Genome-Wide Association Study for Root Morphology and Phosphorus Acquisition Efficiency in Diverse Maize Panels
Abstract
1. Introduction
2. Results
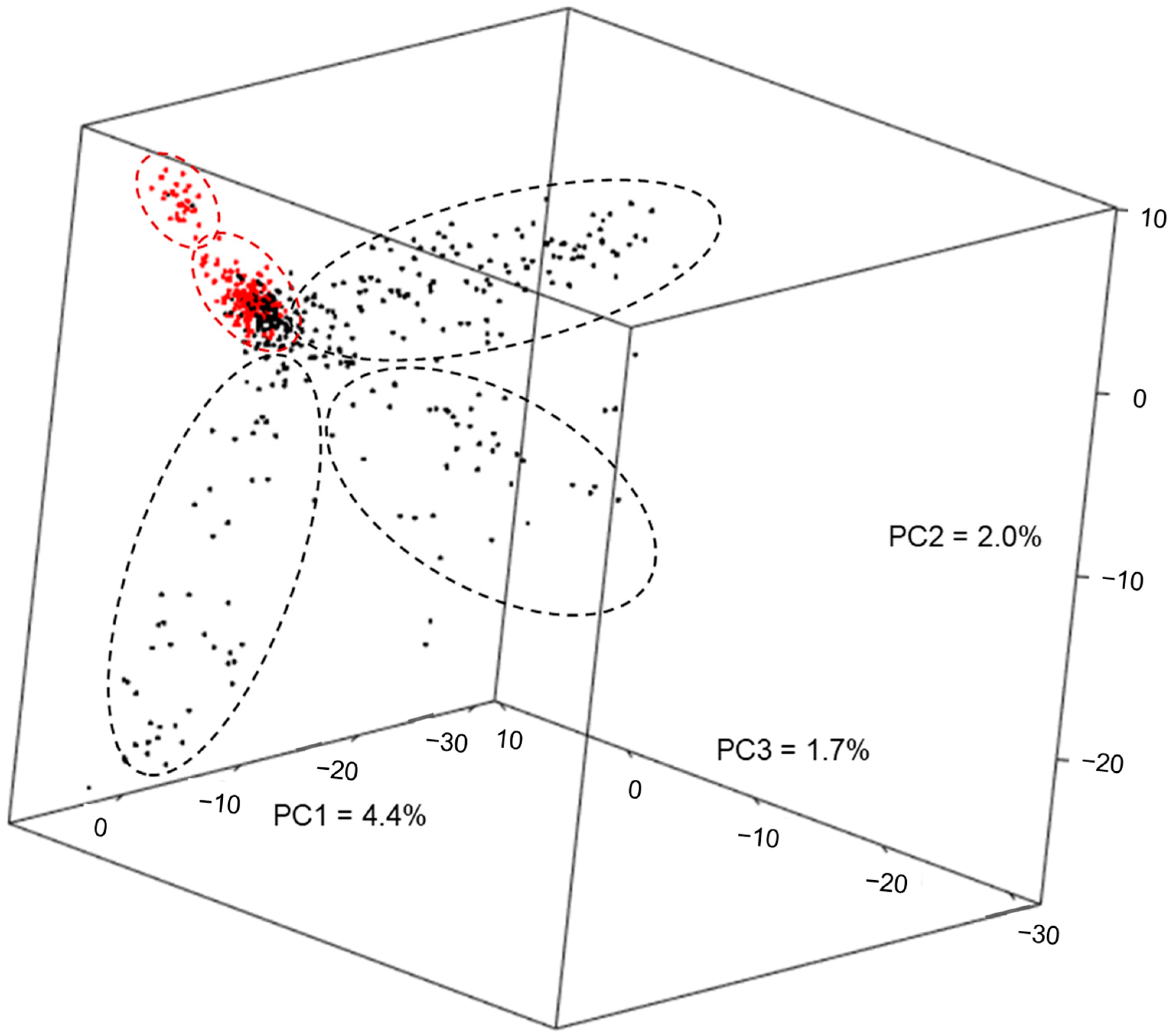
2.1. Genetic Diversity of the Maize Panels—Principal Component Analysis
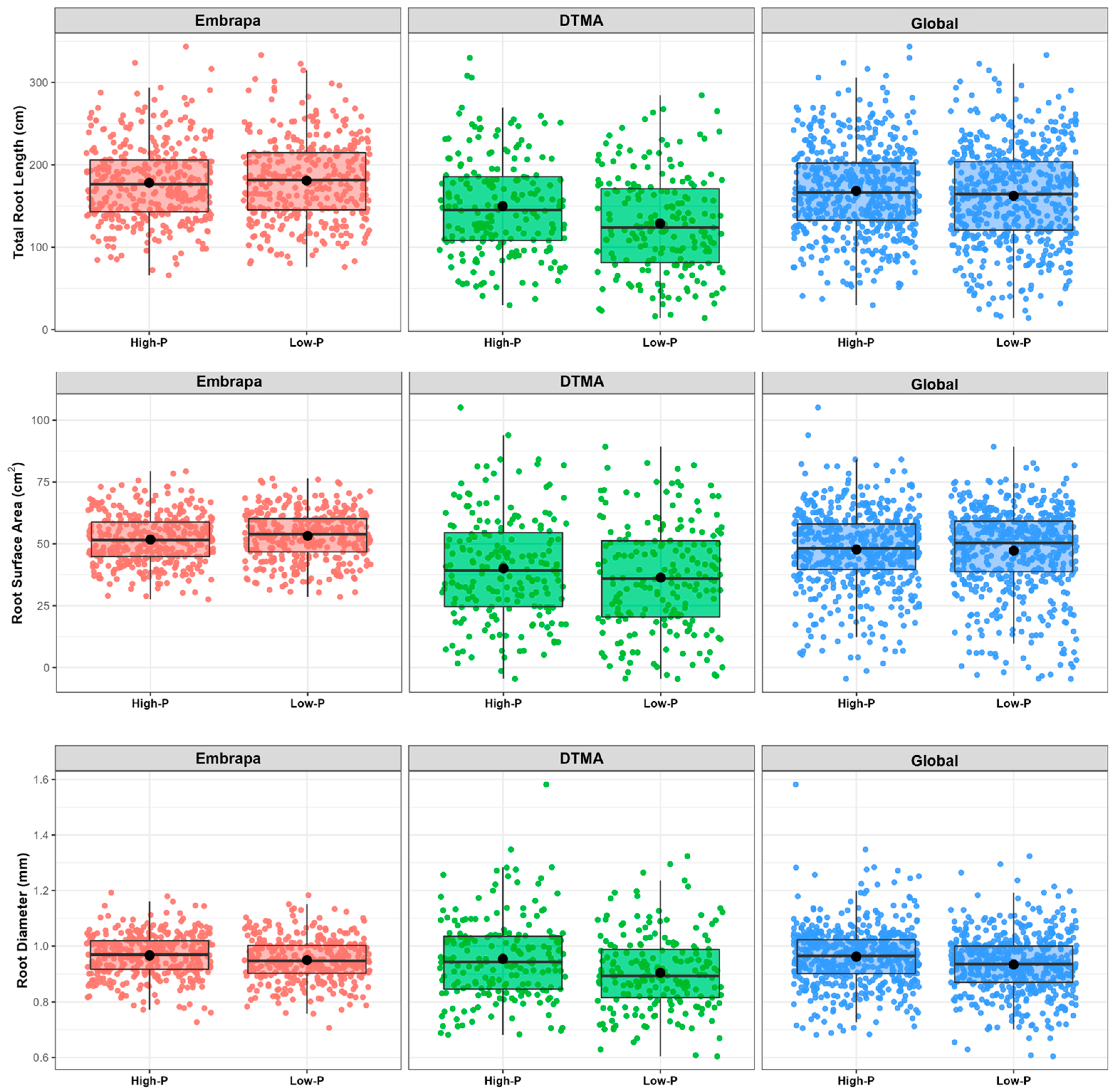
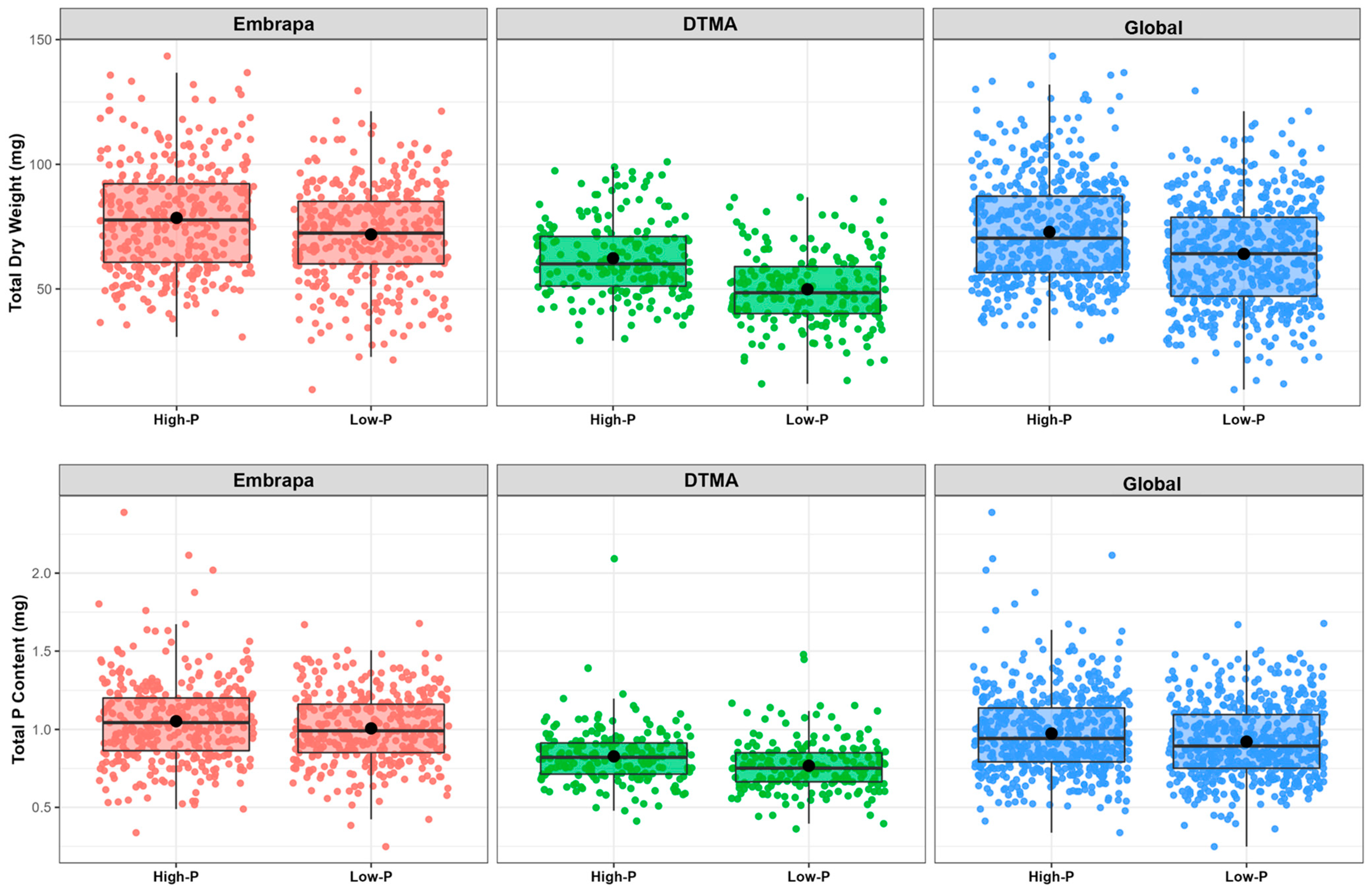
2.2. Phenotypic Characterization of the Maize Association Panel
2.3. Influence of P on Root Morphology and Biomass Accumulation
2.4. Multiple Testing and Type-I Error Corrections for GWAS
2.5. SNPs Significantly Associated with Root Morphology, P Acquisition and Biomass Related-Traits
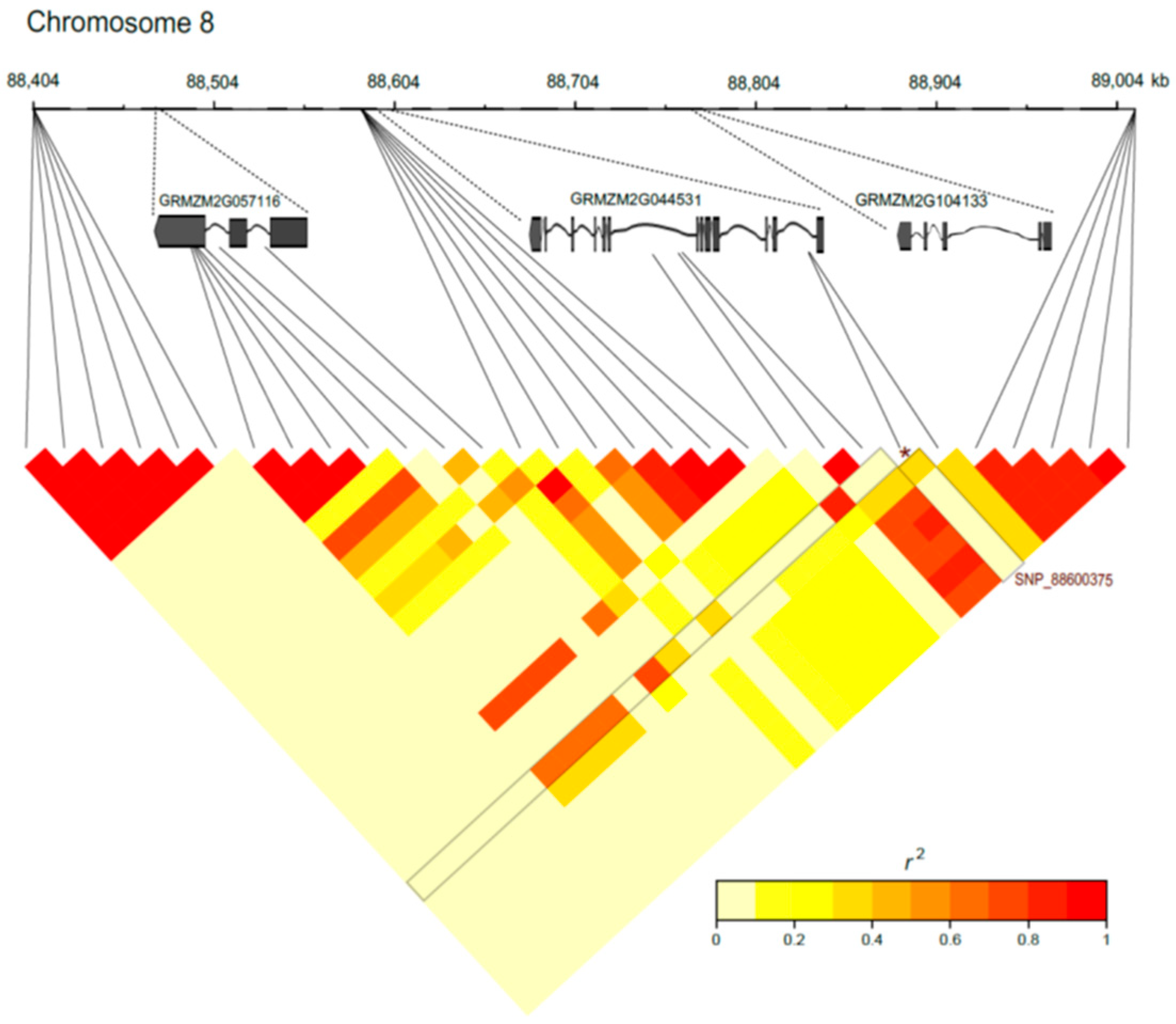
2.6. Exploring the Genomic Region Flanking the SNP S8_8600375
3. Discussion
3.1. Root and Biomass Plasticity under P Deficiency
3.2. Candidate Genes Associated with Root Morphology and P Acquisition-Related Traits
3.3. Exploring SNPs at Bin 8.03
4. Materials and Methods
4.1. Plant Material
4.2. Root Morphology Analysis, Phosphorus and Biomass Quantification
4.3. Experimental Design and Adjusted Means
4.4. SNP Data
4.5. Population Structure and Familial Relatedness
4.6. Linkage Disequilibrium (LD)
4.7. GWAS Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Vance, C.P.; Chiou, T.J. Phosphorus focus editorial. Plant Physiol. 2011, 156, 987–988. [Google Scholar] [CrossRef]
- Hammond, J.P.; Broadley, M.R.; White, P.J. Genetic responses to phosphorus deficiency. Ann. Bot. 2004, 94, 323–332. [Google Scholar] [CrossRef] [PubMed]
- Raghothama, K.G.; Karthikeyan, A.S. Phosphate acquisition. Plant Soil 2005, 274, 37–49. [Google Scholar] [CrossRef]
- Lynch, J.P. Root phenes that reduce the metabolic costs of soil exploration: Opportunities for 21st century agriculture. Plant Cell Environ. 2015, 38, 1775–1784. [Google Scholar] [CrossRef] [PubMed]
- Shen, J.; Yuan, L.; Zhang, J.; Li, H.; Bai, Z.; Chen, X.; Zhang, W.; Zhang, F. Phosphorus dynamics: From soil to plant. Plant Physiol. 2011, 156, 997–1005. [Google Scholar] [CrossRef] [PubMed]
- Baligar, V.C.; Fageria, N.K.; He, Z.L. Nutrient use efficiency in plants. Commun. Soil Sci. Plant Anal. 2001, 32, 921–950. [Google Scholar] [CrossRef]
- Vance, C.P.; Uhde-Stone, C.; Allan, D.L. Phosphorus acquisition and use: Critical adaptations by plants for securing a nonrenewable resource. New Phytol. 2003, 157, 423–447. [Google Scholar] [CrossRef]
- Parentoni, S.N.; De Souza Júnior, C.L. Phosphorus acquisition and internal utilization efficiency in tropical maize genotypes. Pesqui. Agropecu. Bras. 2008, 43, 893–901. [Google Scholar] [CrossRef]
- Zhu, J.; Lynch, J.P. The contribution of lateral rooting to phosphorus acquisition efficiency in maize (Zea mays) seedlings. Funct. Plant Biol. 2004, 31, 949–958. [Google Scholar] [CrossRef]
- Rai, A.; Rai, S.; Rakshit, A. Mycorrhiza-mediated phosphorus use efficiency in plants. Environ. Exp. Biol. 2013, 11, 107–117. [Google Scholar]
- Lynch, J.P. Root phenotypes for improved nutrient capture: An underexploited opportunity for global agriculture. New Phytol. 2019, 223, 548–564. [Google Scholar] [CrossRef]
