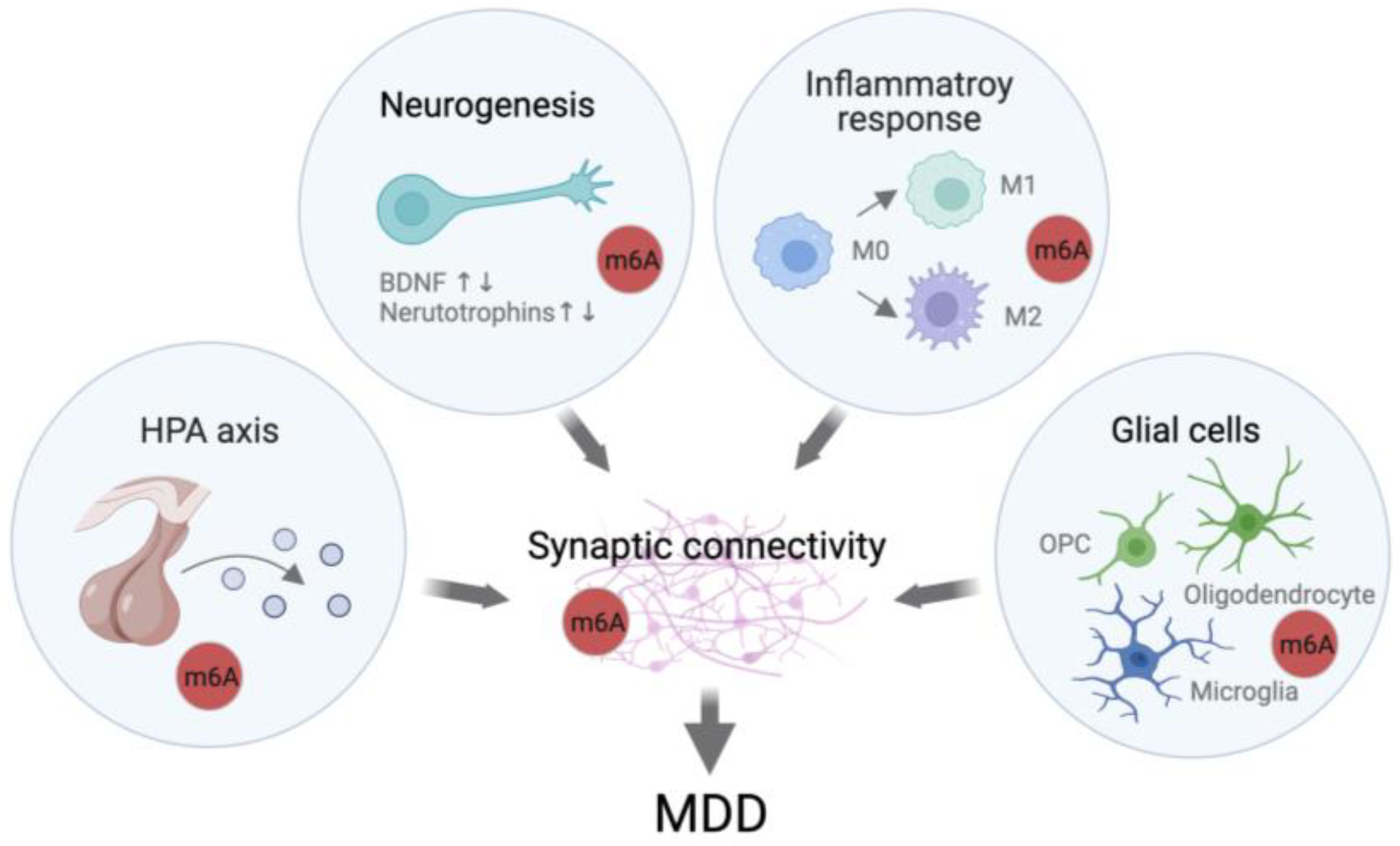
4. Direct Evidence of the Involvement of m6A Regulation in MDD
4.1. Potential Roles of m6A in Stress Response
4.2. KO Models Show Depressive-like/Anxiety-like Behaviors
4.3. Mutation in FTO and ALKBH5 Are Linked to MDD
5. Summary
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Desrosiers, R.; Friderici, K.; Rottman, F. Identification of methylated nucleosides in messenger RNA from Novikoff hepatoma cells. Proc. Natl. Acad. Sci. USA 1974, 71, 3971–3975. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Boccaletto, P.; Stefaniak, F.; Ray, A.; Cappannini, A.; Mukherjee, S.; Purta, E.; Kurkowska, M.; Shirvanizadeh, N.; Destefanis, E.; Groza, P.; et al. MODOMICS: A database of RNA modification pathways. 2021 update. Nucleic Acids Res. 2022, 50, D231–D235. [Google Scholar] [CrossRef] [PubMed]
- Dominissini, D.; Moshitch-Moshkovitz, S.; Schwartz, S.; Salmon-Divon, M.; Ungar, L.; Osenberg, S.; Cesarkas, K.; Jacob-Hirsch, J.; Amariglio, N.; Kupiec, M.; et al. Topology of the human and mouse m6A RNA methylomes revealed by m6A-seq. Nature 2012, 485, 201–206. [Google Scholar] [CrossRef] [PubMed]
- Yang, C.; Hu, Y.; Zhou, B.; Bao, Y.; Li, Z.; Gong, C.; Yang, H.; Wang, S.; Xiao, Y. The role of m6A modification in physiology and disease. Cell Death Dis. 2020, 11, 960. [Google Scholar] [CrossRef] [PubMed]
- Disease, G.B.D.; Injury, I.; Prevalence, C. Global, regional, and national incidence, prevalence, and years lived with disability for 354 diseases and injuries for 195 countries and territories, 1990–2017: A systematic analysis for the Global Burden of Disease Study 2017. Lancet 2018, 392, 1789–1858. [Google Scholar] [CrossRef] [Green Version]
- Sullivan, P.F.; Neale, M.C.; Kendler, K.S. Genetic epidemiology of major depression: Review and meta-analysis. Am. J. Psychiatry 2000, 157, 1552–1562. [Google Scholar] [CrossRef]
- Flint, J.; Kendler, K.S. The genetics of major depression. Neuron 2014, 81, 484–503. [Google Scholar] [CrossRef] [Green Version]
- Duman, R.S.; Aghajanian, G.K. Synaptic dysfunction in depression: Potential therapeutic targets. Science 2012, 338, 68–72. [Google Scholar] [CrossRef] [Green Version]
- Drevets, W.C. Functional anatomical abnormalities in limbic and prefrontal cortical structures in major depression. Prog. Brain Res. 2000, 126, 413–431. [Google Scholar] [CrossRef]
- Stockmeier, C.A.; Mahajan, G.J.; Konick, L.C.; Overholser, J.C.; Jurjus, G.J.; Meltzer, H.Y.; Uylings, H.B.; Friedman, L.; Rajkowska, G. Cellular changes in the postmortem hippocampus in major depression. Biol. Psychiatry 2004, 56, 640–650. [Google Scholar] [CrossRef] [Green Version]
- Rajkowska, G.; Miguel-Hidalgo, J.J.; Wei, J.; Dilley, G.; Pittman, S.D.; Meltzer, H.Y.; Overholser, J.C.; Roth, B.L.; Stockmeier, C.A. Morphometric evidence for neuronal and glial prefrontal cell pathology in major depression. Biol. Psychiatry 1999, 45, 1085–1098. [Google Scholar] [CrossRef] [PubMed]
