Three-Dimensional Cell Co-Culture Liver Models and Their Applications in Pharmaceutical Research
Abstract
1. Introduction
2. The Main Cellular Components of the Liver and Their Functions
3. Potential Advantages of 3D Cell Co-Culture Liver Models for Drug Evaluation
4. Hepatocyte Sources of 3D Cell Co-Culture Liver Models
4.1. Primary Hepatocytes
4.2. Hepatoma Cell Lines
4.3. Stem Cells
5. Common Co-Culture Methods for 3D Cell Co-Culture Liver Models
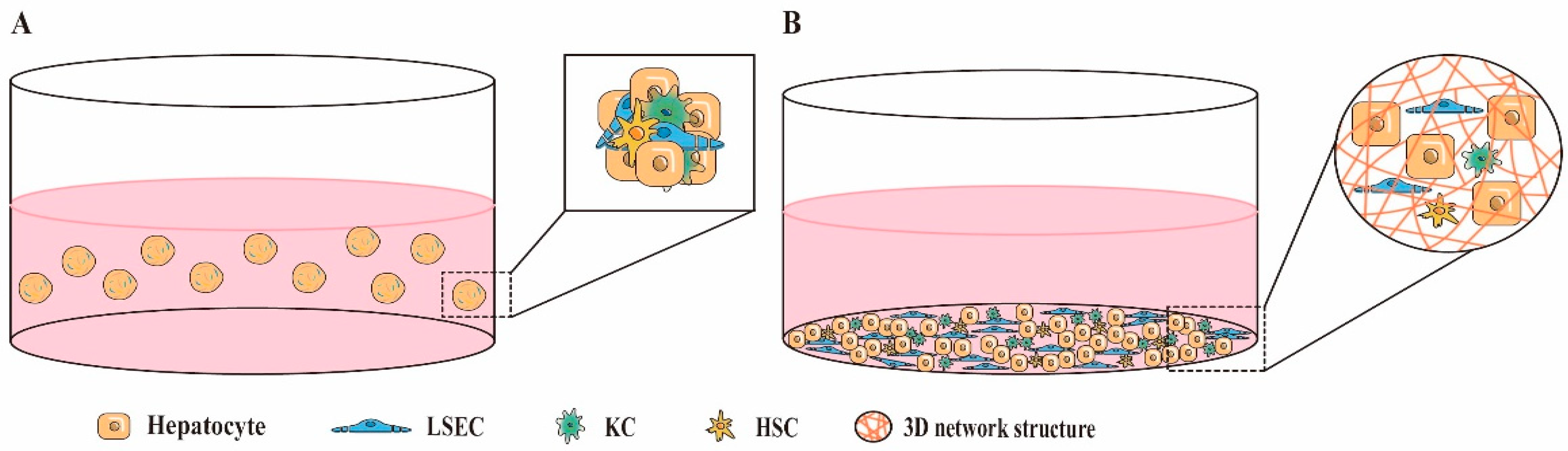
5.1. Direct 3D Co-Culture
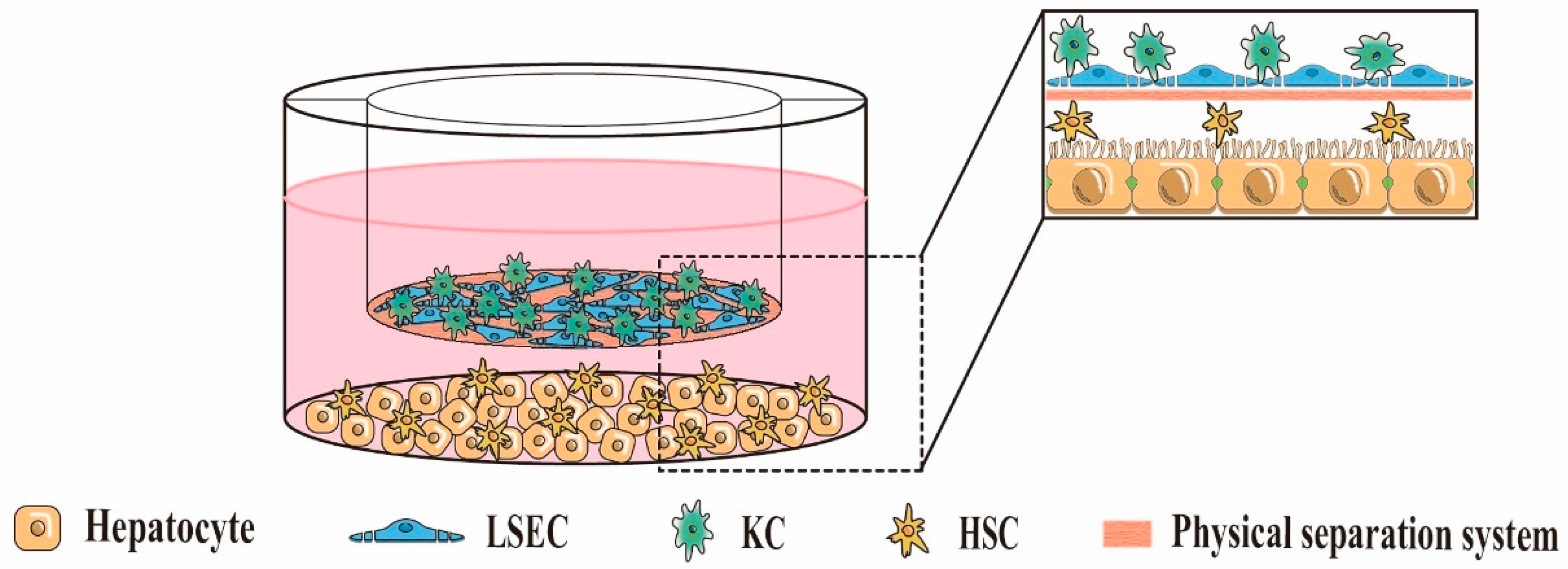
5.2. Indirect 3D Co-Culture
6. Types of 3D Cell Co-Culture Liver Model
6.1. Three-Dimensional Cell Co-Culture Models for Enhanced Hepatocyte Function
6.2. Three-Dimensional Cell Co-Culture Models of Liver Fibrosis
6.3. Three-Dimensional Cell Co-Culture Model of Drug-Induced Liver Injury
7. Conclusions and Application Prospects of 3D Cell Co-Culture Models
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Ma, L.; Wu, Y.; Li, Y.; Aazmi, A.; Zhou, H.; Zhang, B.; Yang, H. Current Advances on 3D-Bioprinted Liver Tissue Models. Adv. Healthc. Mater. 2020, 9, e2001517. [Google Scholar] [CrossRef] [PubMed]
- Zhu, X.; Zhang, B.; He, Y.; Bao, J. Liver Organoids: Formation Strategies and Biomedical Applications. Tissue Eng. Regen. Med. 2021, 18, 573–585. [Google Scholar] [CrossRef] [PubMed]
- Janani, G.; Priya, S.; Dey, S.; Mandal, B.B. Mimicking Native Liver Lobule Microarchitecture In Vitro with Parenchymal and Non-parenchymal Cells Using 3D Bioprinting for Drug Toxicity and Drug Screening Applications. ACS Appl. Mater. Interfaces 2022, 14, 10167–10186. [Google Scholar] [CrossRef] [PubMed]
- Braet, F.; Taatjes, D.J.; Wisse, E. Probing the unseen structure and function of liver cells through atomic force microscopy. Semin. Cell Dev. Biol. 2018, 73, 13–30. [Google Scholar] [CrossRef]
- Calitz, C.; Hamman, J.H.; Fey, S.J.; Wrzesinski, K.; Gouws, C. Recent advances in three-dimensional cell culturing to assess liver function and dysfunction: From a drug biotransformation and toxicity perspective. Toxicol. Mech. Methods 2018, 28, 369–385. [Google Scholar] [CrossRef]
- Shao, T.; Chen, Z.; Rong, J.; Belov, V.; Chen, J.; Jeyarajan, A.; Deng, X.; Fu, H.; Yu, Q.; Rwema, S.H.; et al. [(18)F]MAGL-4-11 positron emission tomography molecular imaging of monoacylglycerol lipase changes in preclinical liver fibrosis models. Acta Pharm. Sin. B 2022, 12, 308–315. [Google Scholar] [CrossRef]
- Mandon, M.; Huet, S.; Dubreil, E.; Fessard, V.; Le Hégarat, L. Three-dimensional HepaRG spheroids as a liver model to study human genotoxicity in vitro with the single cell gel electrophoresis assay. Sci. Rep. 2019, 9, 10548. [Google Scholar] [CrossRef]
- Hiller, T.; Berg, J.; Elomaa, L.; Röhrs, V.; Ullah, I.; Schaar, K.; Dietrich, A.C.; Al-Zeer, M.A.; Kurtz, A.; Hocke, A.C.; et al. Generation of a 3D Liver Model Comprising Human Extracellular Matrix in an Alginate/Gelatin-Based Bioink by Extrusion Bioprinting for Infection and Transduction Studies. Int. J. Mol. Sci. 2018, 19, 3129. [Google Scholar] [CrossRef]
- Tian, Q.Q.; Zhu, Y.T.; Diao, X.X.; Zhang, X.L.; Xu, Y.C.; Jiang, X.R.; Shen, J.S.; Wang, Z.; Zhong, D.F. Species differences in the CYP3A-catalyzed metabolism of TPN729, a novel PDE5 inhibitor. Acta Pharmacol. Sin. 2021, 42, 482–490. [Google Scholar] [CrossRef]
- Wei, J.; Lu, J.; Chen, M.; Xie, S.; Wang, T.; Li, X. 3D spheroids generated on carbon nanotube-functionalized fibrous scaffolds for drug metabolism and toxicity screening. Biomater. Sci. 2019, 8, 426–437. [Google Scholar] [CrossRef]
- Wang, J.; Chen, F.; Liu, L.; Qi, C.; Wang, B.; Yan, X.; Huang, C.; Hou, W.; Zhang, M.Q.; Chen, Y.; et al. Engineering EMT using 3D micro-scaffold to promote hepatic functions for drug hepatotoxicity evaluation. Biomaterials 2016, 91, 11–22. [Google Scholar] [CrossRef]
- Zheng, Y.B.; Ma, L.D.; Wu, J.L.; Wang, Y.M.; Meng, X.S.; Hu, P.; Liang, Q.L.; Xie, Y.Y.; Luo, G.A. Design and fabrication of an integrated 3D dynamic multicellular liver-on-a-chip and its application in hepatotoxicity screening. Talanta 2022, 241, 123262. [Google Scholar] [CrossRef] [PubMed]
- Chen, M.; Borlak, J.; Tong, W. High lipophilicity and high daily dose of oral medications are associated with significant risk for drug-induced liver injury. Hepatology 2013, 58, 388–396. [Google Scholar] [CrossRef] [PubMed]
