Generating Functional and Highly Proliferative Melanocytes Derived from Human Pluripotent Stem Cells: A Promising Tool for Biotherapeutic Approaches to Treat Skin Pigmentation Disorders
Abstract
1. Introduction
2. Results
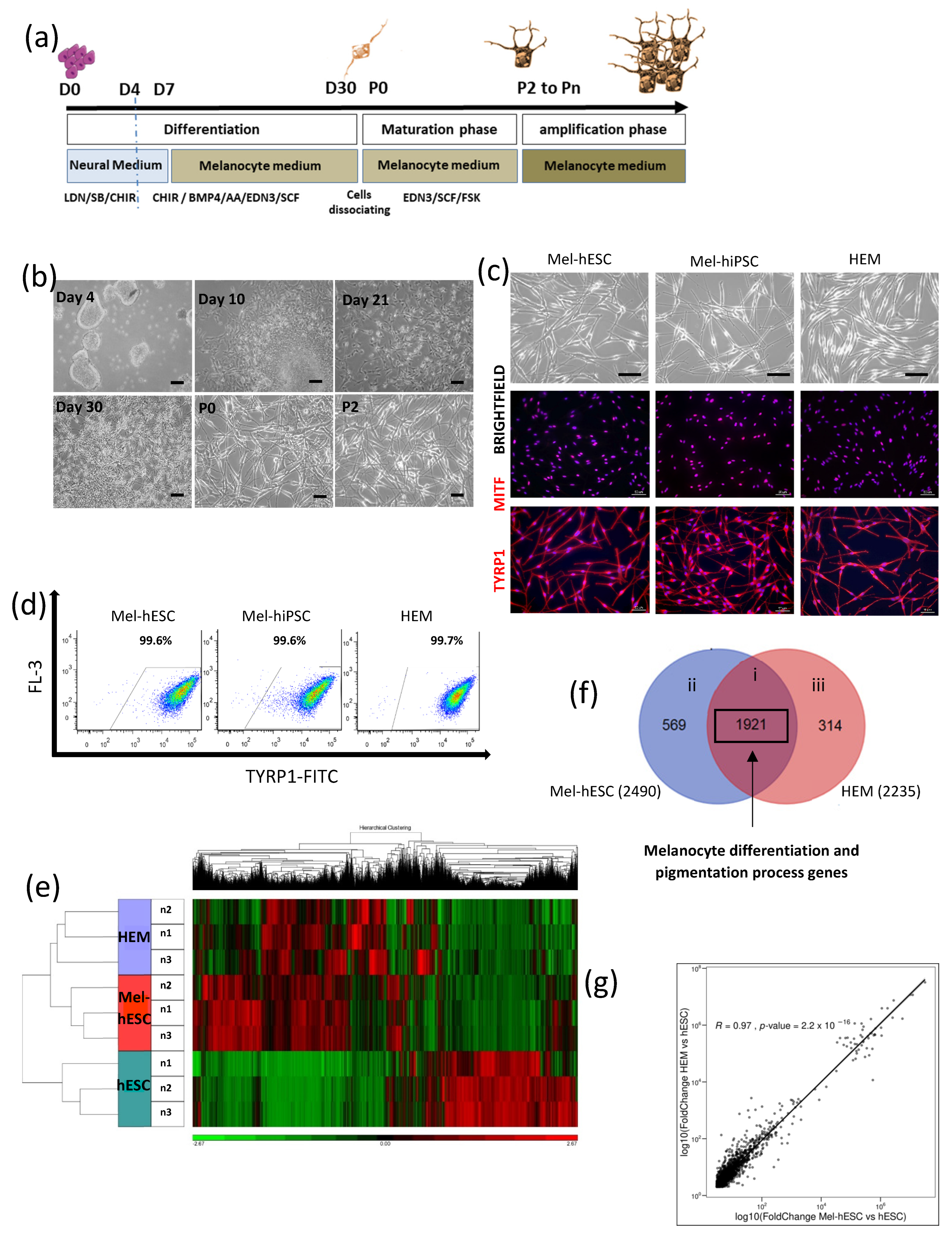
2.1. Characterization of a Pure and Homogenous Population of Melanocytes Derived from hPSC
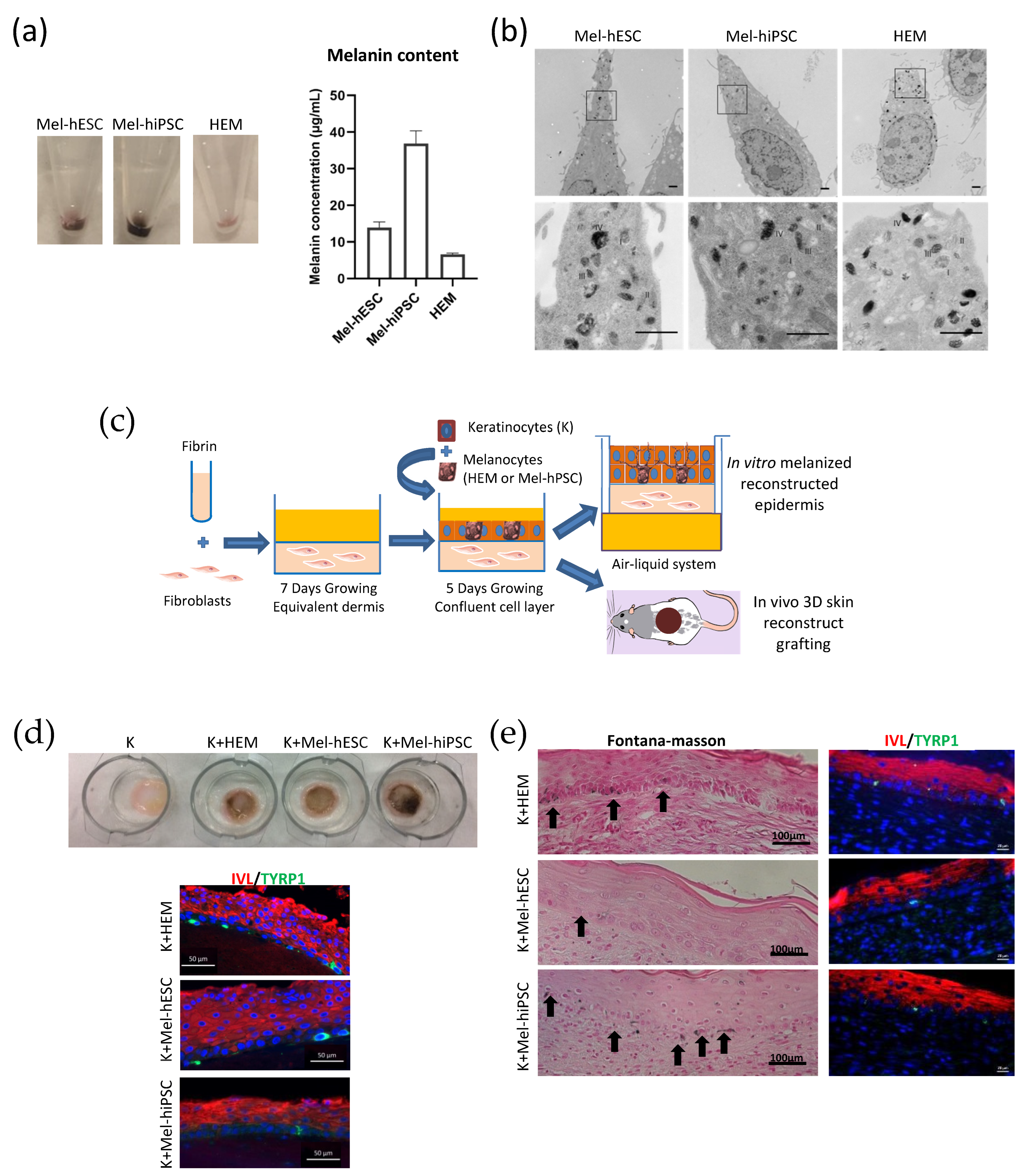
2.2. Functional Characterization of hPSC-Derived Melanocytes
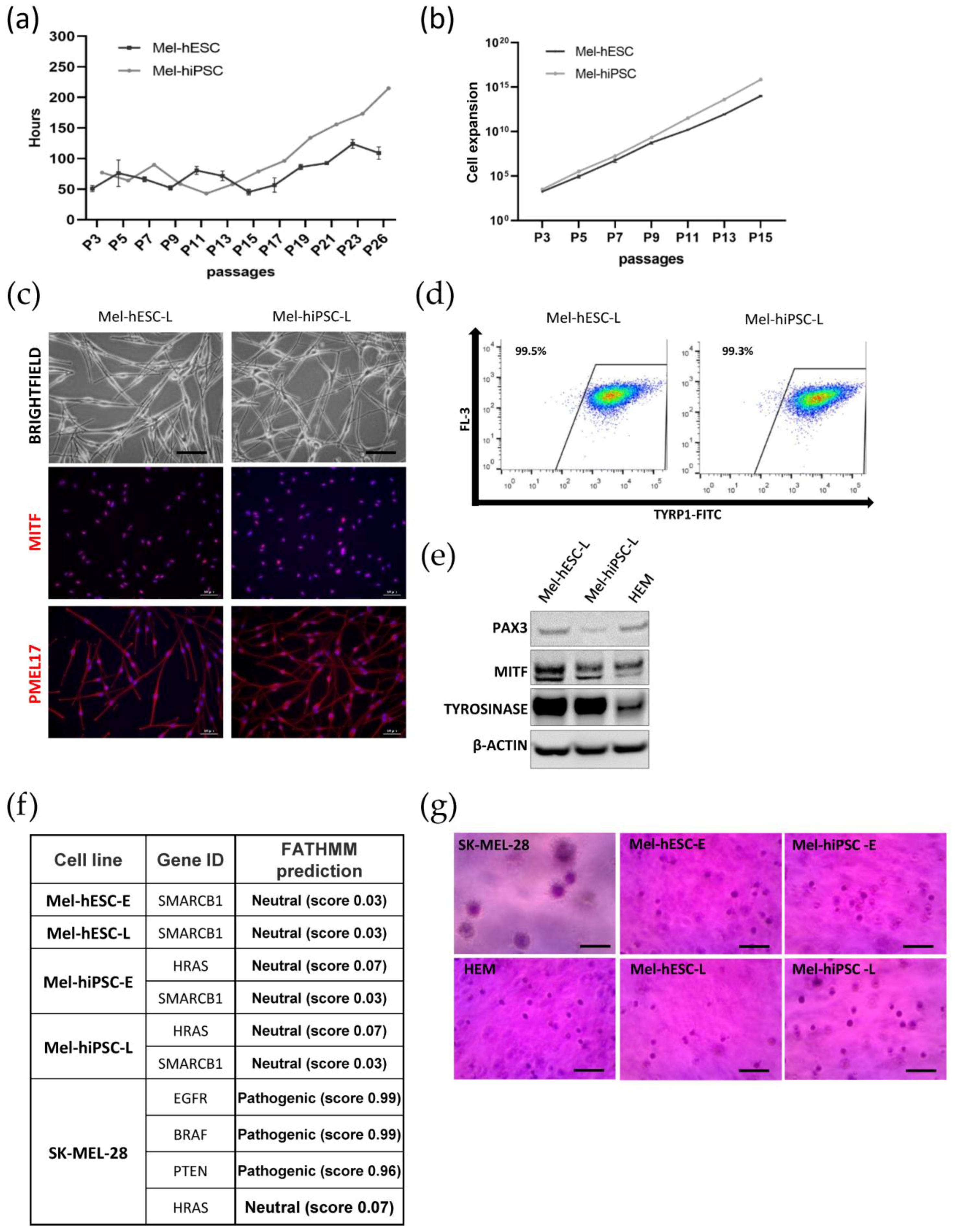
2.3. Long-Term Expansion of hPSC-Derived Melanocytes and Genomic Integrity
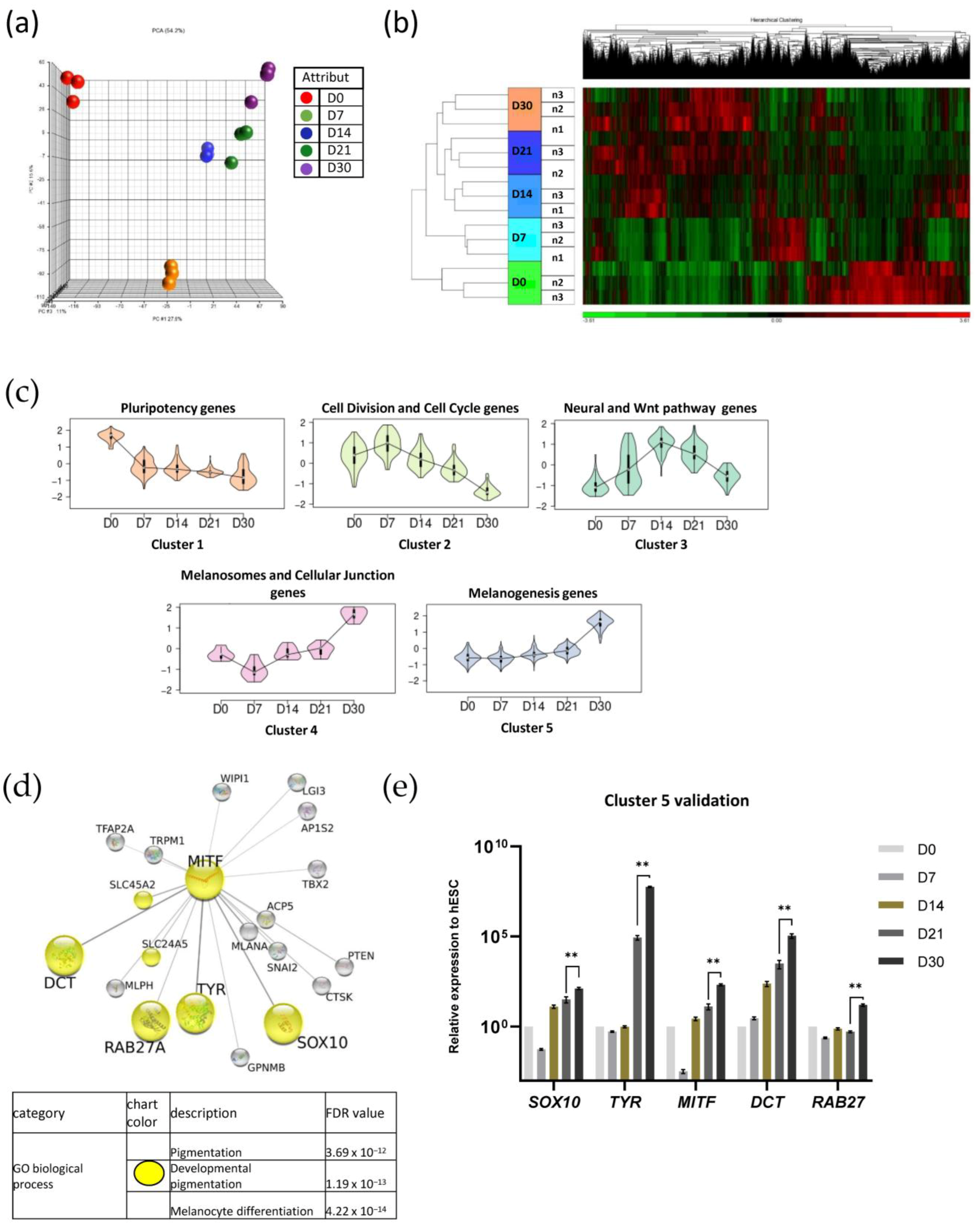
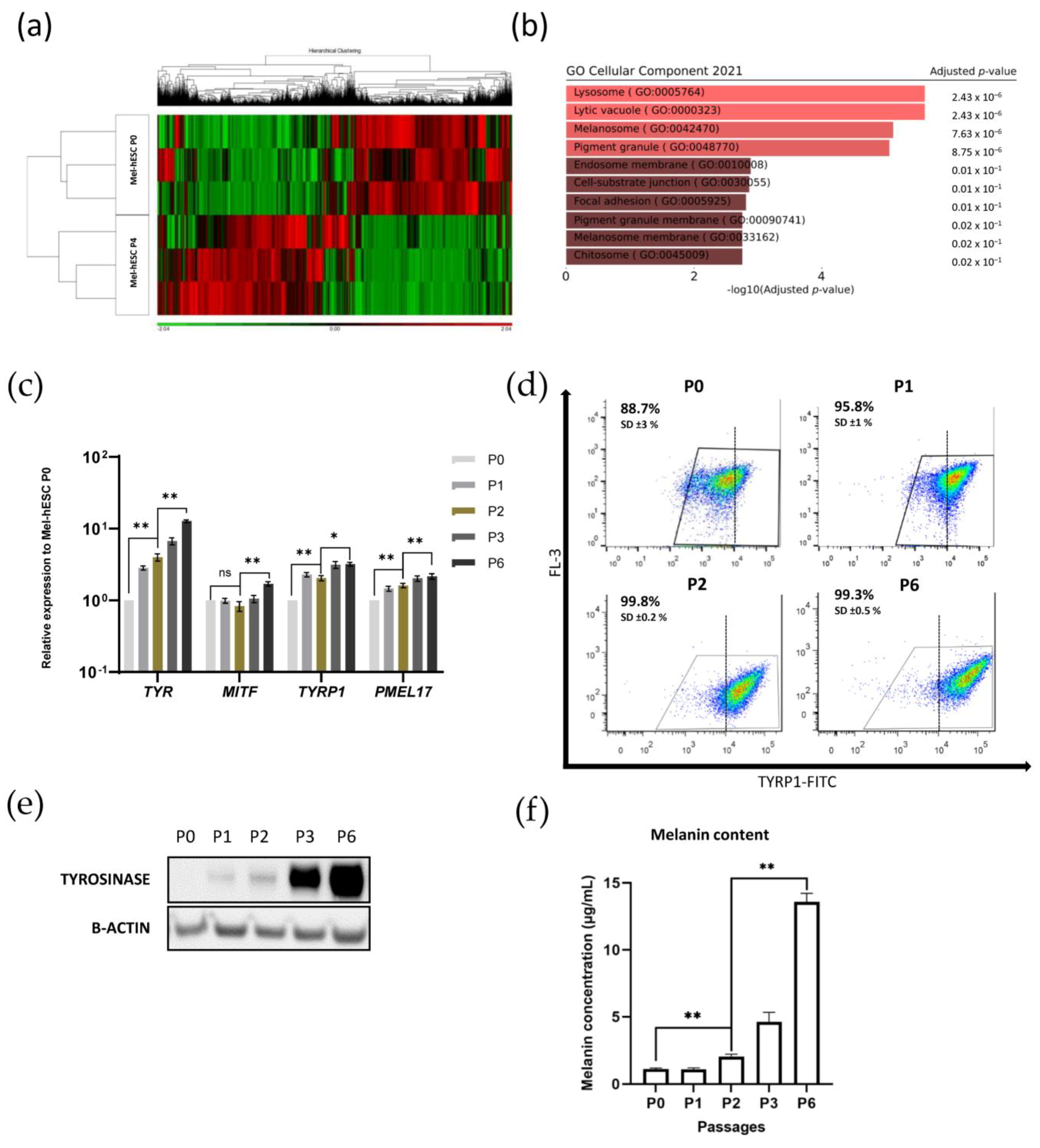
2.4. Functional Genomic Analysis of Melanocyte Differentiation and Maturation Processes
3. Discussion
4. Materials and Methods
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Mort, R.L.; Jackson, I.J.; Patton, E.E. The melanocyte lineage in development and disease. Development 2015, 142, 620–632. [Google Scholar] [CrossRef] [PubMed]
- Feng, Y.; Lu, Y. Advances in vitiligo: Update on therapeutic targets. Front. Immunol. 2022, 13, 986918. [Google Scholar] [CrossRef] [PubMed]
- Kawakami, T. Surgical procedures and innovative approaches for vitiligo regenerative treatment and melanocytorrhagy. J. Dermatol. 2022, 49, 391–401. [Google Scholar] [CrossRef] [PubMed]
- Tang, J.; Li, Q.; Cheng, B.; Jing, L. Primary culture of human face skin melanocytes for the study of hyperpigmentation. Cytotechnology 2014, 66, 891–898. [Google Scholar] [CrossRef]
- Sułkowski, M.; Kot, M.; Badyra, B.; Paluszkiewicz, A.; Płonka, P.M.; Sarna, M.; Michalczyk-Wetula, D.; Zucca, F.A.; Zecca, L.; Majka, M. Highly Effective Protocol for Differentiation of Induced Pluripotent Stem Cells (iPS) into Melanin-Producing Cells. Int. J. Mol. Sci. 2021, 22, 12787. [Google Scholar] [CrossRef]
- Liu, L.-P.; Guo, N.-N.; Li, Y.-M.; Zheng, Y.-W. Generation of Human iMelanocytes from Induced Pluripotent Stem Cells through a Suspension Culture System. STAR Protoc. 2020, 1, 100004. [Google Scholar] [CrossRef]
- Nissan, X.; Larribere, L.; Saidani, M.; Hurbain, I.; Delevoye, C.; Feteira, J.; Lemaitre, G.; Peschanski, M.; Baldeschi, C. Functional melanocytes derived from human pluripotent stem cells engraft into pluristratified epidermis. Proc. Natl. Acad. Sci. USA 2011, 108, 14861–14866. [Google Scholar] [CrossRef]