- Kang, H.J.; Voleti, B.; Hajszan, T.; Rajkowska, G.; Stockmeier, C.A.; Licznerski, P.; Lepack, A.; Majik, M.S.; Jeong, L.S.; Banasr, M.; et al. Decreased expression of synapse-related genes and loss of synapses in major depressive disorder. Nat. Med. 2012, 18, 1413–1417. [Google Scholar] [CrossRef] [PubMed]
- Kaiser, R.H.; Andrews-Hanna, J.R.; Wager, T.D.; Pizzagalli, D.A. Large-Scale Network Dysfunction in Major Depressive Disorder: A Meta-analysis of Resting-State Functional Connectivity. JAMA Psychiatry 2015, 72, 603–611. [Google Scholar] [CrossRef] [PubMed]
- Duman, R.S.; Aghajanian, G.K.; Sanacora, G.; Krystal, J.H. Synaptic plasticity and depression: New insights from stress and rapid-acting antidepressants. Nat. Med. 2016, 22, 238–249. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Turecki, G. The molecular bases of the suicidal brain. Nat. Rev. Neurosci. 2014, 15, 802–816. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Meaney, M.J. Epigenetics and the biological definition of gene x environment interactions. Child Dev. 2010, 81, 41–79. [Google Scholar] [CrossRef] [PubMed]
- Bagot, R.C.; Labonte, B.; Pena, C.J.; Nestler, E.J. Epigenetic signaling in psychiatric disorders: Stress and depression. Dialogues Clin. Neurosci. 2014, 16, 281–295. [Google Scholar] [CrossRef]
- Livneh, I.; Moshitch-Moshkovitz, S.; Amariglio, N.; Rechavi, G.; Dominissini, D. The m6A epitranscriptome: Transcriptome plasticity in brain development and function. Nat. Rev. Neurosci. 2020, 21, 36–51. [Google Scholar] [CrossRef]
- Widagdo, J.; Wong, J.J.L.; Anggono, V. The m6A-epitranscriptome in brain plasticity, learning and memory. Semin. Cell Dev. Biol. 2022, 125, 110–121. [Google Scholar] [CrossRef]
- Huang, R.; Zhang, Y.; Bai, Y.; Han, B.; Ju, M.; Chen, B.; Yang, L.; Wang, Y.; Zhang, H.H.H.; Zhang, H.H.H.; et al. N6-Methyladenosine Modification of Fatty Acid Amide Hydrolase Messenger RNA in Circular RNA STAG1-Regulated Astrocyte Dysfunction and Depressive-like Behaviors. Biol. Psychiatry 2020, 88, 392–404. [Google Scholar] [CrossRef] [Green Version]
- Engel, M.; Eggert, C.; Kaplick, P.M.; Eder, M.; Roh, S.; Tietze, L.; Namendorf, C.; Arloth, J.; Weber, P.; Rex-Haffner, M.; et al. The Role of m6A/m-RNA Methylation in Stress Response Regulation. Neuron 2018, 99, 389–403.e9. [Google Scholar] [CrossRef] [PubMed]
- Zhang, N.; Ding, C.; Zuo, Y.; Peng, Y.; Zuo, L. N6-methyladenosine and Neurological Diseases. Mol. Neurobiol. 2022, 59, 1925–1937. [Google Scholar] [CrossRef] [PubMed]
- Zhang, R.; Zhang, Y.; Guo, F.; Li, S.; Cui, H. RNA N6-Methyladenosine Modifications and Its Roles in Alzheimer’s Disease. Front. Cell. Neurosci. 2022, 16, 820378. [Google Scholar] [CrossRef]
- Meyer, K.D.; Saletore, Y.; Zumbo, P.; Elemento, O.; Mason, C.E.; Jaffrey, S.R. Comprehensive analysis of mRNA methylation reveals enrichment in 3′ UTRs and near stop codons. Cell 2012, 149, 1635–1646. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Linder, B.; Grozhik, A.V.; Olarerin-George, A.O.; Meydan, C.; Mason, C.E.; Jaffrey, S.R. Single-nucleotide-resolution mapping of m6A and m6Am throughout the transcriptome. Nat. Methods 2015, 12, 767–772. [Google Scholar] [CrossRef]