- Babai, S.; Auclert, L.; Le-Louët, H. Safety data and withdrawal of hepatotoxic drugs. Therapie 2021, 76, 715–723. [Google Scholar] [CrossRef]
- Yamada, K.M.; Cukierman, E. Modeling tissue morphogenesis and cancer in 3D. Cell 2007, 130, 601–610. [Google Scholar] [CrossRef] [PubMed]
- Breslin, S.; O’Driscoll, L. Three-dimensional cell culture: The missing link in drug discovery. Drug Discov. Today 2013, 18, 240–249. [Google Scholar] [CrossRef]
- Sarkar, U.; Ravindra, K.C.; Large, E.; Young, C.L.; Rivera-Burgos, D.; Yu, J.; Cirit, M.; Hughes, D.J.; Wishnok, J.S.; Lauffenburger, D.A.; et al. Integrated Assessment of Diclofenac Biotransformation, Pharmacokinetics, and Omics-Based Toxicity in a Three-Dimensional Human Liver-Immunocompetent Coculture System. Drug Metab. Dispos. 2017, 45, 855–866. [Google Scholar] [CrossRef]
- Li, Y.; Zhang, T.; Pang, Y.; Li, L.; Chen, Z.N.; Sun, W. 3D bioprinting of hepatoma cells and application with microfluidics for pharmacodynamic test of Metuzumab. Biofabrication 2019, 11, 034102. [Google Scholar] [CrossRef]
- Ströbel, S.; Kostadinova, R.; Fiaschetti-Egli, K.; Rupp, J.; Bieri, M.; Pawlowska, A.; Busler, D.; Hofstetter, T.; Sanchez, K.; Grepper, S.; et al. A 3D primary human cell-based in vitro model of non-alcoholic steatohepatitis for efficacy testing of clinical drug candidates. Sci. Rep. 2021, 11, 22765. [Google Scholar] [CrossRef]
- Berasain, C.; Avila, M.A. Regulation of hepatocyte identity and quiescence. Cell Mol. Life Sci. 2015, 72, 3831–3851. [Google Scholar] [CrossRef]
- Moradi, E.; Jalili-Firoozinezhad, S.; Solati-Hashjin, M. Microfluidic organ-on-a-chip models of human liver tissue. Acta Biomater. 2020, 116, 67–83. [Google Scholar] [CrossRef] [PubMed]
- Hu, L.; Li, H.; Chi, Z.; He, J. Loss of the RNA-binding protein Rbm15 disrupts liver maturation in zebrafish. J. Biol. Chem. 2020, 295, 11466–11472. [Google Scholar] [CrossRef] [PubMed]
- Kang, Y.B.; Rawat, S.; Cirillo, J.; Bouchard, M.; Noh, H.M. Layered long-term co-culture of hepatocytes and endothelial cells on a transwell membrane: Toward engineering the liver sinusoid. Biofabrication 2013, 5, 045008. [Google Scholar] [CrossRef] [PubMed]
- De Rudder, M.; Dili, A.; Stärkel, P.; Leclercq, I.A. Critical Role of LSEC in Post-Hepatectomy Liver Regeneration and Failure. Int. J. Mol. Sci. 2021, 22, 8053. [Google Scholar] [CrossRef]
- Gracia-Sancho, J.; Caparrós, E.; Fernández-Iglesias, A.; Francés, R. Role of liver sinusoidal endothelial cells in liver diseases. Nat. Rev. Gastroenterol. Hepatol. 2021, 18, 411–431. [Google Scholar] [CrossRef] [PubMed]
- Shetty, S.; Lalor, P.F.; Adams, D.H. Liver sinusoidal endothelial cells-gatekeepers of hepatic immunity. Nat. Rev. Gastroenterol. Hepatol. 2018, 15, 555–567. [Google Scholar] [CrossRef] [PubMed]
- DeLeve, L.D.; Maretti-Mira, A.C. Liver Sinusoidal Endothelial Cell: An Update. Semin. Liver Dis. 2017, 37, 377–387. [Google Scholar] [CrossRef]
- Lafoz, E.; Ruart, M.; Anton, A.; Oncins, A.; Hernández-Gea, V. The Endothelium as a Driver of Liver Fibrosis and Regeneration. Cells 2020, 9, 929. [Google Scholar] [CrossRef]
- Hammoutene, A.; Rautou, P.E. Role of liver sinusoidal endothelial cells in non-alcoholic fatty liver disease. J. Hepatol. 2019, 70, 1278–1291. [Google Scholar] [CrossRef]
- Ezhilarasan, D. Hepatic stellate cells in the injured liver: Perspectives beyond hepatic fibrosis. J. Cell Physiol. 2022, 237, 436–449. [Google Scholar] [CrossRef]
- Terkelsen, M.K.; Bendixen, S.M.; Hansen, D.; Scott, E.A.H.; Moeller, A.F.; Nielsen, R.; Mandrup, S.; Schlosser, A.; Andersen, T.L.; Sorensen, G.L.; et al. Transcriptional Dynamics of Hepatic Sinusoid-Associated Cells After Liver Injury. Hepatology 2020, 72, 2119–2133. [Google Scholar] [CrossRef] [PubMed]
- Krenkel, O.; Tacke, F. Liver macrophages in tissue homeostasis and disease. Nat. Rev. Immunol. 2017, 17, 306–321. [Google Scholar] [CrossRef] [PubMed]
- Brooks, A.; Liang, X.; Zhang, Y.; Zhao, C.X.; Roberts, M.S.; Wang, H.; Zhang, L.; Crawford, D.H.G. Liver organoid as a 3D in vitro model for drug validation and toxicity assessment. Pharmacol. Res. 2021, 169, 105608. [Google Scholar] [CrossRef] [PubMed]
- Tasnim, F.; Singh, N.H.; Tan, E.K.F.; Xing, J.; Li, H.; Hissette, S.; Manesh, S.; Fulwood, J.; Gupta, K.; Ng, C.W.; et al. Tethered primary hepatocyte spheroids on polystyrene multi-well plates for high-throughput drug safety testing. Sci. Rep. 2020, 10, 4768. [Google Scholar] [CrossRef] [PubMed]
