Genomic Identification of CCCH-Type Zinc Finger Protein Genes Reveals the Role of HuTZF3 in Tolerance of Heat and Salt Stress of Pitaya (Hylocereus polyrhizus)
Abstract
:1. Introduction
2. Results
2.1. Identification of the CCCH-Type Zinc Finger Protein Genes from Pitaya
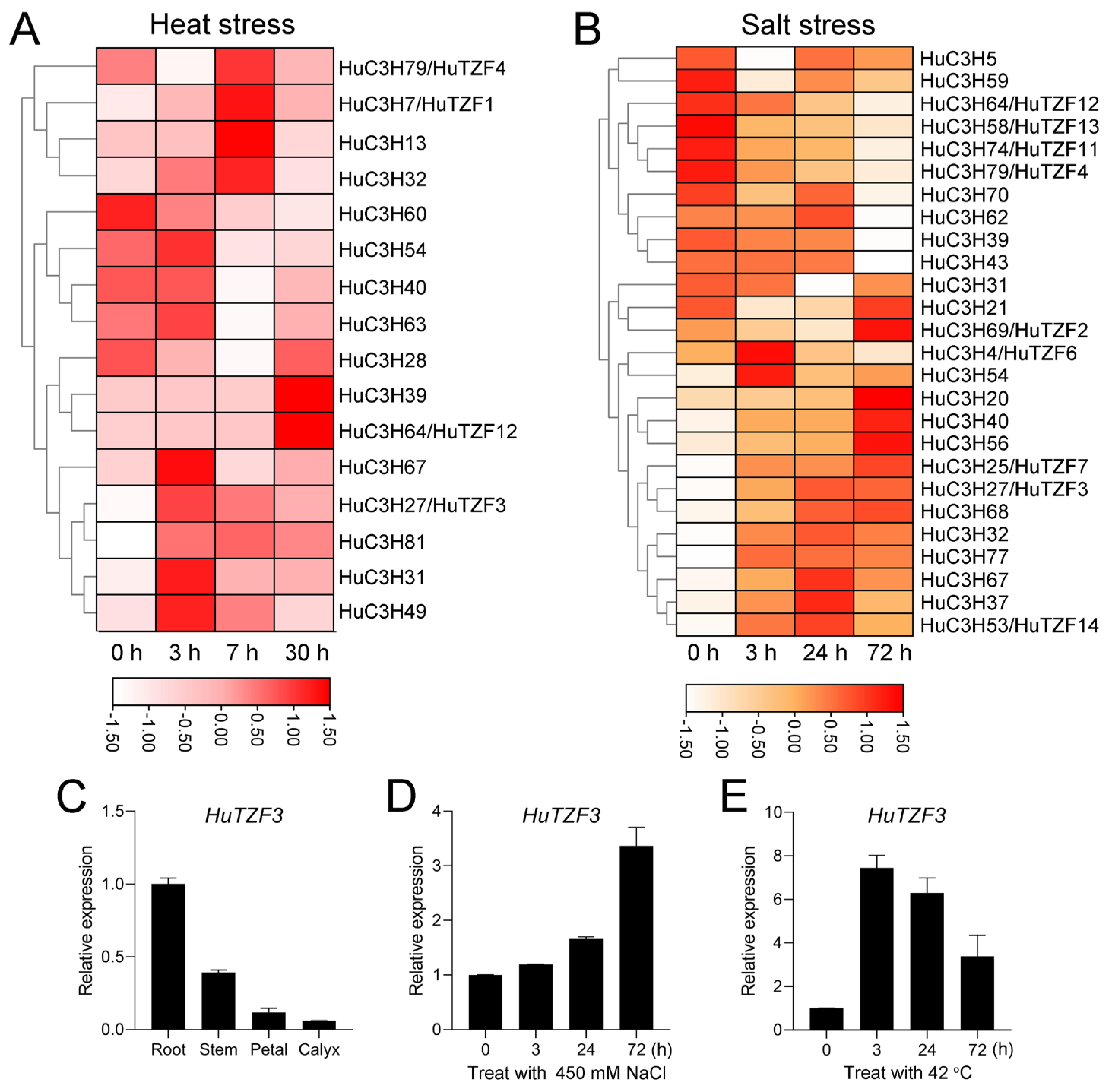
2.2. Identification of HuCCCHs Response to Heat and Salt Stress
2.3. Heterologous Expression of HuTZF3 Improved Salt and Heat Tolerance in Arabidopsis
2.4. Heterologous Expression of HuTZF3 Repressed Burst of Oxidative Stress in Arabidopsis
2.5. HuTZF3 Is Co-Localized with PBs and SGs Markers in Arabidopsis
3. Discussion
4. Materials and Methods
4.1. Plant and Growth Conditions
4.2. Abiotic Stress Treatment
4.3. Sequence Analysis of HuTZF Genes
4.4. RNA Isolation and RT-qPCR Analysis
4.5. Vector Construction and Genetic Transformation
4.6. Histochemical and Physiological Analysis of Oxidative Stress
4.7. Subcellular Localization of HuTZF3
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Tubiello, F.N.; Soussana, J.F.; Howden, S.M. Crop and pasture response to climate change. Proc. Natl. Acad. Sci. USA 2007, 104, 19686–19690. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yamaguchi, T.; Blumwald, E. Developing salt-tolerant crop plants: Challenges and opportunities. Trends Plant Sci. 2005, 10, 615–620. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Zhou, J.F.; Kan, Y.; Shan, J.X.; Ye, W.W.; Dong, N.Q.; Guo, T.; Xiang, Y.H.; Yang, Y.B.; Li, Y.C.; et al. A genetic module at one locus in rice protects chloroplasts to enhance thermotolerance. Science 2022, 376, 1293–1300. [Google Scholar] [CrossRef]
- Zhuang, Y.L.; Zhang, Y.F.; Sun, L.P. Characteristics of fibre-rich powder and antioxidant activity of pitaya (Hylocereus undatus) peels. Int. J. Food Sci. Technol. 2012, 47, 1279–1285. [Google Scholar] [CrossRef]
- Nong, Q.D.; Zhang, M.Y.; Chen, J.T.; Zhang, M.; Cheng, H.P.; Jian, S.G.; Lu, H.F.; Xia, K.F. RNA-Seq de novo assembly of red pitaya (Hylocereus polyrhizus) roots and differential transcriptome analysis in response to salt stress. Trop. Plant Biol. 2019, 12, 55–66. [Google Scholar] [CrossRef]
- Jiao, Z.L.; Xu, W.J.; Nong, Q.D.; Zhang, M.; Jian, S.G.; Lu, H.F.; Chen, J.T.; Zhang, M.Y.; Xia, K.F. An integrative transcriptomic and metabolomic analysis of red pitaya (Hylocereus polyrhizus) seedlings in response to heat stress. Genes 2021, 12, 1714. [Google Scholar] [CrossRef] [PubMed]
- Raveh, E.; Gersani, M.; Nobel, P.S. CO2 uptake and fluorescence responses for a shade-tolerant cactus Hylocereus undatus under current and doubled CO2 concentrations. Physiol. Plant 1995, 93, 505–511. [Google Scholar] [CrossRef]