- López-Bucio, J.; Cruz-Ramírez, A.; Herrera-Estrella, L. The role of nutrient availability in regulating root architecture. Curr. Opin. Plant Biol. 2003, 6, 280–287. [Google Scholar] [CrossRef]
- Ramaekers, L.; Remans, R.; Rao, I.M.; Blair, M.W.; Vanderleyden, J. Strategies for improving phosphorus acquisition efficiency of crop plants. Field Crops Res. 2010, 117, 169–176. [Google Scholar] [CrossRef]
- Mendes, F.F. Guimarães, L.J.M.; Souza, J.C.; Guimarães, P.E.O.; Magalhaes, J.V.; Garcia, A.A.F.; Parentoni, S.N.; Guimaraes, C.T. Genetic architecture of phosphorus use efficiency in tropical maize cultivated in a low-P soil. Crop Sci. 2014, 54, 1530–1538. [Google Scholar] [CrossRef]
- Magalhaes, J.V.; Piñeros, M.A.; Maciel, L.S.; Kochian, L.V. Emerging pleiotropic mechanisms underlying aluminum resistance and phosphorus acquisition on acidic soils. Front. Plant Sci. 2018, 9, 1420. [Google Scholar] [CrossRef] [PubMed]
- Barros, V.A.; Chandnani, R.; de Sousa, S.M.; Maciel, L.S.; Tokizawa, M.; Guimaraes, C.T.; Magalhaes, J.V.; Kochian, L.V. Root adaptation via common genetic factors conditioning tolerance to multiple stresses for crops cultivated on acidic tropical soils. Front. Plant Sci. 2020, 11, 565339. [Google Scholar] [CrossRef] [PubMed]
- Burton, A.L.; Johnson, J.M.; Foerster, J.M.; Hirsch, C.N.; Buell, C.R.; Hanlon, M.T.; Kaeppler, S.M.; Brown, K.M.; Lynch, J.P. QTL mapping and phenotypic variation for root architectural traits in maize (Zea mays L.). Theor. Appl. Genet. 2014, 127, 2293–2311. [Google Scholar] [CrossRef]
- Cai, H.; Chen, F.; Mi, G.; Zhang, F.; Maurer, H.P.; Liu, W.; Reif, J.C.; Yuan, L. Mapping QTLs for root system architecture of maize (Zea mays L.) in the field at different developmental stages. Theor. Appl. Genet. 2012, 125, 1313–1324. [Google Scholar] [CrossRef]
- Kumar, B.; Abdel-Ghani, A.H.; Pace, J.; Reyes-Matamoros, J.; Hochholdinger, F.; Lübberstedt, T. Association analysis of single nucleotide polymorphisms in candidate genes with root traits in maize (Zea mays L.) seedlings. Plant Sci. 2014, 224, 9–19. [Google Scholar] [CrossRef]
- Pace, J.; Gardner, C.; Romay, C.; Ganapathysubramanian, B.; Lübberstedt, T. Genome-wide association analysis of seedling root development in maize (Zea mays L.). BMC Genom. 2015, 16, 47. [Google Scholar] [CrossRef]
- Ren, W.; Zhao, L.; Liang, J.; Wang, L.; Chen, L.; Li, P.; Liu, Z.; Li, X.; Zhang, Z.; Li, J.; et al. Genome-wide dissection of changes in maize root system architecture during modern breeding. Nat. Plants 2022, 8, 1408–1422. [Google Scholar] [CrossRef]
- Song, W.; Wang, B.; Hauck, A.L.; Dong, X.; Li, J.; Lai, J. Genetic dissection of maize seedling root system architecture traits using an ultra-high density bin-map and a recombinant inbred line population. J. Integr. Plant Biol. 2016, 58, 266–279. [Google Scholar] [CrossRef]
- Zurek, P.R.; Topp, C.N.; Benfey, P.N. Quantitative trait locus mapping reveals regions of the maize genome controlling root system architecture. Plant Physiol. 2015, 167, 1487–1496. [Google Scholar] [CrossRef] [PubMed]