- Corrado, B.; De Gregorio, V.; Imparato, G.; Attanasio, C.; Urciuolo, F.; Netti, P.A. A three-dimensional microfluidized liver system to assess hepatic drug metabolism and hepatotoxicity. Biotechnol. Bioeng. 2019, 116, 1152–1163. [Google Scholar] [CrossRef]
- Shiota, J.; Samuelson, L.C.; Razumilava, N. Hepatobiliary Organoids and Their Applications for Studies of Liver Health and Disease: Are We There Yet? Hepatology 2021, 74, 2251–2263. [Google Scholar] [CrossRef]
- Ya, S.; Ding, W.; Li, S.; Du, K.; Zhang, Y.; Li, C.; Liu, J.; Li, F.; Li, P.; Luo, T.; et al. On-Chip Construction of Liver Lobules with Self-Assembled Perfusable Hepatic Sinusoid Networks. ACS Appl. Mater. Interfaces 2021, 13, 32640–32652. [Google Scholar] [CrossRef]
- Lee, H.; Chae, S.; Kim, J.Y.; Han, W.; Kim, J.; Choi, Y.; Cho, D.W. Cell-printed 3D liver-on-a-chip possessing a liver microenvironment and biliary system. Biofabrication 2019, 11, 025001. [Google Scholar] [CrossRef]
- Lu, Y.; Ma, J.; Lin, G. Development of a two-layer transwell co-culture model for the in vitro investigation of pyrrolizidine alkaloid-induced hepatic sinusoidal damage. Food Chem. Toxicol. 2019, 129, 391–398. [Google Scholar] [CrossRef]
- Al Hrout, A.; Cervantes-Gracia, K.; Chahwan, R.; Amin, A. Modelling liver cancer microenvironment using a novel 3D culture system. Sci. Rep. 2022, 12, 8003. [Google Scholar] [CrossRef]
- Maepa, S.W.; Ndlovu, H. Advances in generating liver cells from pluripotent stem cells as a tool for modeling liver diseases. Stem Cells 2020, 38, 606–612. [Google Scholar] [CrossRef]
- Shoemaker, J.T.; Zhang, W.; Atlas, S.I.; Bryan, R.A.; Inman, S.W.; Vukasinovic, J. A 3D Cell Culture Organ-on-a-Chip Platform With a Breathable Hemoglobin Analogue Augments and Extends Primary Human Hepatocyte Functions in vitro. Front. Mol. Biosci. 2020, 7, 568777. [Google Scholar] [CrossRef]
- Underhill, G.H.; Khetani, S.R. Bioengineered Liver Models for Drug Testing and Cell Differentiation Studies. Cell Mol. Gastroenterol. Hepatol. 2018, 5, 426–439.e421. [Google Scholar] [CrossRef]
- Hu, H.; Gehart, H.; Artegiani, B.; LÖpez-Iglesias, C.; Dekkers, F.; Basak, O.; van Es, J.; Chuva de Sousa Lopes, S.M.; Begthel, H.; Korving, J.; et al. Long-Term Expansion of Functional Mouse and Human Hepatocytes as 3D Organoids. Cell 2018, 175, 1591–1606.e1519. [Google Scholar] [CrossRef]
- Mazzocchi, A.; Devarasetty, M.; Huntwork, R.; Soker, S.; Skardal, A. Optimization of collagen type I-hyaluronan hybrid bioink for 3D bioprinted liver microenvironments. Biofabrication 2018, 11, 015003. [Google Scholar] [CrossRef]
- Li, H.; Tang, Y.; Wei, W.; Yin, C.; Tang, F. Effects of saikosaponin-d on CYP3A4 in HepaRG cell and protein-ligand docking study. Basic Clin. Pharmacol. Toxicol. 2021, 128, 661–668. [Google Scholar] [CrossRef] [PubMed]
- Gerets, H.H.; Tilmant, K.; Gerin, B.; Chanteux, H.; Depelchin, B.O.; Dhalluin, S.; Atienzar, F.A. Characterization of primary human hepatocytes, HepG2 cells, and HepaRG cells at the mRNA level and CYP activity in response to inducers and their predictivity for the detection of human hepatotoxins. Cell Biol. Toxicol. 2012, 28, 69–87. [Google Scholar] [CrossRef] [PubMed]
- Sjogren, A.K.; Liljevald, M.; Glinghammar, B.; Sagemark, J.; Li, X.Q.; Jonebring, A.; Cotgreave, I.; Brolén, G.; Andersson, T.B. Critical differences in toxicity mechanisms in induced pluripotent stem cell-derived hepatocytes, hepatic cell lines and primary hepatocytes. Arch. Toxicol. 2014, 88, 1427–1437. [Google Scholar] [CrossRef] [PubMed]
- Lin, J.; Schyschka, L.; Mühl-Benninghaus, R.; Neumann, J.; Hao, L.; Nussler, N.; Dooley, S.; Liu, L.; Stöckle, U.; Nussler, A.K.; et al. Comparative analysis of phase I and II enzyme activities in 5 hepatic cell lines identifies Huh-7 and HCC-T cells with the highest potential to study drug metabolism. Arch. Toxicol. 2012, 86, 87–95. [Google Scholar] [CrossRef]
- Beckwitt, C.H.; Clark, A.M.; Wheeler, S.; Taylor, D.L.; Stolz, D.B.; Griffith, L.; Wells, A. Liver organ on a chip. Exp. Cell Res. 2018, 363, 15–25. [Google Scholar] [CrossRef]