- Jones, J.C.; Sabatini, K.; Liao, X.; Tran, H.T.; Lynch, C.L.; Morey, R.E.; Glenn-Pratola, V.; Boscolo, F.S.; Yang, Q.; Parast, M.M.; et al. Melanocytes derived from transgene-free human induced pluripotent stem cells. J. Investig. Dermatol. 2013, 133, 2104–2108. [Google Scholar] [CrossRef]
- Fang, D.; Leishear, K.; Nguyen, T.K.; Finko, R.; Cai, K.; Fukunaga, M.; Li, L.; Brafford, P.A.; Kulp, A.N.; Xu, X.; et al. Defining the conditions for the generation of melanocytes from human embryonic stem cells. Stem Cells 2006, 24, 1668–1677. [Google Scholar] [CrossRef]
- Kawakami, T.; Okano, T.; Takeuchi, S.; Osumi, K.; Soma, Y.; Itoh, M.; Hirobe, T.; Jimbow, K. Approach for the Derivation of Melanocytes from Induced Pluripotent Stem Cells. J. Investig. Dermatol. 2018, 138, 150–158. [Google Scholar] [CrossRef]
- Chambers, S.M.; Mica, Y.; Lee, G.; Studer, L.; Tomishima, M.J. Dual-SMAD Inhibition/WNT Activation-Based Methods to Induce Neural Crest and Derivatives from Human Pluripotent Stem Cells. Methods Mol. Biol. 2016, 1307, 329–343. [Google Scholar] [PubMed]
- Gomez, G.A.; Prasad, M.S.; Sandhu, N.; Shelar, P.B.; Leung, A.W.; García-Castro, M.I. Human Neural Crest Induction by Temporal Modulation of WNT Activation. Dev. Biol. 2019, 449, 99–106. [Google Scholar] [CrossRef] [PubMed]
- Shannon, P.; Markiel, A.; Ozier, O.; Baliga, N.S.; Wang, J.T.; Ramage, D.; Amin, N.; Schwikowski, B.; Ideker, T. Cytoscape: A software environment for integrated models of biomolecular interaction networks. Genome Res. 2003, 13, 2498–2504. [Google Scholar] [CrossRef] [PubMed]
- Kawa, Y.; Ito, M.; Ono, H.; Asano, M.; Takano, N.; Ooka, S.; Watabe, H.; Hosaka, E.; Baba, T.; Kubota, Y.; et al. Stem cell factor and/or endothelin-3 dependent immortal melanoblast and melanocyte populations derived from mouse neural crest cells. Pigment Cell Res. 2000, 13 (Suppl. 8), 73–80. [Google Scholar] [CrossRef] [PubMed]
- Takeda, K.; Yasumoto, K.; Takada, R.; Takada, S.; Watanabe, K.; Udono, T.; Saito, H.; Takahashi, K.; Shibahara, S. Induction of melanocyte-specific microphthalmia-associated transcription factor by Wnt-3a. J. Biol. Chem. 2000, 275, 14013–14016. [Google Scholar] [CrossRef] [PubMed]
- Guenou, H.; Nissan, X.; Larcher, F.; Feteira, J.; Lemaitre, G.; Saidani, M.; Del Rio, M.; Barrault, C.C.; Bernard, F.-X.; Peschanski, M.; et al. Human embryonic stem-cell derivatives for full reconstruction of the pluristratified epidermis: A preclinical study. Lancet 2009, 374, 1745–1753. [Google Scholar] [CrossRef] [PubMed]