- Garcia, T.M.; Heyduk, K.; Kuzmick, E.; Mayer, J.A. Crassulacean acid metabolism biology. New Phytol. 2014, 204, 738–740. [Google Scholar] [CrossRef] [PubMed]
- Males, J.; Griffiths, H. Stomatal biology of CAM plants. Plant Physiol. 2017, 174, 550–560. [Google Scholar] [CrossRef] [Green Version]
- Winter, K.; Smith, J.A.C. CAM photosynthesis: The acid test. New Phytol. 2022, 233, 599–609. [Google Scholar] [CrossRef]
- Luttge, U. Ability of crassulacean acid metabolism plants to overcome interacting stresses in tropical environments. AoB Plants 2010, 2010, plq005. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fan, Q.J.; Yan, F.X.; Qiao, G.; Zhang, B.X.; Wen, X.P. Identification of differentially-expressed genes potentially implicated in drought response in pitaya (Hylocereus undatus) by suppression subtractive hybridization and cDNA microarray analysis. Gene 2014, 533, 322–331. [Google Scholar] [CrossRef] [PubMed]
- Zhou, J.L.; Wang, Z.; Mao, Y.Y.; Wang, L.J.; Xiao, T.J.; Hu, Y.; Zhang, Y.; Ma, Y.H. Proteogenomic analysis of pitaya reveals cold stress-related molecular signature. PeerJ 2020, 8, e8540. [Google Scholar] [CrossRef] [Green Version]
- Nie, Q.; Gao, G.L.; Fan, Q.J.; Qiao, G.; Wen, X.P.; Liu, T.; Peng, Z.J.; Cai, Y.Q. Isolation and characterization of a catalase gene “HuCAT3” from pitaya (Hylocereus undatus) and its expression under abiotic stress. Gene 2015, 563, 63–71. [Google Scholar] [CrossRef]
- Qu, Y.J.; Nong, Q.D.; Jian, S.G.; Lu, H.F.; Zhang, M.Y.; Xia, K.F. An AP2/ERF gene, HuERF1, from pitaya (Hylocereus undatus) positively regulates salt tolerance. Int. J. Mol. Sci. 2020, 21, 4586. [Google Scholar] [CrossRef] [PubMed]
- Li, A.L.; Wen, Z.; Yang, K.; Wen, X.P. Conserved miR396b-GRF regulation is involved in abiotic stress responses in pitaya (Hylocereus polyrhizus). Int. J. Mol. Sci. 2019, 20, 2501. [Google Scholar] [CrossRef] [Green Version]
- Chen, J.Y.; Xie, F.F.; Cui, Y.Z.; Chen, C.B.; Lu, W.J.; Hu, X.D.; Hua, Q.Z.; Zhao, J.; Wu, Z.J.; Gao, D.; et al. A chromosome-scale genome sequence of pitaya (Hylocereus undatus) provides novel insights into the genome evolution and regulation of betalain biosynthesis. Hort. Res. 2021, 8, 164. [Google Scholar] [CrossRef]
- Takatsuji, H. Zinc-finger transcription factors in plants. Cell. Mol. Life Sci. 1998, 54, 582–596. [Google Scholar] [CrossRef]
- Moore, M.; Ullman, C. Recent developments in the engineering of zinc finger proteins. Brief. Funct. Genomic Proteomic 2003, 1, 342–355. [Google Scholar] [CrossRef] [Green Version]
- Berg, J.M.; Shi, Y.G. The galvanization of biology: A growing appreciation for the roles of zinc. Science 1996, 271, 1081–1085. [Google Scholar] [CrossRef]