- Nelson, L.J.; Morgan, K.; Treskes, P.; Samuel, K.; Henderson, C.J.; LeBled, C.; Homer, N.; Grant, M.H.; Hayes, P.C.; Plevris, J.N. Human Hepatic HepaRG Cells Maintain an Organotypic Phenotype with High Intrinsic CYP450 Activity/Metabolism and Significantly Outperform Standard HepG2/C3A Cells for Pharmaceutical and Therapeutic Applications. Basic Clin. Pharmacol. Toxicol. 2017, 120, 30–37. [Google Scholar] [CrossRef]
- Parent, R.; Marion, M.J.; Furio, L.; Trépo, C.; Petit, M.A. Origin and characterization of a human bipotent liver progenitor cell line. Gastroenterology 2004, 126, 1147–1156. [Google Scholar] [CrossRef] [PubMed]
- Mayati, A.; Moreau, A.; Le Vée, M.; Bruyère, A.; Jouan, E.; Denizot, C.; Parmentier, Y.; Fardel, O. Functional polarization of human hepatoma HepaRG cells in response to forskolin. Sci. Rep. 2018, 8, 16115. [Google Scholar] [CrossRef] [PubMed]
- Le Vee, M.; Jigorel, E.; Glaise, D.; Gripon, P.; Guguen-Guillouzo, C.; Fardel, O. Functional expression of sinusoidal and canalicular hepatic drug transporters in the differentiated human hepatoma HepaRG cell line. Eur. J. Pharm. Sci. 2006, 28, 109–117. [Google Scholar] [CrossRef] [PubMed]
- Dubois-Pot-Schneider, H.; Fekir, K.; Coulouarn, C.; Glaise, D.; Aninat, C.; Jarnouen, K.; Le Guével, R.; Kubo, T.; Ishida, S.; Morel, F.; et al. Inflammatory cytokines promote the retrodifferentiation of tumor-derived hepatocyte-like cells to progenitor cells. Hepatology 2014, 60, 2077–2090. [Google Scholar] [CrossRef] [PubMed]
- Fekir, K.; Dubois-Pot-Schneider, H.; Désert, R.; Daniel, Y.; Glaise, D.; Rauch, C.; Morel, F.; Fromenty, B.; Musso, O.; Cabillic, F.; et al. Retrodifferentiation of Human Tumor Hepatocytes to Stem Cells Leads to Metabolic Reprogramming and Chemoresistance. Cancer Res. 2019, 79, 1869–1883. [Google Scholar] [CrossRef] [PubMed]
- Cerec, V.; Glaise, D.; Garnier, D.; Morosan, S.; Turlin, B.; Drenou, B.; Gripon, P.; Kremsdorf, D.; Guguen-Guillouzo, C.; Corlu, A. Transdifferentiation of hepatocyte-like cells from the human hepatoma HepaRG cell line through bipotent progenitor. Hepatology 2007, 45, 957–967. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Sun, M.; Liu, W.; Li, Y.; Li, M. Stem Cell-Based Therapies for Liver Diseases: An Overview and Update. Tissue Eng. Regen. Med. 2019, 16, 107–118. [Google Scholar] [CrossRef]
- Grandy, R.; Tomaz, R.A.; Vallier, L. Modeling Disease with Human Inducible Pluripotent Stem Cells. Annu. Rev. Pathol. 2019, 14, 449–468. [Google Scholar] [CrossRef]
- Hu, C.; Li, L. In vitro culture of isolated primary hepatocytes and stem cell-derived hepatocyte-like cells for liver regeneration. Protein Cell 2015, 6, 562–574. [Google Scholar] [CrossRef]
- Zhang, L.; Pu, K.; Liu, X.; Bae, S.D.W.; Nguyen, R.; Bai, S.; Li, Y.; Qiao, L. The Application of Induced Pluripotent Stem Cells Against Liver Diseases: An Update and a Review. Front. Med. 2021, 8, 644594. [Google Scholar] [CrossRef] [PubMed]
- Roy-Chowdhury, N.; Wang, X.; Guha, C.; Roy-Chowdhury, J. Hepatocyte-like cells derived from induced pluripotent stem cells. Hepatol. Int. 2017, 11, 54–69. [Google Scholar] [CrossRef] [PubMed]
- Nakamori, D.; Takayama, K.; Nagamoto, Y.; Mitani, S.; Sakurai, F.; Tachibana, M.; Mizuguchi, H. Hepatic maturation of human iPS cell-derived hepatocyte-like cells by ATF5, c/EBPα, and PROX1 transduction. Biochem. Biophys. Res. Commun. 2016, 469, 424–429. [Google Scholar] [CrossRef] [PubMed]
- Poorna, M.R.; Sudhindran, S.; Thampi, M.V.; Mony, U. Differentiation of induced pluripotent stem cells to hepatocyte-like cells on cellulose nanofibril substrate. Colloids Surf. B Biointerfaces 2021, 198, 111466. [Google Scholar] [CrossRef]
- Soret, P.A.; Magusto, J.; Housset, C.; Gautheron, J. In Vitro and In Vivo Models of Non-Alcoholic Fatty Liver Disease: A Critical Appraisal. J. Clin. Med. 2020, 10, 36. [Google Scholar] [CrossRef]
- Segovia-Zafra, A.; Di Zeo-Sánchez, D.E.; López-Gómez, C.; Pérez-Valdés, Z.; García-Fuentes, E.; Andrade, R.J.; Lucena, M.I.; Villanueva-Paz, M. Preclinical models of idiosyncratic drug-induced liver injury (iDILI): Moving towards prediction. Acta Pharm. Sin. B 2021, 11, 3685–3726. [Google Scholar] [CrossRef]
- Jin, M.; Yi, X.; Liao, W.; Chen, Q.; Yang, W.; Li, Y.; Li, S.; Gao, Y.; Peng, Q.; Zhou, S. Advancements in stem cell-derived hepatocyte-like cell models for hepatotoxicity testing. Stem Cell Res. Ther. 2021, 12, 84. [Google Scholar] [CrossRef]
