A Novel E3 Probiotics Formula Restored Gut Dysbiosis and Remodelled Gut Microbial Network and Microbiome Dysbiosis Index (MDI) in Southern Chinese Adult Psoriasis Patients
Abstract
1. Introduction
2. Results
2.1. Study Cohort
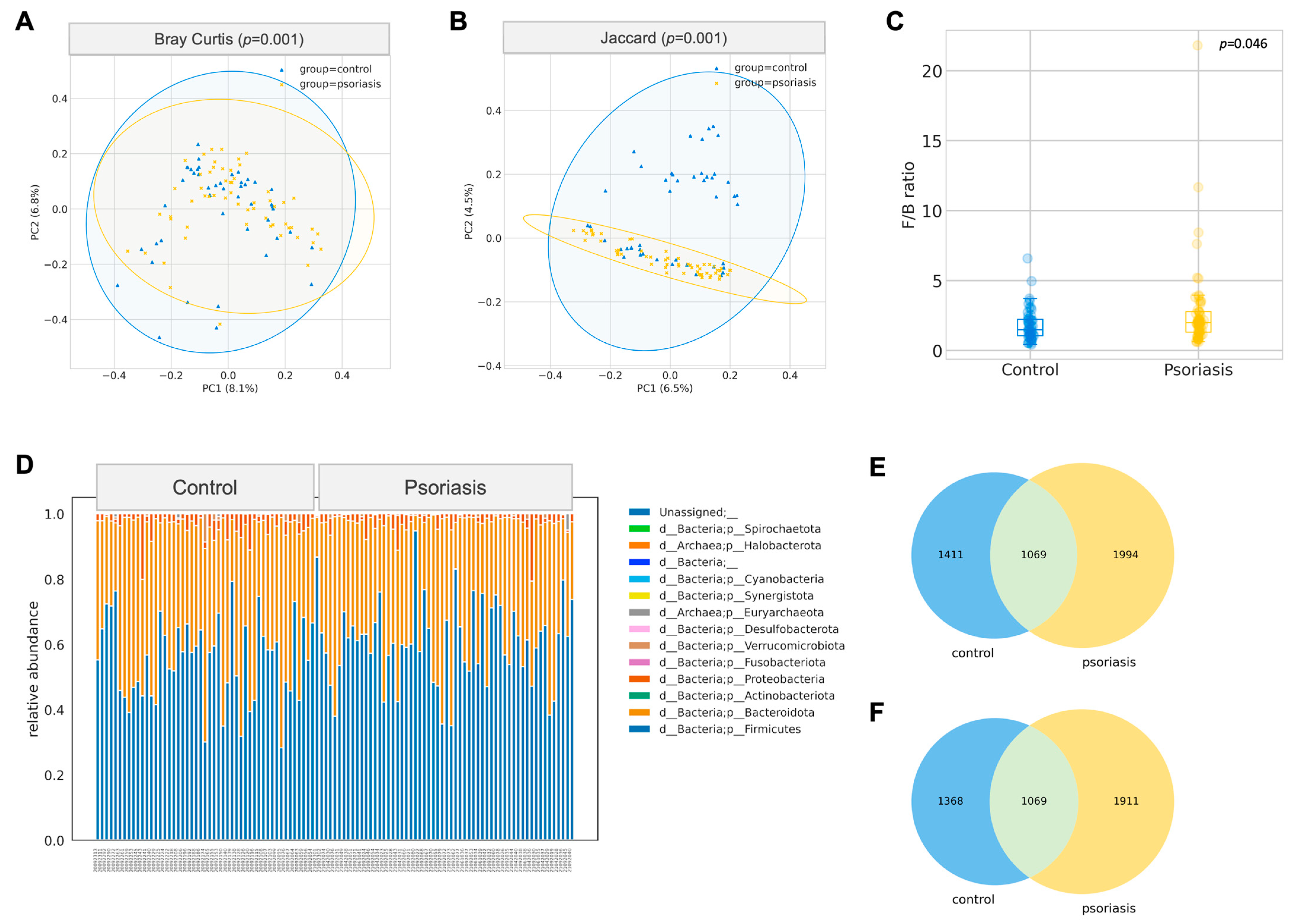
2.2. Significant Difference in Gut Microbiome Composition between Psoriasis and Control Group
2.3. Effect of 8-Week Probiotics Intake in the Gut Microbiome Composition in Psoriasis Group
2.4. Depleted SCFA Related Functional Abundance in Psoriasis Group
2.5. Remodelling of Microbial Co-Occurrence Network
2.6. Development of Psoriasis Specific Machine Learning Based Microbiome Dysbiosis Index (MDI)
2.7. Microbiome Dysbiosis Index (MDI) Correlates with PASI Responsiveness
3. Discussion
4. Materials and Methods
4.1. Subject Recruitment and Study Design
4.2. Library Preparation and 16S rRNA Sequencing
4.3. Probiotic Mixture
4.4. Bioinformatics Analysis
4.5. Quantitative Real Time PCR
4.6. Development of Gut Microbiome Dysbiosis Index (MDI)
4.7. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Rachakonda, T.D.; Schupp, C.W.; Armstrong, A.W. Psoriasis Prevalence among Adults in the United States. J. Am. Acad. Dermatol. 2014, 70, 512–516. [Google Scholar] [CrossRef]
- Global Report on Psoriasis. Available online: https://apps.who.int/iris/handle/10665/204417 (accessed on 22 March 2022).
- Michalek, I.M.; Loring, B.; John, S.M. A Systematic Review of Worldwide Epidemiology of Psoriasis. J. Eur. Acad. Dermatol. Venereol. 2017, 31, 205–212. [Google Scholar] [CrossRef]