- Levy, C.; Khaled, M.; Fisher, D.E. MITF: Master regulator of melanocyte development and melanoma oncogene. Trends Mol. Med. 2006, 12, 406–414. [Google Scholar] [CrossRef]
- Kindl, G.H.; D’Orazio, J.A. Pharmacologic manipulation of skin pigmentation. Pigment Cell Melanoma Res. 2021, 34, 777–785. [Google Scholar] [CrossRef]
- Mica, Y.; Lee, G.; Chambers, S.M.; Tomishima, M.J.; Studer, L. Modeling neural crest induction, melanocyte specification, and disease-related pigmentation defects in hESCs and patient-specific iPSCs. Cell Rep. 2013, 3, 1140–1152. [Google Scholar] [CrossRef]
- Bonaventure, J.; Domingues, M.J.; Larue, L. Cellular and molecular mechanisms controlling the migration of melanocytes and melanoma cells. Pigment Cell Melanoma Res. 2013, 26, 316–325. [Google Scholar] [CrossRef]
- Leung, A.W.; Murdoch, B.; Salem, A.F.; Prasad, M.S.; Gomez, G.A.; García-Castro, M.I. WNT/β-catenin signaling mediates human neural crest induction via a pre-neural border intermediate. Development 2016, 143, 398–410. [Google Scholar] [CrossRef] [PubMed]
- White, R.M.; Zon, L.I. Melanocytes in Development, Regeneration, and Cancer. Cell Stem Cell 2008, 3, 242–252. [Google Scholar] [CrossRef]
- Marubashi, S.; Fukuda, M. Rab7B/42 Is Functionally Involved in Protein Degradation on Melanosomes in Keratinocytes. Cell Struct. Funct. 2020, 45, 45–55. [Google Scholar] [CrossRef] [PubMed]
- Krzewski, K.; Cullinane, A.R. Evidence for defective Rab GTPase-dependent cargo traffic in immune disorders. Exp. Cell Res. 2013, 319, 2360–2367. [Google Scholar] [CrossRef] [PubMed]
- Khaitan, B.K.; Kathuria, S.; Ramam, M. A descriptive study to characterize segmental vitiligo. Indian J. Dermatol. Venereol. Leprol. 2012, 78, 715–721. [Google Scholar] [CrossRef]
- Kubelis-López, D.E.; Zapata-Salazar, N.A.; Said-Fernández, S.L.; Sánchez-Domínguez, C.N.; Salinas-Santander, M.A.; Martínez-Rodríguez, H.G.; Vázquez-Martínez, O.T.; Wollina, U.; Lotti, T.; Ocampo-Candiani, J. Updates and new medical treatments for vitiligo (Review). Exp. Ther. Med. 2021, 22, 797. [Google Scholar] [CrossRef]
- Kim, J.Y.; Nam, Y.; Rim, Y.A.; Ju, J.H. Review of the Current Trends in Clinical Trials Involving Induced Pluripotent Stem Cells. Stem Cell Rev. Rep. 2022, 18, 142–154. [Google Scholar] [CrossRef]