- Topp, C.N.; Iyer-Pascuzzi, A.S.; Anderson, J.T.; Lee, C.R.; Zurek, P.R.; Symonova, O.; Zheng, Y.; Bucksch, A.; Mileyko, Y.; Galkovskyi, T.; et al. 3D phenotyping and quantitative trait locus mapping identify core regions of the rice genome controlling root architecture. Proc. Natl. Acad. Sci. USA 2013, 110, E1695–E1704. [Google Scholar] [CrossRef] [PubMed]
- Zhu, J.; Mickelson, S.M.; Kaeppler, S.M.; Lynch, J.P. Detection of quantitative trait loci for seminal root traits in maize (Zea mays L.) seedlings grown under differential phosphorus levels. Theor. Appl. Genet. 2006, 113, 1–10. [Google Scholar] [CrossRef]
- Zhu, J.; Kaeppler, S.M.; Lynch, J.P. Mapping of QTLs for lateral root branching and length in maize (Zea mays L.) under differential phosphorus supply. Theor. Appl. Genet. 2005, 111, 688–695. [Google Scholar] [CrossRef]
- Zhu, J.; Kaeppler, S.M.; Lynch, J.P. Mapping of QTL controlling root hair length in maize (Zea mays L.) under phosphorus deficiency. Plant Soil 2005, 270, 299–310. [Google Scholar] [CrossRef]
- Azevedo, G.C.; Cheavegatti-Gianotto, A.; Negri, B.F.; Hufnagel, B.; Silva, L.C.; Magalhaes, J.V.; Garcia, A.A.F.; Lana, U.G.; de Sousa, S.M.; Guimaraes, C.T. Multiple interval QTL mapping and searching for PSTOL1 homologs associated with root morphology, biomass accumulation and phosphorus content in maize seedlings under low-P. BMC Plant Biol. 2015, 15, 172. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.; Xu, L.; Cai, Y.; Xu, J. QTL mapping of phosphorus efficiency and relative biologic characteristics in maize (Zea mays L.) at two sites. Plant Soil 2008, 313, 251–266. [Google Scholar] [CrossRef]
- Gamuyao, R.; Chin, J.H.; Pariasca-Tanaka, J.; Pesaresi, P.; Catausan, S.; Dalid, C.; Slamet-Loedin, I.; Tecson-Mendoza, E.M.; Wissuwa, M.; Heuer, S. The protein kinase Pstol1 from traditional rice confers tolerance of phosphorus deficiency. Nature 2012, 488, 535–539. [Google Scholar] [CrossRef] [PubMed]
- Anthony, R.G.; Henriques, R.; Helfer, A.; Mészáros, T.; Rios, G.; Testerink, C.; Munnik, T.; Deák, M.; Koncz, C.; Bögre, L. A protein kinase target of a PDK1 signalling pathway is involved in root hair growth in Arabidopsis. EMBO J. 2004, 23, 572–581. [Google Scholar] [CrossRef] [PubMed]
- Camehl, I.; Drzewiecki, C.; Vadassery, J.; Shahollari, B.; Sherameti, I.; Forzani, C.; Munnik, T.; Hirt, H.; Oelmüller, R. The OXI1 kinase pathway mediates Piriformospora indica-induced growth promotion in Arabidopsis. PLoS Pathog. 2011, 7, e1002051. [Google Scholar] [CrossRef]
- Oyama, T.; Shimura, Y.; Okada, K. The IRE gene encodes a protein kinase homologue and modulates root hair growth in Arabidopsis. Plant J. 2002, 30, 289–299. [Google Scholar] [CrossRef]
- Santner, A.A.; Watson, J.C. The WAG1 and WAG2 protein kinases negatively regulate root waving in Arabidopsis. Plant J. 2006, 45, 752–764. [Google Scholar] [CrossRef]
- Yu, J.; Buckler, E.S. Genetic association mapping and genome organization of maize. Curr. Opin. Biotechnol. 2006, 17, 155–160. [Google Scholar] [CrossRef]
- Yan, J.; Warburton, M.; Crouch, J. Association mapping for enhancing maize (Zea mays L.) genetic improvement. Crop Sci. 2011, 51, 433–449. [Google Scholar] [CrossRef]