- He, P.C.; He, C. m6A RNA methylation: From mechanisms to therapeutic potential. EMBO J. 2021, 40, e105977. [Google Scholar] [CrossRef]
- Yue, Y.; Liu, J.; Cui, X.; Cao, J.; Luo, G.; Zhang, Z.; Cheng, T.; Gao, M.; Shu, X.; Ma, H.; et al. VIRMA mediates preferential m6A mRNA methylation in 3′UTR and near stop codon and associates with alternative polyadenylation. Cell Discov. 2018, 4, 10. [Google Scholar] [CrossRef] [Green Version]
- Patil, D.P.; Chen, C.K.; Pickering, B.F.; Chow, A.; Jackson, C.; Guttman, M.; Jaffrey, S.R. m6A RNA methylation promotes XIST-mediated transcriptional repression. Nature 2016, 537, 369–373. [Google Scholar] [CrossRef] [Green Version]
- Knuckles, P.; Lence, T.; Haussmann, I.U.; Jacob, D.; Kreim, N.; Carl, S.H.; Masiello, I.; Hares, T.; Villasenor, R.; Hess, D.; et al. Zc3h13/Flacc is required for adenosine methylation by bridging the mRNA-binding factor Rbm15/Spenito to the m6A machinery component Wtap/Fl(2)d. Genes Dev. 2018, 32, 415–429. [Google Scholar] [CrossRef] [Green Version]
- Wen, J.; Lv, R.; Ma, H.; Shen, H.; He, C.; Wang, J.; Jiao, F.; Liu, H.; Yang, P.; Tan, L.; et al. Zc3h13 Regulates Nuclear RNA m6A Methylation and Mouse Embryonic Stem Cell Self-Renewal. Mol. Cell 2018, 69, 1028–1038.e6. [Google Scholar] [CrossRef] [Green Version]
- Fu, Y.; Dominissini, D.; Rechavi, G.; He, C. Gene expression regulation mediated through reversible m6A RNA methylation. Nat. Rev. Genet. 2014, 15, 293–306. [Google Scholar] [CrossRef]
- Xiao, W.; Adhikari, S.; Dahal, U.; Chen, Y.S.; Hao, Y.J.; Sun, B.F.; Sun, H.Y.; Li, A.; Ping, X.L.; Lai, W.Y.; et al. Nuclear m6A Reader YTHDC1 Regulates mRNA Splicing. Mol. Cell 2016, 61, 507–519. [Google Scholar] [CrossRef] [Green Version]
- Roundtree, I.A.; Luo, G.Z.; Zhang, Z.; Wang, X.; Zhou, T.; Cui, Y.; Sha, J.; Huang, X.; Guerrero, L.; Xie, P.; et al. YTHDC1 mediates nuclear export of N6-methyladenosine methylated mRNAs. eLife 2017, 6, e31311. [Google Scholar] [CrossRef]
- Mao, Y.; Dong, L.; Liu, X.M.; Guo, J.; Ma, H.; Shen, B.; Qian, S.B. m6A in mRNA coding regions promotes translation via the RNA helicase-containing YTHDC2. Nat. Commun. 2019, 10, 5332. [Google Scholar] [CrossRef] [Green Version]
- Kretschmer, J.; Rao, H.; Hackert, P.; Sloan, K.E.; Hobartner, C.; Bohnsack, M.T. The m6A reader protein YTHDC2 interacts with the small ribosomal subunit and the 5′-3′ exoribonuclease XRN1. RNA 2018, 24, 1339–1350. [Google Scholar] [CrossRef] [Green Version]
- Du, H.; Zhao, Y.; He, J.; Zhang, Y.; Xi, H.; Liu, M.; Ma, J.; Wu, L. YTHDF2 destabilizes m6A-containing RNA through direct recruitment of the CCR4-NOT deadenylase complex. Nat. Commun. 2016, 7, 12626. [Google Scholar] [CrossRef]