- Wang, D.; Guo, Y.H.; Wu, C.G.; Yang, G.D.; Li, Y.Y.; Zheng, C.C. Genome-wide analysis of CCCH zinc finger family in Arabidopsis and rice. BMC Genom. 2008, 9, 44. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bogamuwa, S.P.; Jang, J.C. Tandem CCCH zinc finger proteins in plant growth, development and stress response. Plant Cell Physiol. 2014, 55, 1367–1375. [Google Scholar] [CrossRef] [PubMed]
- Becerra, C.; Jahrmann, T.; Puigdomènech, P.; Vicient, C.M. Ankyrin repeat-containing proteins in Arabidopsis: Characterization of a novel and abundant group of genes coding ankyrin-transmembrane proteins. Gene 2004, 340, 111–121. [Google Scholar] [CrossRef]
- Qu, J.; Kang, S.G.; Wang, W.; Musier-Forsyth, K.; Jang, J.C. Arabidopsis thaliana tandem zinc finger 1 (AtTZF1) protein in RNA binding and decay. Plant J. 2014, 78, 452–467. [Google Scholar] [CrossRef] [Green Version]
- Ivanov, P.; Kedersha, N.; Anderson, P. Stress granules and processing bodies in translational control. Cold Spring Harb. Perspect. Biol. 2019, 11, a032813. [Google Scholar] [CrossRef]
- Decker, C.J.; Parker, R. P-bodies and stress granules: Possible roles in the control of translation and mRNA degradation. Cold Spring Harb. Perspect. Biol. 2012, 4, a012286. [Google Scholar] [CrossRef] [Green Version]
- Lin, P.C.; Pomeranz, M.C.; Jikumaru, Y.; Kang, S.G.; Hah, C.; Fujioka, S.; Kamiya, Y.; Jang, J.C. The Arabidopsis tandem zinc finger protein AtTZF1 affects ABA- and GA-mediated growth, stress and gene expression responses. Plant J. 2011, 65, 253–268. [Google Scholar] [CrossRef]
- Pomeranz, M.C.; Hah, C.; Lin, P.C.; Kang, S.G.; Finer, J.J.; Blackshear, P.J.; Jang, J.C. The Arabidopsis tandem zinc finger protein AtTZF1 traffics between the nucleus and cytoplasmic foci and binds both DNA and RNA. Plant Physiol. 2010, 152, 151–165. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lee, S.J.; Jung, H.J.; Kang, H.S.; Kim, S.Y. Arabidopsis zinc finger proteins AtC3H49/AtTZF3 and AtC3H20/AtTZF2 are involved in ABA and JA responses. Plant Cell Physiol. 2012, 53, 673–686. [Google Scholar] [CrossRef]
- Jan, A.; Maruyama, K.; Todaka, D.; Kidokoro, S.; Abo, M.; Yoshimura, E.; Shinozaki, K.; Nakashima, K.; Yamaguchi-Shinozaki, K. OsTZF1, a CCCH-tandem zinc finger protein, confers delayed senescence and stress tolerance in rice by regulating stress-related genes. Plant Physiol. 2013, 161, 1202–1216. [Google Scholar] [CrossRef] [Green Version]
- Guo, C.M.; Chen, L.L.; Cui, Y.C.; Tang, M.; Guo, Y.; Yi, Y.; Li, Y.; Liu, L.Q.; Chen, L. RNA binding protein OsTZF7 traffics between the nucleus and processing podies/stress granules and positively regulates drought stress in rice. Front. Plant Sci. 2022, 13, 802337. [Google Scholar] [CrossRef]