- Cairns, J.E.; Crossa, J.; Zaidi, P.H.; Grudloyma, P.; Sanchez, C.; Luis Araus, J.; Thaitad, S.; Makumbi, D.; Magorokosho, C.; Bänziger, M.; et al. Identification of drought, heat, and combined drought and heat tolerant donors in maize. Crop Sci. 2013, 53, 1335–1346. [Google Scholar] [CrossRef]
- Chen, J.; Xu, L. The candidate QTLs affecting phosphorus absorption efficiency and root weight in maize (Zea mays L.). Front. Agric. Chin. 2011, 5, 456–462. [Google Scholar] [CrossRef]
- Xu, C.; Zhang, H.; Sun, J.; Guo, Z.; Zou, C.; Li, W.X.; Xie, C.; Huang, C.; Xu, R.; Liao, H.; et al. Genome-wide association study dissects yield components associated with low-phosphorus stress tolerance in maize. Theor. Appl. Genet. 2018, 131, 1699–1714. [Google Scholar] [CrossRef]
- Flint-Garcia, S.A.; Thornsberry, J.M.; Buckler, E.S. Structure of linkage disequilibrium in plants. Annu. Rev. Plant Biol. 2003, 54, 357–374. [Google Scholar] [CrossRef] [PubMed]
- Romay, M.C.; Millard, M.J.; Glaubitz, J.C.; Peiffer, J.A.; Swarts, K.L.; Casstevens, T.M.; Elshire, R.J.; Acharya, C.B.; Mitchell, S.E.; Flint-Garcia, S.A.; et al. Comprehensive genotyping of the USA national maize inbred seed bank. Genome Biol. 2013, 14, R55. [Google Scholar] [CrossRef] [PubMed]
- Nair, S.K.; Babu, R.; Magorokosho, C.; Mahuku, G.; Semagn, K.; Beyene, Y.; Das, B.; Makumbi, D.; Lava Kumar, P.; Olsen, M.; et al. Fine mapping of Msv1, a major QTL for resistance to Maize Streak Virus leads to development of production markers for breeding pipelines. Theor. Appl. Genet. 2015, 128, 1839–1854. [Google Scholar] [CrossRef] [PubMed]
- Svistoonoff, S.; Creff, A.; Reymond, M.; Sigoillot-Claude, C.; Ricaud, L.; Blanchet, A.; Nussaume, L.; Desnos, T. Root tip contact with low-phosphate media reprograms plant root architecture. Nat. Genet. 2007, 39, 792–796. [Google Scholar] [CrossRef] [PubMed]
- Wen, Z.; Li, H.; Shen, Q.; Tang, X.; Xiong, C.; Li, H.; Pang, J.; Ryan, M.H.; Lambers, H.; Shen, J. Tradeoffs among root morphology, exudation and mycorrhizal symbioses for phosphorus-acquisition strategies of 16 crop species. New Phytol. 2019, 223, 882–895. [Google Scholar] [CrossRef]
- Ryan, M.H.; Tibbett, M.; Edmonds-Tibbett, T.; Suriyagoda, L.D.B.; Lambers, H.; Cawthray, G.R.; Pang, J. Carbon trading for phosphorus gain: The balance between rhizosphere carboxylates and arbuscular mycorrhizal symbiosis in plant phosphorus acquisition. Plant Cell Environ. 2012, 35, 2170–2180. [Google Scholar] [CrossRef]
- Lambers, H.; Shane, M.W.; Cramer, M.D.; Pearse, S.J.; Veneklaas, E.J. Root structure and functioning for efficient acquisition of phosphorus: Matching morphological and physiological traits. Ann. Bot. 2006, 98, 693–713. [Google Scholar] [CrossRef]
- King, W.L.; Yates, C.F.; Guo, J.; Fleishman, S.M.; Trexler, R.V.; Centinari, M.; Bell, T.H.; Eissenstat, D.M. The hierarchy of root branching order determines bacterial composition, microbial carrying capacity and microbial filtering. Commun. Biol. 2021, 4, 483. [Google Scholar] [CrossRef]