- Shi, H.; Wang, X.; Lu, Z.; Zhao, B.S.; Ma, H.; Hsu, P.J.; Liu, C.; He, C. YTHDF3 facilitates translation and decay of N6-methyladenosine-modified RNA. Cell Res. 2017, 27, 315–328. [Google Scholar] [CrossRef] [Green Version]
- Wang, X.; Zhao, B.S.; Roundtree, I.A.; Lu, Z.; Han, D.; Ma, H.; Weng, X.; Chen, K.; Shi, H.; He, C. N6-methyladenosine modulates messenger RNA translation efficiency. Cell 2015, 161, 1388–1399. [Google Scholar] [CrossRef] [Green Version]
- Zaccara, S.; Jaffrey, S.R. A Unified Model for the Function of YTHDF Proteins in Regulating m6A-Modified mRNA. Cell 2020, 181, 1582–1595.e18. [Google Scholar] [CrossRef]
- Hazra, D.; Chapat, C.; Graille, M. m6A mRNA Destiny: Chained to the rhYTHm by the YTH-Containing Proteins. Genes 2019, 10, 49. [Google Scholar] [CrossRef] [Green Version]
- Hsu, P.J.; Shi, H.; Zhu, A.C.; Lu, Z.; Miller, N.; Edens, B.M.; Ma, Y.C.; He, C. The RNA-binding protein FMRP facilitates the nuclear export of N6-methyladenosine-containing mRNAs. J. Biol. Chem. 2019, 294, 19889–19895. [Google Scholar] [CrossRef] [PubMed]
- Liu, N.; Dai, Q.; Zheng, G.; He, C.; Parisien, M.; Pan, T. N6-methyladenosine-dependent RNA structural switches regulate RNA-protein interactions. Nature 2015, 518, 560–564. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Alarcon, C.R.; Goodarzi, H.; Lee, H.; Liu, X.; Tavazoie, S.; Tavazoie, S.F. HNRNPA2B1 Is a Mediator of m6A-Dependent Nuclear RNA Processing Events. Cell 2015, 162, 1299–1308. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Huang, H.; Weng, H.; Sun, W.; Qin, X.; Shi, H.; Wu, H.; Zhao, B.S.; Mesquita, A.; Liu, C.; Yuan, C.L.; et al. Recognition of RNA N6-methyladenosine by IGF2BP proteins enhances mRNA stability and translation. Nat. Cell Biol. 2018, 20, 285–295. [Google Scholar] [CrossRef]
- Chang, M.; Lv, H.; Zhang, W.; Ma, C.; He, X.; Zhao, S.; Zhang, Z.W.; Zeng, Y.X.; Song, S.; Niu, Y.; et al. Region-specific RNA m6A methylation represents a new layer of control in the gene regulatory network in the mouse brain. Open Biol. 2017, 7. [Google Scholar] [CrossRef] [Green Version]
- Xiong, X.; Hou, L.; Park, Y.P.; Molinie, B.; Gregory, R.I.; Kellis, M.; Consortium, G.T.; Gregory, R.I.; Kellis, M. Genetic drivers of m6A methylation in human brain, lung, heart and muscle. Nat. Genet. 2021, 53, 1156–1165. [Google Scholar] [CrossRef]
- Xu, H.; Dzhashiashvili, Y.; Shah, A.; Kunjamma, R.B.; Weng, Y.L.; Elbaz, B.; Fei, Q.; Jones, J.S.; Li, Y.I.; Zhuang, X.; et al. m6A mRNA Methylation Is Essential for Oligodendrocyte Maturation and CNS Myelination. Neuron 2020, 105, 293–309.e5. [Google Scholar] [CrossRef]
- Wu, R.; Li, A.; Sun, B.; Sun, J.G.; Zhang, J.; Zhang, T.; Chen, Y.; Xiao, Y.; Gao, Y.; Zhang, Q.; et al. A novel m6A reader Prrc2a controls oligodendroglial specification and myelination. Cell Res. 2019, 29, 23–41. [Google Scholar] [CrossRef]
- Merkurjev, D.; Hong, W.T.; Iida, K.; Oomoto, I.; Goldie, B.J.; Yamaguti, H.; Ohara, T.; Kawaguchi, S.Y.; Hirano, T.; Martin, K.C.; et al. Synaptic N6-methyladenosine (m6A) epitranscriptome reveals functional partitioning of localized transcripts. Nat. Neurosci. 2018, 21, 1004–1014. [Google Scholar] [CrossRef]