- Chen, C.J.; Chen, H.; Zhang, Y.; Thomas, H.R.; Frank, M.H.; He, Y.H.; Xia, R. TBtools: An integrative toolkit developed for interactive analyses of big biological data. Mol. Plant 2020, 13, 1194–1202. [Google Scholar] [CrossRef] [PubMed]
- Peng, X.J.; Zhao, Y.; Cao, J.G.; Zhang, W.; Jiang, H.Y.; Li, X.Y.; Ma, Q.; Zhu, S.W.; Cheng, B.J. CCCH-type zinc finger family in maize: Genome-wide identification, classification and expression profiling under abscisic acid and drought treatments. PLoS ONE 2012, 7, e40120. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Guo, Y.H.; Yu, Y.P.; Wang, D.; Wu, C.A.; Yang, G.D.; Huang, J.G.; Zheng, C.C. GhZFP1, a novel CCCH-type zinc finger protein from cotton, enhances salt stress tolerance and fungal disease resistance in transgenic tobacco by interacting with GZIRD21A and GZIPR5. New Phytol. 2009, 183, 62–75. [Google Scholar] [CrossRef]
- Chai, G.H.; Hu, R.B.; Zhang, D.Y.; Qi, G.; Zuo, R.; Cao, Y.P.; Chen, P.; Kong, Y.Z.; Zhou, G.K. Comprehensive analysis of CCCH zinc finger family in poplar (Populus trichocarpa). BMC Genom. 2012, 13, 253. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hu, X.; Zuo, J.F. The CCCH zinc finger family of soybean (Glycine max L.): Genome-wide identification, expression, domestication, GWAS and haplotype analysis. BMC Genom. 2021, 22, 511. [Google Scholar] [CrossRef]
- Selvaraj, M.G.; Jan, A.; Ishizaki, T.; Valencia, M.; Dedicova, B.; Maruyama, K.; Ogata, T.; Todaka, D.; Yamaguchi-Shinozaki, K.; Nakashima, K.; et al. Expression of the CCCH-tandem zinc finger protein gene OsTZF5 under a stress-inducible promoter mitigates the effect of drought stress on rice grain yield under field conditions. Plant Biotechnol. J. 2020, 18, 1711–1721. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Seong, S.Y.; Shim, J.S.; Bang, S.W.; Kim, J.K. Overexpression of OsC3H10, a CCCH-zinc finger, improves drought tolerance in rice by regulating stress-related genes. Plants 2020, 9, 1298. [Google Scholar] [CrossRef]
- Erickson, S.L.; Lykke-Andersen, J. Cytoplasmic mRNP granules at a glance. J. Cell. Sci. 2011, 124, 293–297. [Google Scholar] [CrossRef] [Green Version]
- Buchan, J.R. mRNP granules. assembly, function, and connections with disease. RNA Biol. 2014, 11, 1019–1030. [Google Scholar] [CrossRef] [Green Version]
- Anderson, P.; Kedersha, N. RNA granules: Post-transcriptional and epigenetic modulators of gene expression. Nat. Rev. Mol. Cell Biol. 2009, 10, 430–436. [Google Scholar] [CrossRef] [PubMed]
- Maruri-Lopez, I.; Figueroa, N.E.; Hernandez-Sanchez, I.E.; Chodasiewicz, M. Plant stress granules: Trends and beyond. Front. Plant Sci. 2021, 12, 722643. [Google Scholar] [CrossRef]