- Lynch, J.P. Steep, cheap and deep: An ideotype to optimize water and N acquisition by maize root systems. Ann. Bot. 2013, 112, 347–357. [Google Scholar] [CrossRef]
- Bengough, A.G.; McKenzie, B.M.; Hallett, P.D.; Valentine, T.A. Root elongation, water stress, and mechanical impedance: A review of limiting stresses and beneficial root tip traits. J. Exp. Bot. 2011, 62, 59–68. [Google Scholar] [CrossRef]
- McCormack, M.L.; Dickie, I.A.; Eissenstat, D.M.; Fahey, T.J.; Fernandez, C.W.; Guo, D.; Helmisaari, H.S.; Hobbie, E.A.; Iversen, C.M.; Jackson, R.B.; et al. Redefining fine roots improves understanding of below-ground contributions to to terrestrial biosphere processes. New Phytol. 2015, 20, 505–518. [Google Scholar] [CrossRef]
- Zhang, H.W.; Huang, Y.; Ye, X.S.; Xu, F. Genotypic variation in phosphorus acquisition from sparingly soluble P sources is related to root morphology and root exudates in Brassica napus. Sci. Chin. Life Sci. 2011, 54, 1134–1142. [Google Scholar] [CrossRef]
- Hufnagel, B.; de Sousa, S.M.; Assis, L.; Guimaraes, C.T.; Leiser, W.; Azevedo, G.C.; Negri, B.; Larson, B.G.; Shaff, J.E.; Pastina, M.M.; et al. Duplicate and conquer: Multiple homologs of PHOSPHORUS-STARVATION TOLERANCE1 enhance phosphorus acquisition and sorghum performance on low-phosphorus soils. Plant Physiol. 2014, 166, 659–677. [Google Scholar] [CrossRef]
- Bernardino, K.C.; de Menezes, C.B.; de Sousa, S.M.; Guimarães, C.T.; Carneiro, P.C.S.; Schaffert, R.E.; Kochian, L.V.; Hufnagel, B.; Pastina, M.M.; Magalhaes, J.V. Association mapping and genomic selection for sorghum adaptation to tropical soils of Brazil in a sorghum multiparental random mating population. Theor. Appl. Genet. 2021, 134, 295–312. [Google Scholar] [CrossRef] [PubMed]
- Bernardino, K.C.; Pastina, M.M.; Menezes, C.B.; De Sousa, S.M.; Maciel, L.S.; Carvalho, G.C.; Guimarães, C.T.; Barros, B.A.; Silva, L.C.; Carneiro, P.C.S.; et al. The genetic architecture of phosphorus efficiency in sorghum involves pleiotropic QTL for root morphology and grain yield under low phosphorus availability in the soil. BMC Plant Biol. 2019, 19, 1–15. [Google Scholar] [CrossRef] [PubMed]
- Gao, S.Q.; Chen, G.D.; Hu, D.Y.; Zhang, X.Z.; Li, T.X.; Liu, S.H.; Liu, C.J. A major quantitative trait locus controlling phosphorus utilization efficiency under different phytate-P conditions at vegetative stage in barley. J. Integr. Agric. 2018, 17, 285–295. [Google Scholar] [CrossRef]
- Kong, X.; Lv, W.; Zhang, D.; Jiang, S.; Zhang, S.; Li, D. Genome-wide identification and analysis of expression profiles of maize mitogen-activated protein kinase kinase kinase. PLoS ONE 2013, 8, e57714. [Google Scholar] [CrossRef]
- Shi, Z.; Wen, B.; Song, W.; Liu, Y.; Zhou, M.; Wang, J.; Zhao, J.; Ren, W. Genome-wide identification and characterization of the MAPKKK MKK and MPK families in.pdf. Plant Genome 2022, 15, e20216. [Google Scholar] [CrossRef] [PubMed]
- Mollier, A.; Pellerin, S. Maize root system growth and development as influenced by phosphorus deficiency. J. Exp. Bot. 1999, 50, 487–497. [Google Scholar] [CrossRef]