- Flamand, M.N.; Meyer, K.D. m6A and YTHDF proteins contribute to the localization of select neuronal mRNAs. Nucleic Acids. Res. 2022, 1, 13–14. [Google Scholar] [CrossRef]
- Madugalle, S.U.; Meyer, K.; Wang, D.O.; Bredy, T.W. RNA N6-Methyladenosine and the Regulation of RNA Localization and Function in the Brain. Trends Neurosci. 2020, 43, 1011–1023. [Google Scholar] [CrossRef]
- Shi, H.; Zhang, X.; Weng, Y.L.; Lu, Z.; Liu, Y.; Lu, Z.; Li, J.; Hao, P.; Zhang, Y.; Zhang, F.; et al. m6A facilitates hippocampus-dependent learning and memory through YTHDF1. Nature 2018, 563, 249–253. [Google Scholar] [CrossRef]
- Yu, J.; Chen, M.; Huang, H.; Zhu, J.J.; Song, H.; Zhu, J.J.; Park, J.; Ji, S.J. Dynamic m6A modification regulates local translation of mRNA in axons. Nucleic Acids Res. 2018, 46, 1412–1423. [Google Scholar] [CrossRef] [Green Version]
- Yu, J.; She, Y.; Yang, L.; Zhuang, M.; Han, P.; Liu, J.; Lin, X.; Wang, N.; Chen, M.; Jiang, C.; et al. The m 6 A Readers YTHDF1 and YTHDF2 Synergistically Control Cerebellar Parallel Fiber Growth by Regulating Local Translation of the Key Wnt5a Signaling Components in Axons. Adv. Sci. 2021, 8, e2101329. [Google Scholar] [CrossRef]
- Zhang, Z.; Wang, M.; Xie, D.; Huang, Z.; Zhang, L.; Yang, Y.; Ma, D.; Li, W.; Zhou, Q.; Yang, Y.G.; et al. METTL3-mediated N6-methyladenosine mRNA modification enhances long-term memory consolidation. Cell Res. 2018, 28, 1050–1061. [Google Scholar] [CrossRef] [Green Version]
- Widagdo, J.; Zhao, Q.Y.; Kempen, M.J.; Tan, M.C.; Ratnu, V.S.; Wei, W.; Leighton, L.; Spadaro, P.A.; Edson, J.; Anggono, V.; et al. Experience-Dependent Accumulation of N6-Methyladenosine in the Prefrontal Cortex Is Associated with Memory Processes in Mice. J. Neurosci. 2016, 36, 6771–6777. [Google Scholar] [CrossRef] [Green Version]
- Walters, B.J.; Mercaldo, V.; Gillon, C.J.; Yip, M.; Neve, R.L.; Boyce, F.M.; Frankland, P.W.; Josselyn, S.A. The Role of The RNA Demethylase FTO (Fat Mass and Obesity-Associated) and mRNA Methylation in Hippocampal Memory Formation. Neuropsychopharmacology 2017, 42, 1502–1510. [Google Scholar] [CrossRef] [Green Version]
- Spychalaid, A.; Rü, U. FTO affects hippocampal function by regulation of BDNF processing. PLoS ONE 2019, 14, e0211937. [Google Scholar] [CrossRef]
- Han, M.; Liu, Z.; Xu, Y.; Liu, X.; Wang, D.; Li, F.; Wang, Y.; Bi, J. Abnormality of m6A mRNA Methylation Is Involved in Alzheimer’s Disease. Front. Neurosci. 2020, 14, 98. [Google Scholar] [CrossRef]
- Li, H.; Ren, Y.; Mao, K.; Hua, F.; Yang, Y.; Wei, N.; Yue, C.; Li, D.; Zhang, H. FTO is involved in Alzheimer’s disease by targeting TSC1-mTOR-Tau signaling. Biochem. Biophys. Res. Commun. 2018, 498, 234–239. [Google Scholar] [CrossRef]