- Madrid, M.; Sumen, C.; Aivio, S.; Saklayen, N. Autologous Induced Pluripotent Stem Cell-Based Cell Therapies: Promise, Progress, and Challenges. Curr. Protoc. 2021, 1, e88. [Google Scholar] [CrossRef]
- Liu, L.-P.; Li, Y.-M.; Guo, N.-N.; Li, S.; Ma, X.; Zhang, Y.-X.; Gao, Y.; Huang, J.-L.; Zheng, D.-X.; Wang, L.-Y.; et al. Therapeutic Potential of Patient iPSC-Derived iMelanocytes in Autologous Transplantation. Cell Rep. 2019, 27, 455–466.e5. [Google Scholar] [CrossRef]
- Kawakami, Y.; Eliyahu, S.; Jennings, C.; Sakaguchi, K.; Kang, X.; Southwood, S.; Robbins, P.F.; Sette, A.; Appella, E.; Rosenberg, S.A. Recognition of multiple epitopes in the human melanoma antigen gp100 by tumor-infiltrating T lymphocytes associated with in vivo tumor regression. J. Immunol. 1995, 154, 3961–3968. [Google Scholar] [CrossRef]
- Ellis, J.M.; Henson, V.; Slack, R.; Ng, J.; Hartzman, R.J.; Katovich Hurley, C. Frequencies of HLA-A2 alleles in five U.S. population groups. Predominance Of A*02011 and identification of HLA-A*0231. Hum. Immunol. 2000, 61, 334–340. [Google Scholar] [CrossRef] [PubMed]
- Zhang, D.; Hong, W.; Fu, L.; Wei, X.; Xu, A. A randomized controlled study of the effects of different modalities of narrow-band ultraviolet B therapy on the outcome of cultured autologous melanocytes transplantation in treating vitiligo. Dermatol. Surg. 2014, 40, 420–426. [Google Scholar] [CrossRef]
- Ju, H.J.; Bae, J.M.; Lee, R.W.; Kim, S.H.; Parsad, D.; Pourang, A.; Hamzavi, I.; Shourick, J.; Ezzedine, K. Surgical Interventions for Patients with Vitiligo: A Systematic Review and Meta-analysis. JAMA Dermatol. 2021, 157, 307–316. [Google Scholar] [CrossRef]
- Valmori, D.; Fonteneau, J.F.; Lizana, C.M.; Gervois, N.; Liénard, D.; Rimoldi, D.; Jongeneel, V.; Jotereau, F.; Cerottini, J.C.; Romero, P. Enhanced generation of specific tumor-reactive CTL in vitro by selected Melan-A/MART-1 immunodominant peptide analogues. J. Immunol. 1998, 160, 1750–1758. [Google Scholar] [CrossRef] [PubMed]
- Song, Y.H.; Connor, E.; Li, Y.; Zorovich, B.; Balducci, P.; Maclaren, N. The role of tyrosinase in autoimmune vitiligo. Lancet 1994, 344, 1049–1052. [Google Scholar] [CrossRef] [PubMed]
- Cui, T.; Yi, X.; Guo, S.; Zhou, F.; Liu, L.; Li, C.; Li, K.; Gao, T. Identification of Novel HLA-A*0201-Restricted CTL Epitopes in Chinese Vitiligo Patients. Sci. Rep. 2016, 6, 36360. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.-B.; Li, M.; Chen, H.; Zhong, S.-Q.; Yang, S.; Du, W.-D.; Hao, J.-H.; Zhang, T.-S.; Zhang, X.-J.; Zeegers, M.P. Association of vitiligo with HLA-A2: A meta-analysis. J. Eur. Acad. Dermatol. Venereol. 2007, 21, 205–213. [Google Scholar] [CrossRef]