- Abu-Absi, S.F.; Hansen, L.K.; Hu, W.S. Three-dimensional co-culture of hepatocytes and stellate cells. Cytotechnology 2004, 45, 125–140. [Google Scholar] [CrossRef]
- Pingitore, P.; Sasidharan, K.; Ekstrand, M.; Prill, S.; Lindén, D.; Romeo, S. Human Multilineage 3D Spheroids as a Model of Liver Steatosis and Fibrosis. Int. J. Mol. Sci. 2019, 20, 1629. [Google Scholar] [CrossRef]
- Dutta, D.; Heo, I.; Clevers, H. Disease Modeling in Stem Cell-Derived 3D Organoid Systems. Trends Mol. Med. 2017, 23, 393–410. [Google Scholar] [CrossRef]
- Li, Y.; Yang, X.; Plummer, R.; Hayashi, Y.; Deng, X.S.; Nie, Y.Z.; Taniguchi, H. Human Pluripotent Stem Cell-Derived Hepatocyte-Like Cells and Organoids for Liver Disease and Therapy. Int. J. Mol. Sci. 2021, 22, 471. [Google Scholar] [CrossRef] [PubMed]
- Azar, J.; Bahmad, H.F.; Daher, D.; Moubarak, M.M.; Hadadeh, O.; Monzer, A.; Al Bitar, S.; Jamal, M.; Al-Sayegh, M.; Abou-Kheir, W. The Use of Stem Cell-Derived Organoids in Disease Modeling: An Update. Int. J. Mol. Sci. 2021, 22, 7667. [Google Scholar] [CrossRef] [PubMed]
- Ho, B.X.; Pek, N.M.Q.; Soh, B.S. Disease Modeling Using 3D Organoids Derived from Human Induced Pluripotent Stem Cells. Int. J. Mol. Sci. 2018, 19, 936. [Google Scholar] [CrossRef]
- Zhang, X.; Jiang, T.; Chen, D.; Wang, Q.; Zhang, L.W. Three-dimensional liver models: State of the art and their application for hepatotoxicity evaluation. Crit. Rev. Toxicol. 2020, 50, 279–309. [Google Scholar] [CrossRef] [PubMed]
- Thanapirom, K.; Caon, E.; Papatheodoridi, M.; Frenguelli, L.; Al-Akkad, W.; Zhenzhen, Z.; Vilia, M.G.; Pinzani, M.; Mazza, G.; Rombouts, K. Optimization and Validation of a Novel Three-Dimensional Co-Culture System in Decellularized Human Liver Scaffold for the Study of Liver Fibrosis and Cancer. Cancers 2021, 13, 4936. [Google Scholar] [CrossRef]
- Wei, G.; Wang, J.; Lv, Q.; Liu, M.; Xu, H.; Zhang, H.; Jin, L.; Yu, J.; Wang, X. Three-dimensional coculture of primary hepatocytes and stellate cells in silk scaffold improves hepatic morphology and functionality in vitro. J. Biomed. Mater. Res. A 2018, 106, 2171–2180. [Google Scholar] [CrossRef]
- Bardsley, K.; Deegan, A.J.; El Haj, A.; Yang, Y. Current State-of-the-Art 3D Tissue Models and Their Compatibility with Live Cell Imaging. Adv. Exp. Med. Biol. 2017, 1035, 3–18. [Google Scholar] [CrossRef]
- Salloum, S.; Holmes, J.A.; Jindal, R.; Bale, S.S.; Brisac, C.; Alatrakchi, N.; Lidofsky, A.; Kruger, A.J.; Fusco, D.N.; Luther, J.; et al. Exposure to human immunodeficiency virus/hepatitis C virus in hepatic and stellate cell lines reveals cooperative profibrotic transcriptional activation between viruses and cell types. Hepatology 2016, 64, 1951–1968. [Google Scholar] [CrossRef]
- Liu, J.; Li, P.; Wang, L.; Li, M.; Ge, Z.; Noordam, L.; Lieshout, R.; Verstegen, M.M.A.; Ma, B.; Su, J.; et al. Cancer-Associated Fibroblasts Provide a Stromal Niche for Liver Cancer Organoids That Confers Trophic Effects and Therapy Resistance. Cell Mol. Gastroenterol. Hepatol. 2021, 11, 407–431. [Google Scholar] [CrossRef]
- Cuvellier, M.; Ezan, F.; Oliveira, H.; Rose, S.; Fricain, J.C.; Langouët, S.; Legagneux, V.; Baffet, G. 3D culture of HepaRG cells in GelMa and its application to bioprinting of a multicellular hepatic model. Biomaterials 2021, 269, 120611. [Google Scholar] [CrossRef]
- Taymour, R.; Kilian, D.; Ahlfeld, T.; Gelinsky, M.; Lode, A. 3D bioprinting of hepatocytes: Core-shell structured co-cultures with fibroblasts for enhanced functionality. Sci. Rep. 2021, 11, 5130. [Google Scholar] [CrossRef] [PubMed]
- Van Grunsven, L.A. 3D in vitro models of liver fibrosis. Adv. Drug Deliv. Rev. 2017, 121, 133–146. [Google Scholar] [CrossRef] [PubMed]
- Dellaquila, A.; Le Bao, C.; Letourneur, D.; Simon-Yarza, T. In Vitro Strategies to Vascularize 3D Physiologically Relevant Models. Adv. Sci. 2021, 8, e2100798. [Google Scholar] [CrossRef]
- Bircsak, K.M.; DeBiasio, R.; Miedel, M.; Alsebahi, A.; Reddinger, R.; Saleh, A.; Shun, T.; Vernetti, L.A.; Gough, A. A 3D microfluidic liver model for high throughput compound toxicity screening in the OrganoPlate®. Toxicology 2021, 450, 152667. [Google Scholar] [CrossRef] [PubMed]
- Du, Y.; Li, N.; Yang, H.; Luo, C.; Gong, Y.; Tong, C.; Gao, Y.; Lü, S.; Long, M. Mimicking liver sinusoidal structures and functions using a 3D-configured microfluidic chip. Lab A Chip 2017, 17, 782–794. [Google Scholar] [CrossRef]