- Shafik, A.M.; Zhang, F.; Guo, Z.; Dai, Q.; Pajdzik, K.; Li, Y.; Kang, Y.; Yao, B.; Wu, H.; He, C.; et al. N6-methyladenosine dynamics in neurodevelopment and aging, and its potential role in Alzheimer’s disease. Genome Biol. 2021, 22, 1–19. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Chen, Y.; Song, Y. Regulatory mechanism of FTO in Parkinson’s disease cell model. FASEB J. 2021, 35. [Google Scholar] [CrossRef]
- Pupak, A.; Singh, A.; Sancho-Balsells, A.; Alcalá-Vida, R.; Espina, M.; Giralt, A.; Martí, E.; Ørom, U.A.V.; Ginés, S.; Brito, V. Altered m6A RNA methylation contributes to hippocampal memory deficits in Huntington’s disease mice. Cell. Mol. Life Sci. CMLS 2022, 79, 416. [Google Scholar] [CrossRef]
- Liu, Y.; Zhang, H. RNA m6A Modification Changes in Postmortem Nucleus Accumbens of Subjects with Alcohol Use Disorder: A Pilot Study. Genes 2022, 13, 958. [Google Scholar] [CrossRef] [PubMed]
- Reis, A.L.M.; Hammond, J.M.; Stevanovski, I.; Arnold, J.C.; McGregor, I.S.; Deveson, I.W.; Gururajan, A. Sex-specific transcriptomic and epitranscriptomic signatures of PTSD-like fear acquisition. iScience 2022, 25, 104861. [Google Scholar] [CrossRef]
- Takeuchi, T.; Duszkiewicz, A.J.; Morris, R.G. The synaptic plasticity and memory hypothesis: Encoding, storage and persistence. Philos. Trans. R. Soc. B 2014, 369, 20130288. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, L.; Zang, L.; Zhang, F.; Chen, J.; Shen, H.; Shu, L.; Liang, F.; Feng, C.; Chen, D.; Tao, H.; et al. Fat mass and obesity-associated (FTO) protein regulates adult neurogenesis. Hum. Mol. Genet. 2017, 26, 2398–2411. [Google Scholar] [CrossRef] [Green Version]
- Cao, Y.; Zhuang, Y.; Chen, J.; Xu, W.; Shou, Y.; Huang, X.; Shu, Q.; Li, X. Dynamic effects of Fto in regulating the proliferation and differentiation of adult neural stem cells of mice. Hum. Mol. Genet 2020, 29, 727–735. [Google Scholar] [CrossRef]
- Sun, L.; Ma, L.; Zhang, H.; Cao, Y.; Wang, C.; Hou, N.; Huang, N.; von Deneen, K.M.; Zhao, C.; Shi, Y.; et al. FTO deficiency reduces anxiety- and depression-like behaviors in mice via alterations in gut microbiota. Theranostics 2019, 9, 721–733. [Google Scholar] [CrossRef]
- Chen, J.; Zhang, Y.J.Y.C.; Huang, C.; Shen, H.; Sun, B.; Cheng, X.; Zhang, Y.J.Y.C.; Yang, Y.G.Y.; Shu, Q.; Yang, Y.G.Y.; et al. m6A Regulates Neurogenesis and Neuronal Development by Modulating Histone Methyltransferase Ezh2. Genom. Proteom. Bioinform. 2019, 17, 154–168. [Google Scholar] [CrossRef]
- Li, Q.; Wen, S.; Ye, W.; Zhao, S.; Liu, X. The potential roles of m6A modification in regulating the inflammatory response in microglia. J. Neuroinflammation 2021, 18, 149. [Google Scholar] [CrossRef]
- Gu, X.; Zhang, Y.; Li, D.; Cai, H.; Cai, L.; Xu, Q. N6-methyladenosine demethylase FTO promotes M1 and M2 macrophage activation. Cell. Signal. 2020, 69, 109553. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Liu, Z.; Tang, H.; Shen, Y.; Gong, Z.; Xie, N.; Zhang, X.; Wang, W.; Kong, W.; Zhou, Y.; et al. The N6-methyladenosine (m6A)-forming enzyme METTL3 facilitates M1 macrophage polarization through the methylation of STAT1 mRNA. Am. J. Physiol. Cell Physiol. 2019, 317, C762–C775. [Google Scholar] [CrossRef] [PubMed]