- Mandelcorn-Monson, R.L.; Shear, N.H.; Yau, E.; Sambhara, S.; Barber, B.H.; Spaner, D.; DeBenedette, M.A. Cytotoxic T lymphocyte reactivity to gp100, MelanA/MART-1, and tyrosinase, in HLA-A2-positive vitiligo patients. J. Investig. Dermatol. 2003, 121, 550–556. [Google Scholar] [CrossRef]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef]
- Martínez-Santamaría, L.; Guerrero-Aspizua, S.; Del Río, M. Skin bioengineering: Preclinical and clinical applications. Actas Dermosifiliogr. 2012, 103, 5–11. [Google Scholar] [CrossRef]
- Dobin, A.; Davis, C.A.; Schlesinger, F.; Drenkow, J.; Zaleski, C.; Jha, S.; Batut, P.; Chaisson, M.; Gingeras, T.R. STAR: Ultrafast universal RNA-seq aligner. Bioinformatics 2013, 29, 15–21. [Google Scholar] [CrossRef]
- Li, H.; Handsaker, B.; Wysoker, A.; Fennell, T.; Ruan, J.; Homer, N.; Marth, G.; Abecasis, G.; Durbin, R.; 1000 Genome Project Data Processing Subgroup. The Sequence Alignment/Map format and SAMtools. Bioinformatics 2009, 25, 2078–2079. [Google Scholar] [CrossRef] [PubMed]
- Anders, S.; Pyl, P.T.; Huber, W. HTSeq—A Python framework to work with high-throughput sequencing data. Bioinformatics 2015, 31, 166–169. [Google Scholar] [CrossRef] [PubMed]
- Sander, J.; Yosef, N. ImpulseDE: Detection of DE genes in time series data using impulse models. Bioinformatics 2017, 33, 757–759. [Google Scholar] [CrossRef] [PubMed]
| Clusters Description | |||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Cluster 1 | Cluster 2 | Cluster 3 | Cluster 4 | Cluster 5 | Cluster 6 | Cluster 7 | Cluster 8 | ||||||
| LAS1L | CLDN3 | LIG3 | FOXB1 | TNFRSF9 | HSPA5 | ATP10A | CEBPD | HSBP1L1 | GAS2 | NPAS2 | HIST1H4B | HMGA1 | TPX2 |
| CRLF1 | MYO5B | TACC3 | AFF1 | C6orf186 | CD81 | HSPB6 | LGALS3BP | SERPINF1 | ENDOD1 | PDGFD | HNRNPU | HIST1H2BD | CBX5 |
| CD9 | WDR81 | FLT4 | HES3 | OPRK1 | MYL12B | TBXA2R | SLC16A6 | TBC1D14 | ZC3H12C | JUNB | HIST1H2BI | HIST1H3I | SFRP1 |
| KAL1 | PCSK9 | POLD1 | CBX2 | LTBP4 | CD63 | GAS7 | PMP22 | BCAN | OXSM | PTEN | HIST1H1C | HIST1H2AH | FBL |
| NLRP2 | KRT8 | SUGP2 | MSL2 | GALNTL1 | MGST3 | CYTH3 | ST3GAL4 | TMCC2 | CCDC3 | ISG20 | HIST1H2BM | SMCHD1 | CENPF |