- Ahmed, H.M.M.; Salerno, S.; Morelli, S.; Giorno, L.; De Bartolo, L. 3D liver membrane system by co-culturing human hepatocytes, sinusoidal endothelial and stellate cells. Biofabrication 2017, 9, 025022. [Google Scholar] [CrossRef]
- Otsuka, H.; Sasaki, K.; Okimura, S.; Nagamura, M.; Nakasone, Y. Micropatterned co-culture of hepatocyte spheroids layered on non-parenchymal cells to understand heterotypic cellular interactions. Sci. Technol. Adv. Mater. 2013, 14, 065003. [Google Scholar] [CrossRef]
- Wu, Y.; Wenger, A.; Golzar, H.; Tang, X.S. 3D bioprinting of bicellular liver lobule-mimetic structures via microextrusion of cellulose nanocrystal-incorporated shear-thinning bioink. Sci. Rep. 2020, 10, 20648. [Google Scholar] [CrossRef]
- Ding, B.S.; Nolan, D.J.; Butler, J.M.; James, D.; Babazadeh, A.O.; Rosenwaks, Z.; Mittal, V.; Kobayashi, H.; Shido, K.; Lyden, D.; et al. Inductive angiocrine signals from sinusoidal endothelium are required for liver regeneration. Nature 2010, 468, 310–315. [Google Scholar] [CrossRef]
- Alghuwainem, A.; Alshareeda, A.T.; Alsowayan, B. Scaffold-Free 3-D Cell Sheet Technique Bridges the Gap between 2-D Cell Culture and Animal Models. Int. J. Mol. Sci. 2019, 20, 4926. [Google Scholar] [CrossRef]
- Nahmias, Y.; Casali, M.; Barbe, L.; Berthiaume, F.; Yarmush, M.L. Liver endothelial cells promote LDL-R expression and the uptake of HCV-like particles in primary rat and human hepatocytes. Hepatology 2006, 43, 257–265. [Google Scholar] [CrossRef]
- Ardalani, H.; Sengupta, S.; Harms, V.; Vickerman, V.; Thomson, J.A.; Murphy, W.L. 3-D culture and endothelial cells improve maturity of human pluripotent stem cell-derived hepatocytes. Acta Biomater. 2019, 95, 371–381. [Google Scholar] [CrossRef] [PubMed]
- German, C.L.; Madihally, S.V. Type of endothelial cells affects HepaRG cell acetaminophen metabolism in both 2D and 3D porous scaffold cultures. J. Appl. Toxicol. 2019, 39, 461–472. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Li, Y.; Liu, Q.; Huang, Y.; Li, R.; Wu, T.; Zhang, Z.; Zhou, J.; Huang, H.; Tang, Q.; et al. Sirt6 Alleviated Liver Fibrosis by Deacetylating Conserved Lysine 54 on Smad2 in Hepatic Stellate Cells. Hepatology 2021, 73, 1140–1157. [Google Scholar] [CrossRef]
- Prestigiacomo, V.; Weston, A.; Suter-Dick, L. Rat multicellular 3D liver microtissues to explore TGF-β1 induced effects. J. Pharmacol. Toxicol. Methods 2020, 101, 106650. [Google Scholar] [CrossRef]
- Mannaerts, I.; Eysackers, N.; Anne van Os, E.; Verhulst, S.; Roosens, T.; Smout, A.; Hierlemann, A.; Frey, O.; Leite, S.B.; van Grunsven, L.A. The fibrotic response of primary liver spheroids recapitulates in vivo hepatic stellate cell activation. Biomaterials 2020, 261, 120335. [Google Scholar] [CrossRef]
- Leite, S.B.; Roosens, T.; El Taghdouini, A.; Mannaerts, I.; Smout, A.J.; Najimi, M.; Sokal, E.; Noor, F.; Chesne, C.; van Grunsven, L.A. Novel human hepatic organoid model enables testing of drug-induced liver fibrosis in vitro. Biomaterials 2016, 78, 1–10. [Google Scholar] [CrossRef]
- Gaul, S.; Leszczynska, A.; Alegre, F.; Kaufmann, B.; Johnson, C.D.; Adams, L.A.; Wree, A.; Damm, G.; Seehofer, D.; Calvente, C.J.; et al. Hepatocyte pyroptosis and release of inflammasome particles induce stellate cell activation and liver fibrosis. J. Hepatol. 2021, 74, 156–167. [Google Scholar] [CrossRef]
- Fabregat, I.; Moreno-Càceres, J.; Sánchez, A.; Dooley, S.; Dewidar, B.; Giannelli, G.; Ten Dijke, P. TGF-β signalling and liver disease. FEBS J. 2016, 283, 2219–2232. [Google Scholar] [CrossRef] [PubMed]
- Kisseleva, T.; Brenner, D. Molecular and cellular mechanisms of liver fibrosis and its regression. Nat. Rev. Gastroenterol. Hepatol. 2021, 18, 151–166. [Google Scholar] [CrossRef]
- Yu, H.; Jiang, X.; Dong, F.; Zhang, F.; Ji, X.; Xue, M.; Yang, F.; Chen, J.; Hu, X.; Bao, Z. Lipid accumulation-induced hepatocyte senescence regulates the activation of hepatic stellate cells through the Nrf2-antioxidant response element pathway. Exp. Cell Res. 2021, 405, 112689. [Google Scholar] [CrossRef]
- Marrone, G.; Shah, V.H.; Gracia-Sancho, J. Sinusoidal communication in liver fibrosis and regeneration. J. Hepatol. 2016, 65, 608–617. [Google Scholar] [CrossRef] [PubMed]