- Yu, R.; Li, Q.; Feng, Z.; Cai, L.; Xu, Q. m6A Reader YTHDF2 Regulates LPS-Induced Inflammatory Response. Int. J. Mol. Sci. 2019, 20, 1323. [Google Scholar] [CrossRef] [Green Version]
- Chokkalla, A.K.; Mehta, S.L.; Kim, T.; Chelluboina, B.; Kim, J.; Vemuganti, R. Transient Focal Ischemia Significantly Alters the m6A Epitranscriptomic Tagging of RNAs in the Brain. Stroke 2019, 50, 2912–2921. [Google Scholar] [CrossRef] [PubMed]
- Loos, R.J.; Bouchard, C. FTO: The first gene contributing to common forms of human obesity. Obes. Rev. 2008, 9, 246–250. [Google Scholar] [CrossRef]
- Luppino, F.S.; de Wit, L.M.; Bouvy, P.F.; Stijnen, T.; Cuijpers, P.; Penninx, B.W.; Zitman, F.G. Overweight, obesity, and depression: A systematic review and meta-analysis of longitudinal studies. Arch. Gen. Psychiatry 2010, 67, 220–229. [Google Scholar] [CrossRef] [PubMed]
- Liu, S.; Xiu, J.; Zhu, C.; Meng, K.; Li, C.; Han, R.; Du, T.; Li, L.; Xu, L.; Liu, R.; et al. Fat mass and obesity-associated protein regulates RNA methylation associated with depression-like behavior in mice. Nat. Commun. 2021, 12, 6937. [Google Scholar] [CrossRef] [PubMed]
- Joshi, K.; Wang, D.O.; Gururajan, A. The m6A-methylome in major depression: A bioinformatic analysis of publicly available datasets. Psychiatry Res. Commun. 2022, 2, 100089. [Google Scholar] [CrossRef]
- Wang, X.-L.; Wei, X.; Yuan, J.-J.; Mao, Y.-Y.; Wang, Z.-Y.; Xing, N.; Gu, H.-W.; Lin, C.-H.; Wang, W.-T.; Zhang, W.; et al. Downregulation of Fat Mass and Obesity-Related Protein in the Anterior Cingulate Cortex participates in Anxiety- and Depression-Like Behaviors Induced by Neuropathic Pain. Front. Cell. Neurosci. 2022, 16, 884296. [Google Scholar] [CrossRef]
- Wu, P.-F.F.; Han, Q.-Q.Q.; Chen, F.-F.F.; Shen, T.-T.T.; Li, Y.-H.H.; Cao, Y.; Chen, J.-G.G.; Wang, F. Erasing m6A-dependent transcription signature of stress-sensitive genes triggers antidepressant actions. Neurobiol. Stress 2021, 15, 100390. [Google Scholar] [CrossRef]
- Choudhry, Z.; Sengupta, S.M.; Grizenko, N.; Thakur, G.A.; Fortier, M.E.; Schmitz, N.; Joober, R. Association between obesity-related gene FTO and ADHD. Obesity 2013, 21, E738–E744. [Google Scholar] [CrossRef] [PubMed]
- Rivera, M.; Locke, A.E.; Corre, T.; Czamara, D.; Wolf, C.; Ching-Lopez, A.; Milaneschi, Y.; Kloiber, S.; Cohen-Woods, S.; Rucker, J.; et al. Interaction between the FTO gene, body mass index and depression: Meta-analysis of 13701 individuals. Br. J. Psychiatry 2017, 211, 70–76. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Samaan, Z.; Anand, S.S.; Zhang, X.; Desai, D.; Rivera, M.; Pare, G.; Thabane, L.; Xie, C.; Gerstein, H.; Engert, J.C.; et al. The protective effect of the obesity-associated rs9939609 A variant in fat mass- and obesity-associated gene on depression. Mol. Psychiatry 2013, 18, 1281–1286. [Google Scholar] [CrossRef]