| CDH3 | XKR6 | MCM10 | UNC119B | CHRDL1 | MMP14 | NFIX | ELMOD1 | TRPM1 | ACSL1 | RCAN2 | HIST1H2BE | SNRPF | |
| PRKCZ | ZIK1 | GLI2 | LMNB2 | COTL1 | COL1A2 | LRRC23 | CLEC2B | SORT1 | TXNDC11 | IL16 | G3BP1 | ||
| SRRT | BNC2 | TFAP4 | ZNF678 | FZD3 | PPIB | MVP | LTBR | CABLES1 | DAB2 | EHBP1L1 | RCOR2 | ||
| KDM2B | ZNF483 | KIF4A | GINS3 | LGI1 | CTNNB1 | SNAI2 | GPR133 | EMP1 | C9orf150 | CSPG4 | UBE2C | ||
| CYP26A1 | PSMD2 | SRPK1 | FIGN | DBX1 | SILV | GPR124 | COL12A1 | RAB33A | FAM134B | YIF1A | HIST1H2AC | ||
| ITPR3 | GSG2 | DSP | IQGAP3 | ZBTB16 | RUNX3 | NCOA7 | P2RX4 | C10orf90 | A2M | HIST1H2BG | |||
| CECR2 | GLDC | NUP93 | HIST1H1B | PARD3B | ABCC2 | SMOC2 | CD164 | ADAMTS1 | KLHL38 | HIST1H4D | |||
| SALL4 | ARID3B | DNMBP | KIF18B | HOXB1 | ELN | LOX | AGT | FGD5 | NUPR1 | HIST1H2BH | |||
| HAGHL | FGD6 | MAP2K6 | LIN28B | NPPB | HEXB | PDE4D | FAM129A | ACSS1 | RTTN | HIST1H3A | |||
| CTSH | ERCC6L | NCAPG | TCF4 | WFIKKN1 | CCDC90A | PDGFRB | SERPINE2 | CA10 | FIBIN | HIST1H2AL | |||
| ESRP1 | SEMA4D | PTGES3 | WNK3 | DNAJB1 | HERPUD1 | CPEB4 | CYP27A1 | MARVELD1 | DPP7 | ||||
| GSDMD | ARID2 | LMNB1 | HIST1H4I | WASF3 | BRP44L | ROPN1B | ARMC9 | GOLGA7B | LRRN4CL | ||||
| LIG1 | HIST1H3E | UXS1 | HIST1H4L | NAV1 | IDH3G | PLSCR4 | LMO7 | CD109 | GBA | ||||
| SIPA1L3 | SIPA1L1 | ARID3A | SMOC1 | SOX3 | RAB27A | STAM2 | SETDB2 | FBXO32 | C7orf41 | ||||
| UNC5B | ZNF770 | EPHA4 | LGR4 | LGR5 | WIPI1 | KCNJ13 | GPNMB | FZD1 | MAB21L1 | ||||
| SLC9A3R1 | WDHD1 | C1orf109 | CCDC85C | ATBF1 | ST6GALNAC2 | MLPH | NDUFB5 | C9orf91 | GREM2 | ||||
| PVRL1 | DAXX | ARID1A | SMN2 | BOC | SEL1L | PLCL1 | ABI1 | TNFRSF14 | SLC9A9 | ||||
| SLC38A1 | EIF5AL1 | KDM3B | NEURL1B | SFRP2 | CYBRD1 | SLC1A4 | STXBP1 | TRIM63 | AP1S2 | ||||
| NUP155 | LYPLA1 | HIST1H2AM | SCUBE3 | CDH19 | TRAK2 | NOV | CYB5R1 | NTM | |||||
| GALNT3 | NCAPH | KIFC1 | CDKN2B | SLC6A15 | SCAMP3 | TFAP2A | SPON2 | TMEM119 | |||||
| MSH6 | ZWINT | CRB2 | PDE8A | GBP1 | IRF4 | S100B | SOCS3 | ||||||
| HDAC1 | CIT | AP000654.2 | PTGS2 | RAB32 | CPEB2 | LMNA | TMEM173 | ||||||
| OLFML3 | PPAT | TMEM132D | TYR | KLF9 | SULF1 | C19orf28 | NDUFA4L2 | ||||||
| TTF2 | SMO | WNT7A | GPR137B | ITGB1BP1 | THBS1 | JOSD2 | SHC4 | ||||||
| RCAN3 | CNN1 | DPYSL5 | FAP | NR4A3 | IFI44 | GRASP | KCNQ5 | ||||||
| PLXDC2 | NUP210 | KLHDC8A | CEACAM1 | RHOQ | ANXA7 | NFIA | IRS2 | ||||||