- Zegzouti, H.; Li, W.; Lorenz, T.C.; Xie, M.; Payne, C.T.; Smith, K.; Glenny, S.; Payne, G.S.; Christensen, S.K. Structural and functional insights into the regulation of Arabidopsis AGC VIIIa kinases. J. Biol. Chem. 2006, 281, 35520–35530. [Google Scholar] [CrossRef]
- Eulgem, T.; Rushton, P.J.; Robatzek, S.; Somssich, I.E. The WRKY superfamily of plant transcription factors. Trends Plant Sci. 2000, 5, 199–206. [Google Scholar] [CrossRef]
- Marè, C.; Mazzucotelli, E.; Crosatti, C.; Francia, E.; Stanca, A.M.; Cattivelli, L. Hv-WRKY38: A new transcription factor involved in cold- and drought-response in barley. Plant Mol. Biol. 2004, 55, 399–416. [Google Scholar] [CrossRef] [PubMed]
- Mattiello, L.; Kirst, M.; da Silva, F.R.; Jorge, R.A.; Menossi, M. Transcriptional profile of maize roots under acid soil growth. BMC Plant Biol. 2010, 10, 196. [Google Scholar] [CrossRef] [PubMed]
- Melo, J.O.; Martins, L.G.C.; Barros, B.A.; Pimenta, M.R.; Lana, U.G.P.; Duarte, C.E.M.; Pastina, M.M.; Guimaraes, C.T.; Schaffert, R.E.; Kochian, L.V.; et al. Repeat variants for the SbMATE transporter protect sorghum roots from aluminum toxicity by transcriptional interplay in cis and trans. Proc. Natl. Acad. Sci. USA 2019, 116, 313–318. [Google Scholar] [CrossRef] [PubMed]
- Seki, M.; Narusaka, M.; Ishida, J.; Nanjo, T.; Fujita, M.; Oono, Y.; Kamiya, A.; Nakajima, M.; Enju, A.; Sakurai, T.; et al. Monitoring the expression profiles of 7000 Arabidopsis genes under drought, cold and high-salinity stresses using a full-length cDNA microarray. Plant J. 2002, 31, 279–292. [Google Scholar] [CrossRef] [PubMed]
- Wei, K.F.; Chen, J.; Chen, Y.F.; Wu, L.J.; Xie, D.X. Molecular phylogenetic and expression analysis of the complete WRKY transcription factor family in maize. DNA Res. 2012, 19, 153–164. [Google Scholar] [CrossRef]
- Zhang, T.; Tan, D.; Zhang, L.; Zhang, X.; Han, Z. Phylogenetic analysis and drought-responsive expression profiles of the WRKY transcription factor family in maize. Agri Gene 2017, 3, 99–108. [Google Scholar] [CrossRef]
- Parentoni, S.N.; De Souza, C.L.; Alves, V.M.C.; Gama, E.E.G.; Coelho, A.M.; Oliveira, A.C.; Guimarães, P.E.O.; Guimarães, C.T.; Vasconcelos, M.J.V.; Pacheco, C.A.P.; et al. Inheritance and breeding strategies for phosphorus efficiency in tropical maize (Zea mays L.). Maydica 2010, 55, 1–15. [Google Scholar]
- De Sousa, S.M.; Clark, R.T.; Mendes, F.F.; Oliveira, A.C.; Vasconcelos, M.J.V.; Parentoni, S.N.; Kochian, L.V.; Guimarães, C.T.; Magalhães, J.V. A role for root morphology and related candidate genes in P acquisition efficiency in maize. Funct. Plant Biol. 2012, 39, 925–935. [Google Scholar] [CrossRef]
- Magnavaca, R.; Gardner, C.O.; Clark, R.B. Inheritance of aluminum toleranee in maize. In Genetic Aspects of Plant Mineral Nutrition; Gabelman, W.H., Loughman, B.C., Eds.; Martinus Nijhoff Publishers: Dordrecht, The Netherlands, 1987; Volume 27, pp. 201–212. [Google Scholar]
- Clark, R.T.; Famoso, A.N.; Zhao, K.; Shaff, J.E.; Craft, E.J.; Bustamante, C.D.; McCouch, S.R.; Aneshansley, D.J.; Kochian, L.V. High-throughput two-dimensional root system phenotyping platform facilitates genetic analysis of root growth and development. Plant Cell Environ. 2013, 36, 454–466. [Google Scholar] [CrossRef]