- Guzikowski, A.R.; Chen, Y.S.; Zid, B.M. Stress-induced mRNP granules: Form and function of processing bodies and stress granules. Wiley Interdiscip. Rev. RNA 2019, 10, e1524. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.B.; Li, F.P.; Xie, F.F.; Chen, J.X.; Hua, Q.Z.; Chen, J.Y.; Wu, Z.J.; Zhang, Z.K.; Zhang, R.; Zhao, J.T.; et al. Pitaya genome and multiomics database (PGMD): A comprehensive and integrative resource of Selenicereus undatus. Genes 2022, 13, 745. [Google Scholar] [CrossRef]
- Nong, Q.D.; Yang, Y.C.; Zhang, M.Y.; Zhang, M.; Chen, J.T.; Jian, S.G.; Lu, H.F.; Xia, K.F. RNA-seq-based selection of reference genes for RT-qPCR analysis of pitaya. FEBS Open Bio 2019, 9, 1403–1412. [Google Scholar] [CrossRef] [Green Version]
- Clough, S.J.; Bent, A.F. Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 1998, 16, 735–743. [Google Scholar] [CrossRef] [Green Version]
- Kumar, D.; Yusuf, M.A.; Singh, P.; Sardar, M.; Sarin, N.B. Histochemical detection of superoxide and H2O2 accumulation in Brassica juncea seedlings. Bio-Protocol 2014, 4, e1108. [Google Scholar] [CrossRef]
- Gutierrez-Beltran, E.; Moschou, P.N.; Smertenko, A.P.; Bozhkov, P.V. Tudor staphylococcal nuclease links formation of stress granules and processing bodies with mRNA catabolism in Arabidopsis. Plant Cell 2015, 27, 926–943. [Google Scholar] [CrossRef] [Green Version]





Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Xu, W.; Jian, S.; Li, J.; Wang, Y.; Zhang, M.; Xia, K. Genomic Identification of CCCH-Type Zinc Finger Protein Genes Reveals the Role of HuTZF3 in Tolerance of Heat and Salt Stress of Pitaya (Hylocereus polyrhizus). Int. J. Mol. Sci. 2023, 24, 6359. https://doi.org/10.3390/ijms24076359
Xu W, Jian S, Li J, Wang Y, Zhang M, Xia K. Genomic Identification of CCCH-Type Zinc Finger Protein Genes Reveals the Role of HuTZF3 in Tolerance of Heat and Salt Stress of Pitaya (Hylocereus polyrhizus). International Journal of Molecular Sciences. 2023; 24(7):6359. https://doi.org/10.3390/ijms24076359
Chicago/Turabian StyleXu, Weijuan, Shuguang Jian, Jianyi Li, Yusang Wang, Mingyong Zhang, and Kuaifei Xia. 2023. "Genomic Identification of CCCH-Type Zinc Finger Protein Genes Reveals the Role of HuTZF3 in Tolerance of Heat and Salt Stress of Pitaya (Hylocereus polyrhizus)" International Journal of Molecular Sciences 24, no. 7: 6359. https://doi.org/10.3390/ijms24076359
APA StyleXu, W., Jian, S., Li, J., Wang, Y., Zhang, M., & Xia, K. (2023). Genomic Identification of CCCH-Type Zinc Finger Protein Genes Reveals the Role of HuTZF3 in Tolerance of Heat and Salt Stress of Pitaya (Hylocereus polyrhizus). International Journal of Molecular Sciences, 24(7), 6359. https://doi.org/10.3390/ijms24076359