- Milaneschi, Y.; Lamers, F.; Mbarek, H.; Hottenga, J.J.; Boomsma, D.I.; Penninx, B.W.J.H. The effect of FTO rs9939609 on major depression differs across MDD subtypes. Mol. Psychiatry 2014, 19, 960–962. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yao, Y.; Wen, Y.; Du, T.; Sun, N.; Deng, H.; Ryan, J.; Rao, S. Meta-analysis indicates that SNP rs9939609 within FTO is not associated with major depressive disorder (MDD) in Asian population. J. Affect. Disord. 2016, 193, 27–30. [Google Scholar] [CrossRef]
- Du, T.; Rao, S.; Wu, L.; Ye, N.; Liu, Z.; Hu, H.; Xiu, J.; Shen, Y.; Xu, Q. An association study of the m6A genes with major depressive disorder in Chinese Han population. J. Affect. Disord. 2015, 183, 279–286. [Google Scholar] [CrossRef]
- Reitz, C.; Tosto, G.; Mayeux, R.; Luchsinger, J.A.; Group, N.-L.N.F.S.; Alzheimer’s Disease Neuroimaging, I. Genetic variants in the Fat and Obesity Associated (FTO) gene and risk of Alzheimer’s disease. PLoS ONE 2012, 7, e50354. [Google Scholar] [CrossRef]
- Keller, L.; Xu, W.; Wang, H.X.; Winblad, B.; Fratiglioni, L.; Graff, C. The obesity related gene, FTO, interacts with APOE, and is associated with Alzheimer’s disease risk: A prospective cohort study. J. Alzheimers Dis. 2011, 23, 461–469. [Google Scholar] [CrossRef]
| Mice | Brain Region | Expression | Paradigm | Behavior | Citation |
|---|---|---|---|---|---|
| C57BL/6 | HIPP | FTO Knockdown | - | Anxiety-like behaviors - | [21] |
| HIPP | METTL3 Knockdown | - | Anxiety-like behaviors - | ||
| C57BL/6 | HIPP | FTO Knockdown | - | Anxiety-like behavior ↑ | [58] |
| C57BL/6 | Global | FTO Knockdown | - | Depressive-like behaviors ↓ Anxiety-like behaviors ↓ | [69] |
| - | Global | FTO Knockdown | Chronic unpredictable mild stress | Unable to tolerate stress stimulation | |
| Global | FTO Heterozygous (HZ) | Chronic unpredictable mild stress | Stress susceptible ↓ | ||
| C57BL/6 | HIPP | FTO Knockdown | - | Depressive-like behaviors ↑ Anxiety-like behaviors- | [78] |
| HIPP | FTO Knockout | - | Depressive-like behaviors ↑ Anxiety-like behaviors- | ||
| HIPP | FTO Overexpression | Chronic unpredictable mild stress | Depressive-like behaviors ↓ | ||
| C57BL/6 | ACC | FTO Knockdown | Neuropathic pain | Depressive-like behaviors ↓ Anxiety-like behaviors ↓ | [80] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mitsuhashi, H.; Nagy, C. Potential Roles of m6A and FTO in Synaptic Connectivity and Major Depressive Disorder. Int. J. Mol. Sci. 2023, 24, 6220. https://doi.org/10.3390/ijms24076220
Mitsuhashi H, Nagy C. Potential Roles of m6A and FTO in Synaptic Connectivity and Major Depressive Disorder. International Journal of Molecular Sciences. 2023; 24(7):6220. https://doi.org/10.3390/ijms24076220
Chicago/Turabian StyleMitsuhashi, Haruka, and Corina Nagy. 2023. "Potential Roles of m6A and FTO in Synaptic Connectivity and Major Depressive Disorder" International Journal of Molecular Sciences 24, no. 7: 6220. https://doi.org/10.3390/ijms24076220