| PLS1 | PIK3C2B | PTX3 | STX7 | IFIT3 | ARHGAP24 | TM2D1 | THBS2 | ||||||
| HNRNPA2B1 | CDCA8 | MAMDC2 | DCT | MLANA | CPNE8 | LHX8 | C6orf1 | ||||||
| STIL | C9orf100 | CENPV | EPDR1 | C5orf32 | C1RL | DDR2 | MITF | ||||||
| PAIP2B | HIST1H2AB | HOXB9 | PPP1R15A | TGFBI | PHLDA1 | KCNF1 | CYP26C1 | ||||||
| RRBP1 | MDC1 | SNCG | COQ9 | GLT8D2 | FBLN5 | GPR155 | TRPV2 | ||||||
| EGLN3 | MAP2K5 | NRIP3 | P2RX7 | SOCS2 | BCL2A1 | IGFBP7 | THSD4 | ||||||
| MLLT4 | SHROOM3 | IRX5 | SLC17A6 | SORBS3 | DNAJA4 | IFI16 | FAM69C | ||||||
| ASS1 | LRIG3 | IRX3 | GADD45B | TBX2 | ABHD2 | ARHGEF3 | DNER | ||||||
| C19orf66 | STON2 | NRIP1 | PALM | PDZRN3 | TGFB1I1 | EMCN | CHM | ||||||
| NOL11 | C15orf42 | EXT1 | MMP11 | GJA3 | SAT2 | SLC45A2 | SLC24A5 | ||||||
| LIN28A | ZNF710 | HIST2H2BE | SLC7A4 | UBL3 | TTYH2 | EDIL3 | PARVB | ||||||
| CC2D1A | MED9 | COL25A1 | LGALS1 | LRRC39 | NFIC | SHH | LRRK2 | ||||||
| HRSP12 | POU2F1 | FAT4 | SOX10 | INHBA | TMEM50B | BRI3 | APOD | ||||||
| NASP | SOX13 | VEPH1 | TIMP3 | SRGN | EMP3 | CTSB | TDRD7 | ||||||
| SYT6 | SKP2 | HOXC6 | TSPO | BICC1 | ECM1 | NOS3 | MME | ||||||
| DSC2 | LIX1 | ELOVL2 | ATP6V1D | BHLHE41 | CTSK | CRYL1 | CASP4 | ||||||
| HEY2 | ADAMTS19 | DLL1 | CINP | ZC3H13 | DUSP10 | C10orf10 | ANXA4 | ||||||
| USP44 | ZMYM4 | POU3F3 | CTSZ | RAB38 | LYST | MFAP4 | METTL9 | ||||||
| LECT1 | NR6A1 | BIRC7 | F13A1 | ARL8A | NNMT | ANXA6 | |||||||
| SORL1 | QTRTD1 | TRIB3 | SERPINB6 | MANF | SLFN5 | SLC6A17 | |||||||
| FRAS1 | UTRN | PLP2 | SBDS | EPHA5 | STAT6 | LRP10 | |||||||
| PTPRF | SKA1 | RENBP | SIX1 | SNCA | AKTIP | UAP1L1 | |||||||
| MCL1 | SFRS15 | ASB9 | MASP1 | DDIT4L | SLC3A2 | DACT3 | |||||||
| CDCA5 | PPP1R9A | ACP5 | ZFP36 | OSMR | LTBP3 | GSTK1 | |||||||
| LPCAT1 | IGF2BP1 | PLA2G15 | VGF | CXCL14 | LGI3 | MBP | |||||||
| SLC16A1 | DBF4B | PGCP | DNAJB9 | TBC1D7 | SLC16A4 | CHSY3 | |||||||
| DUSP2 | C1orf187 | NIPAL2 | LRRC17 | LRRTM2 | NSG1 | MMP17 | |||||||
| C21orf45 | C1orf106 | RAB2A | ISLR | DAAM2 | C6orf145 | C1orf85 | |||||||
| ACTC1 | PPM1L | ASAH1 | RNASE1 | ARHGAP18 | ATF5 | EGFL6 | |||||||
| FGFR4 | MAD2L1 | CAV2 | GDF15 | C6orf192 | IL1RAPL1 | SHISA4 | |||||||
| PDPN | USP49 | PCOLCE | NDFIP1 | SDK1 | SLC33A1 | TGM2 | |||||||
| PKDCC | PLEKHA7 | AHR | UBE2D2 | TACC1 | IL8 | CARD16 | |||||||
| PQLC3 | HIC2 | AEBP1 | TMEM204 | EBAG9 | HMP19 | CFI | |||||||
| ZNF589 | IRX2 | SFXN3 | LGALS3 | NFIB | CRTAP | ||||||||