- Ye, Q.; Zhou, Y.; Zhao, C.; Xu, L.; Ping, J. Salidroside Inhibits CCl(4)-Induced Liver Fibrosis in Mice by Reducing Activation and Migration of HSC Induced by Liver Sinusoidal Endothelial Cell-Derived Exosomal SphK1. Front. Pharmacol. 2021, 12, 677810. [Google Scholar] [CrossRef] [PubMed]
- Wu, X.; Shu, L.; Zhang, Z.; Li, J.; Zong, J.; Cheong, L.Y.; Ye, D.; Lam, K.S.L.; Song, E.; Wang, C.; et al. Adipocyte Fatty Acid Binding Protein Promotes the Onset and Progression of Liver Fibrosis via Mediating the Crosstalk between Liver Sinusoidal Endothelial Cells and Hepatic Stellate Cells. Adv. Sci. 2021, 8, e2003721. [Google Scholar] [CrossRef] [PubMed]
- Wu, H.; Chen, G.; Wang, J.; Deng, M.; Yuan, F.; Gong, J. TIM-4 interference in Kupffer cells against CCL4-induced liver fibrosis by mediating Akt1/Mitophagy signalling pathway. Cell Prolif. 2020, 53, e12731. [Google Scholar] [CrossRef]
- Ge, X.; Arriazu, E.; Magdaleno, F.; Antoine, D.J.; Dela Cruz, R.; Theise, N.; Nieto, N. High Mobility Group Box-1 Drives Fibrosis Progression Signaling via the Receptor for Advanced Glycation End Products in Mice. Hepatology 2018, 68, 2380–2404. [Google Scholar] [CrossRef] [PubMed]
- Luangmonkong, T.; Suriguga, S.; Mutsaers, H.A.M.; Groothuis, G.M.M.; Olinga, P.; Boersema, M. Targeting Oxidative Stress for the Treatment of Liver Fibrosis. Rev. Physiol. Biochem. Pharmacol. 2018, 175, 71–102. [Google Scholar] [CrossRef]
- Chen, Q.; Xue, Y.; Sun, J. Kupffer cell-mediated hepatic injury induced by silica nanoparticles in vitro and in vivo. Int. J. Nanomed. 2013, 8, 1129–1140. [Google Scholar] [CrossRef]
- Tasnim, F.; Xing, J.; Huang, X.; Mo, S.; Wei, X.; Tan, M.H.; Yu, H. Generation of mature kupffer cells from human induced pluripotent stem cells. Biomaterials 2019, 192, 377–391. [Google Scholar] [CrossRef]
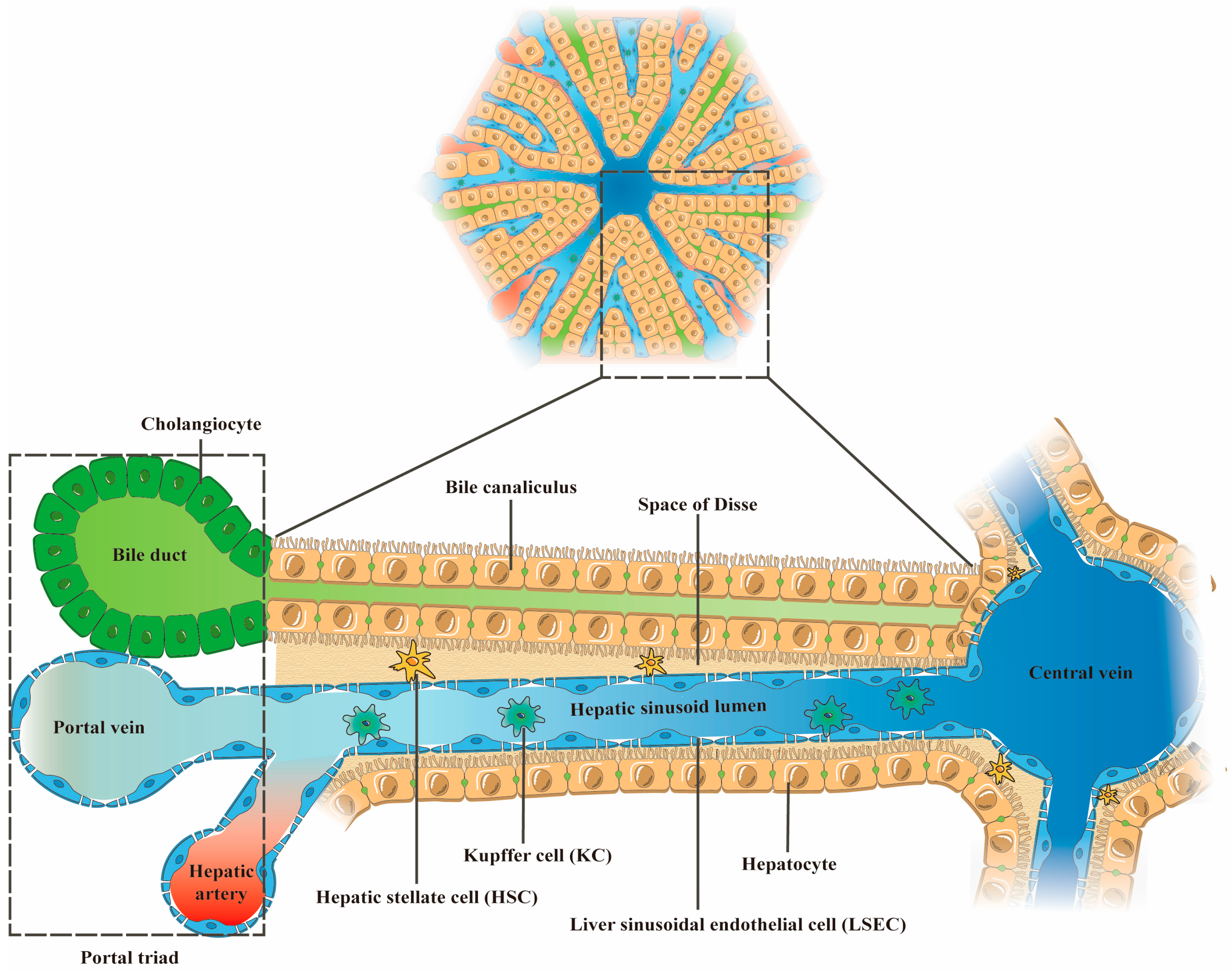
| Cell Type | Number (% Total Liver Cells) | Volume (% Total) | Structural Features (Physiological State/Pathological State) | Functions (Physiological State) | Reference |
|---|---|---|---|---|---|
| Hepatocytes | 60% | 80% | Physiological state: Rectangle; Sinusoidal surface with microvilli structure | Participate in the metabolism, synthesis, and secretion of exogenous and endogenous substances; Lipid storage; Transformation of toxic substances. | [20,22,23] |
| Liver sinusoidal endothelial cells (LSECs) | 15–20% | 3% | Physiological state: long spindle; Lacking basement membrane; Possessing sieve-like fenestrae structure through the cell Pathological state: Basement membrane formation; Fenestration structure reduced or disappeared | Regulate vascular tone; Secrete NO; Present antigen; Filter toxins and antigenic substances in portal blood. | [24,25,26,27,28,29] |
| Hepatic stellate cells (HSCs) | 3–8% | 1.5% | Physiological state: Polygon; With raised cytoplasmic pseudopods Pathological state: Myofibroblast-like phenotype | Store and release vitamin A in the body; Maintain the regeneration ability of liver tissue; Regulate the immune function of the liver; Maintain normal sinusoidal tone and liver stiffness by secreting pro-inflammatory and anti-inflammatory cytokines, as well as extracellular matrix. | [21,25,30,31] |
| Kupffer cells (KCs) | 8–12% | 2% | Physiological state: Elongate or nearly rounded; Morphological variability Pathological state: the M1 phenotype | Modulate liver immune response; Maintain hepatic iron, cholesterol and bilirubin metabolism; Remove pathogens, toxins, senescent red blood cells and platelets from the blood. | [4,32] |
| 3D Cell Co-Culture Method | 3D Cell Co-Culture System | Introduction | Advantages | Disadvantages | Reference |
|---|---|---|---|---|---|
| Direct 3D co-culture (Physical contact and paracrine signaling interactions exist between heterotypic cells) | Self-aggregating multicellular spheroids | Liver cells self-aggregate into cell spheroids in ultra-low adhesion plates or using the suspension drop technique | Easy to operate; Low cost; High throughput | Hypoxia and necrosis of cells in the center of spheroid; Difficultly in controlling the size of the spheroid; Cells lack the support of exogenous matrix; The spheroid is loose; Uncontrollable spatial arrangement of cells; Cell separation is a tedious process | [65,66,67,68,69] |