- Payne, R.W. GenStat. Wiley Interdiscip. Rev. Comput. Stat. 2009, 1, 255–258. [Google Scholar] [CrossRef]
- Akaike, H. A new look at the statistical model identification. IEEE Trans. Automat. Control 1974, 19, 716–723. [Google Scholar] [CrossRef]
- Saghai-Maroof, M.A.; Soliman, K.M.; Jorgensen, R.A.; Allard, R.W. Ribosomal DNA spacer-length polymorphisms in barley: Mendelian inheritance, chromosomal location, and population dynamics. Proc. Natl. Acad. Sci. USA 1984, 81, 8014–8018. [Google Scholar] [CrossRef] [PubMed]
- Elshire, R.J.; Glaubitz, J.C.; Sun, Q.; Poland, J.A.; Kawamoto, K.; Buckler, E.S.; Mitchell, S.E. A robust, simple genotyping-by-sequencing (GBS) approach for high diversity species. PLoS ONE 2011, 6, e19379. [Google Scholar] [CrossRef]
- Glaubitz, J.C.; Casstevens, T.M.; Lu, F.; Harriman, J.; Elshire, R.J.; Sun, Q.; Buckler, E.S. TASSEL-GBS: A high capacity genotyping by sequencing analysis pipeline. PLoS ONE 2014, 9, e90346. [Google Scholar] [CrossRef] [PubMed]
- Bradbury, P.J.; Zhang, Z.; Kroon, D.E.; Casstevens, T.M.; Ramdoss, Y.; Buckler, E.S. TASSEL: Software for association mapping of complex traits in diverse samples. Bioinformatics 2007, 23, 2633–2635. [Google Scholar] [CrossRef]
- Hintze, J.L.; Nelson, R.D. Violin plots: A box plot-density trace synergism. Am. Stat. 1998, 52, 181–184. [Google Scholar]
- R Core Team. R: A language and environment for statistical computing. In R Foundation for Statistical Computing; The R Foundation: Boston, MA, USA, 2015; pp. 1–48. [Google Scholar]
- Johnson, R.C.; Nelson, G.W.; Troyer, J.L.; Lautenberger, J.A.; Kessing, B.D.; Winkler, C.A.; O’Brien, S.J. Accounting for multiple comparisons in a genome-wide association study (GWAS). BMC Genom. 2010, 11, 724. [Google Scholar] [CrossRef] [PubMed]

| Trait | Acronym | P Level | Mean | Min | Max | CV | h2 | ||
|---|---|---|---|---|---|---|---|---|---|
| Root Length (cm) | RL | Low | 162.5 | 32.4 | 479.4 | 1807 | 1935 | 39.7 | 0.71 |
| High | 168.4 | 20.8 | 515.5 | 1858 | 2017 | 44.7 | 0.71 | ||
| Root Surface Area (cm2) | SA | Low | 47.2 | 12.0 | 200.8 | 60.2 | 175.2 | 55.0 | 0.54 |
| High | 47.7 | 9.2 | 154.0 | 93.9 | 165.3 | 48.4 | 0.65 | ||
| Root Diameter (mm) | RD | Low | 0.93 | 0.37 | 2.15 | 0.005 | 0.810 | 33.4 | 0.70 |
| High | 0.96 | 0.43 | 2.25 | 0.006 | 0.704 | 32.5 | 0.70 | ||
| Total Seedling Dry Weight (mg) | TDW | Low | 64.1 | 11.5 | 166.3 | 198.0 | 227.6 | 36.3 | 0.70 |
| High | 72.9 | 15.3 | 220.3 | 263.4 | 327.6 | 40.9 | 0.70 | ||
| Total P Content (mg) | PCont | Low | 0.92 | 0.10 | 1.32 | 0.02 | 0.017 | 36.4 | 0.49 |
| High | 0.97 | 0.07 | 2.39 | 0.03 | 0.019 | 41.1 | 0.56 |
| Traits | RL | SA | RD | TDW | PCont |
|---|---|---|---|---|---|
| RL | 0.46 * | 0.67 * | −0.36 * | 0.54 * | 0.22 * |
| SA | 0.75 * | 0.72 * | 0.38 * | 0.18 * | −0.05 ns |
| RD | −0.02 ns | 0.60 * | 0.92 * | −0.39 * | −0.29 * |
| TDW | 0.34 * | 0.14 * | −0.18 * | 0.54 * | 0.70 * |
| PCont | −0.14 * | −0.27 * | −0.27 * | 0.58 * | 0.42 * |
| Trait | Effects of P within Panels | Genetic Variance a | |||||