| CAMKV | PPID | DNAJC12 | MATN3 | GSN | SEMA3E | ||||||||
| Gene Name | Forward | Reverse |
|---|---|---|
| 18S | GAGGATGAGGTGGAACGTGT | TCTTCAGTCGCTCCAGGTCT |
| S0X10 | AGCCCAGGTGAAGACAGAGA | ATAGGGTCCTGAGGGCTG AT |
| MITF | GCTCACAGCGTGTATTTTTCC | TCTCTTTGGCCAGTGCTCTT |
| TYRP1 | AGCAGTAGTTGGCGCTTTGT | TCAGTGAGGAGAGGCTGGTT |
| TYR | TTGTACTGCCTGCTGTGGAG | CAGGAACCTCTGCCTGAAAG |
| PMEL17 | TGGACAGAAGCCCAGAGACT | GCAATACCTTTTGGCTTCCA |
| DCT | GGTTCCTTTCTTCCCTCCAG | AACCAAAGCCACCAGTGTTC |
| RAB27A | GGGCAGGAGAGGTTTCGTAG | TGCATGCATCTGTAGCTGGC |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Saidani, M.; Darle, A.; Jarrige, M.; Polveche, H.; El Kassar, L.; Julié, S.; Bessou-Touya, S.; Holic, N.; Lemaitre, G.; Martinat, C.; et al. Generating Functional and Highly Proliferative Melanocytes Derived from Human Pluripotent Stem Cells: A Promising Tool for Biotherapeutic Approaches to Treat Skin Pigmentation Disorders. Int. J. Mol. Sci. 2023, 24, 6398. https://doi.org/10.3390/ijms24076398
Saidani M, Darle A, Jarrige M, Polveche H, El Kassar L, Julié S, Bessou-Touya S, Holic N, Lemaitre G, Martinat C, et al. Generating Functional and Highly Proliferative Melanocytes Derived from Human Pluripotent Stem Cells: A Promising Tool for Biotherapeutic Approaches to Treat Skin Pigmentation Disorders. International Journal of Molecular Sciences. 2023; 24(7):6398. https://doi.org/10.3390/ijms24076398
Chicago/Turabian StyleSaidani, Manoubia, Annabelle Darle, Margot Jarrige, Hélène Polveche, Lina El Kassar, Séverine Julié, Sandrine Bessou-Touya, Nathalie Holic, Gilles Lemaitre, Cécile Martinat, and et al. 2023. "Generating Functional and Highly Proliferative Melanocytes Derived from Human Pluripotent Stem Cells: A Promising Tool for Biotherapeutic Approaches to Treat Skin Pigmentation Disorders" International Journal of Molecular Sciences 24, no. 7: 6398. https://doi.org/10.3390/ijms24076398
APA StyleSaidani, M., Darle, A., Jarrige, M., Polveche, H., El Kassar, L., Julié, S., Bessou-Touya, S., Holic, N., Lemaitre, G., Martinat, C., Baldeschi, C., & Allouche, J. (2023). Generating Functional and Highly Proliferative Melanocytes Derived from Human Pluripotent Stem Cells: A Promising Tool for Biotherapeutic Approaches to Treat Skin Pigmentation Disorders. International Journal of Molecular Sciences, 24(7), 6398. https://doi.org/10.3390/ijms24076398