| 3D liver organoids | Stem cells differentiated by multiple lineages or hepatocyte-like cells derived from them are co-cultured with non-parenchymal cells to form self-organizing 3D structures resembling natural liver structures | Complex structure and communication similar to the natural liver; Multiple liver cells can be derived simultaneously; Long-term expansion; Genetic background can be preserved; Genes can be manipulated; High throughput | Cell maturity heterogeneity; Specific reagents are required to induce cell differentiation; Low liver phenotype and functional maturity; Uncontrollable spatial arrangement of cells; Poor repeatability; High cost; Time-consuming | [70,71,72,73] | |
| 3D scaffold co-culture of liver cells | Cells grow attached to porous 3D network scaffolds composed of natural materials (collagen, Matrigel, etc.) or synthetic materials (self-assembling peptides, polystyrene, etc.) | Controllable size and shape; Presence of in vivo-like biochemical and biomechanical microenvironment; Adjustable mechanical and degradation properties of synthetic scaffolds; Existence of cell–ECM interactions | Difficulty in cell-scaffold separation; Difficulty in live cell imaging; Hypoxia and necrosis of cells in the center of the scaffold; Uncontrollable spatial arrangement of cells; Unknown composition and batch-to-batch variation of natural scaffolds; Biocompatibility and cytotoxicity issues with synthetic scaffolds | [67,74,75,76,77] | |
| Indirect 3D co-culture (Paracrine signaling interactions exist between cells, but no physical contact) | 3D liver cell co-culture system based on Transwell chambers | 3D layered co-culture of cells using Transwell chambers as a physical separation system | Mimics the layered structure of natural liver sinusoids; Easy to study intercellular paracrine interactions independently; Avoids unnecessary intercellular contact; Controllable spatial arrangement of cells; Easy separation of co-cultured cells for individual analysis; High throughput | High cost; Lack of physical contact between heterotypic cells | [23,78,79] |
| Direct 3D co-culture/Indirect 3D co-culture | 3D bioprinting co-culture of liver cells | Using printing technologies such as Laser based-, Inkjet based- and bio-extrusion, biological materials (bio-ink) that act as extracellular matrix and living cells can be precisely located layer by layer, to form 3D tissue | High throughput; Excellent stability; Enables precise control of model structure and spatial arrangement of cells | High cost; Low resolution; Cell sedimentation during printing; Limited cell density; Scarcity of available bio-ink materials; The printing process can cause cell damage (phototoxicity and crush damage); The viscosity of bio-ink affects the printing performance; Deformation of scaffolds over time | [80,81,82,83] |
| Microfluidic multicellular liver chips | Cells are grown in a microarray with the continuous medium flow and microstructural features of liver lobules | Similar physiological environment to liver lobules; Continuous culture-medium perfusion; Physiologically related oxygen and nutrient gradients; Continuous oxygen and metabolic waste delivery; Reproducing physiological shear stress; Controllable spatial arrangement of cells | Lack of physical contact between heterotypic cells; High cost; Operating complexity; High technical requirements; Low cell recovery rate | [12,66,84,85] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ma, Y.; Hu, L.; Tang, J.; Guo, W.; Feng, Y.; Liu, Y.; Tang, F. Three-Dimensional Cell Co-Culture Liver Models and Their Applications in Pharmaceutical Research. Int. J. Mol. Sci. 2023, 24, 6248. https://doi.org/10.3390/ijms24076248
Ma Y, Hu L, Tang J, Guo W, Feng Y, Liu Y, Tang F. Three-Dimensional Cell Co-Culture Liver Models and Their Applications in Pharmaceutical Research. International Journal of Molecular Sciences. 2023; 24(7):6248. https://doi.org/10.3390/ijms24076248
Chicago/Turabian StyleMa, Yinping, Lei Hu, Jianhua Tang, Weiwei Guo, Yujie Feng, Yanmiao Liu, and Fushan Tang. 2023. "Three-Dimensional Cell Co-Culture Liver Models and Their Applications in Pharmaceutical Research" International Journal of Molecular Sciences 24, no. 7: 6248. https://doi.org/10.3390/ijms24076248
APA StyleMa, Y., Hu, L., Tang, J., Guo, W., Feng, Y., Liu, Y., & Tang, F. (2023). Three-Dimensional Cell Co-Culture Liver Models and Their Applications in Pharmaceutical Research. International Journal of Molecular Sciences, 24(7), 6248. https://doi.org/10.3390/ijms24076248