|---|---|---|---|---|---|---|---|
| Global | Embrapa | DTMA | Embrapa | DTMA | |||
| High-P | Low-P | High-P | Low-P | ||||
| Root Length (cm) | 3.27 * | −6.37 * | 16.79 ** | 1529.56 (160.84) | 1368.54 (147.86) | 2595.15 (353.17) | 2700.39 (353.06) |
| Root Surface Area (cm2) | 2.81 *** | −2.42 *** | 10.11 *** | 69.97 (7.30) | 43.64 (5.11) | 311.36 (44.05) | 348.23 (47.04) |
| Root Diameter (mm) | 0.07 *** | 0.07 *** | 0.11 ns | 0.003 (0.0004) | 0.003 (0.0004) | 0.02 (0.002) | 0.01 (0.001) |
| Total Dry Weight (mg) | 9.89 *** | 8.29 *** | 13.04 ns | 319.55 (34.42) | 221.47 (23.23) | 185.61 (24.27) | 147.87 (21.92) |
| Total P Content (mg) | 0.02 *** | 0.02 *** | 0.02 ns | 0.04 (0.005) | 0.02 (0.004) | 0.02 (0.002) | 0.01 (0.003) |
| Trait | P level | SNP | Chr (bin) | Position (Mb) | −log10(p-Value) | Alleles | MAF | Predicted Gene |
|---|---|---|---|---|---|---|---|---|
| RD | high | S4_3751192 | 4.01 | 37.51 | 6.44 | A/C | 0.21 | GRMZM2G037472 |
| RL | low | S8_88600375 | 8.03 | 88.60 | 6.30 | C/A | 0.13 | GRMZM2G044531 |
| TDW | low | S6_34607369 | 6.01 | 34.61 | 6.37 | T/C | 0.25 | NA |
| low | S9_137746077 | 9.06 | 137.75 | 6.43 | G/A | 0.48 | GRMZM2G378852 | |
| high | S10_77284783 | 10.03 | 77.28 | 6.41 | G/C | 0.09 | GRMZM2G110145 | |
| PCont | high | S8_21326857 | 8.03 | 21.33 | 6.34 | G/A | 0.22 | NA |
| high | S9_143192439 | 9.06 | 143.19 | 6.11 | T/A | 0.47 | GRMZM2G104618 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ribeiro, C.A.G.; de Sousa Tinoco, S.M.; de Souza, V.F.; Negri, B.F.; Gault, C.M.; Pastina, M.M.; Magalhaes, J.V.; Guimarães, L.J.M.; de Barros, E.G.; Buckler, E.S.; et al. Genome-Wide Association Study for Root Morphology and Phosphorus Acquisition Efficiency in Diverse Maize Panels. Int. J. Mol. Sci. 2023, 24, 6233. https://doi.org/10.3390/ijms24076233
Ribeiro CAG, de Sousa Tinoco SM, de Souza VF, Negri BF, Gault CM, Pastina MM, Magalhaes JV, Guimarães LJM, de Barros EG, Buckler ES, et al. Genome-Wide Association Study for Root Morphology and Phosphorus Acquisition Efficiency in Diverse Maize Panels. International Journal of Molecular Sciences. 2023; 24(7):6233. https://doi.org/10.3390/ijms24076233
Chicago/Turabian StyleRibeiro, Carlos Alexandre Gomes, Sylvia Morais de Sousa Tinoco, Vander Fillipe de Souza, Barbara França Negri, Christine Marie Gault, Maria Marta Pastina, Jurandir Vieira Magalhaes, Lauro José Moreira Guimarães, Everaldo Gonçalves de Barros, Edward S. Buckler, and et al. 2023. "Genome-Wide Association Study for Root Morphology and Phosphorus Acquisition Efficiency in Diverse Maize Panels" International Journal of Molecular Sciences 24, no. 7: 6233. https://doi.org/10.3390/ijms24076233
APA StyleRibeiro, C. A. G., de Sousa Tinoco, S. M., de Souza, V. F., Negri, B. F., Gault, C. M., Pastina, M. M., Magalhaes, J. V., Guimarães, L. J. M., de Barros, E. G., Buckler, E. S., & Guimaraes, C. T. (2023). Genome-Wide Association Study for Root Morphology and Phosphorus Acquisition Efficiency in Diverse Maize Panels. International Journal of Molecular Sciences, 24(7), 6233. https://doi.org/10.3390/ijms24076233