- LCQ9: Support for Psoriasis Patients. Available online: https://www.info.gov.hk/gia/general/202010/21/P2020102100320.htm (accessed on 22 March 2022).
- HKSH Healthcare. Available online: https://www.hksh-healthcare.com/en/clinical-services/dermatology-centre/psoriasis.php (accessed on 22 March 2022).
- Armstrong, A.W.; Read, C. Pathophysiology, Clinical Presentation, and Treatment of Psoriasis: A Review. JAMA 2020, 323, 1945–1960. [Google Scholar] [CrossRef] [PubMed]
- Elmets, C.A.; Korman, N.J.; Prater, E.F.; Wong, E.B.; Rupani, R.N.; Kivelevitch, D.; Armstrong, A.W.; Connor, C.; Cordoro, K.M.; Davis, D.M.R.; et al. Joint AAD–NPF Guidelines of Care for the Management and Treatment of Psoriasis with Topical Therapy and Alternative Medicine Modalities for Psoriasis Severity Measures. J. Am. Acad. Dermatol. 2021, 84, 432–470. [Google Scholar] [CrossRef]
- Ferreira, B.I.R.C.; da Costa Abreu, J.L.P.; dos Reis, J.P.G.; da Costa Figueiredo, A.M. Psoriasis and Associated Psychiatric Disorders: A Systematic Review on Etiopathogenesis and Clinical Correlation. J. Clin. Aesthetic Dermatol. 2016, 9, 36. [Google Scholar]
- Singh, S.; Taylor, C.; Kornmehl, H.; Armstrong, A.W. Psoriasis and Suicidality: A Systematic Review and Meta-Analysis. J. Am. Acad. Dermatol. 2017, 77, 425–440.e2. [Google Scholar] [CrossRef]
- Dowlatshahi, E.A.; Wakkee, M.; Arends, L.R.; Nijsten, T. The Prevalence and Odds of Depressive Symptoms and Clinical Depression in Psoriasis Patients: A Systematic Review and Meta-Analysis. J. Investig. Dermatol. 2014, 134, 1542–1551. [Google Scholar] [CrossRef] [PubMed]
- Dey, A.K.; Joshi, A.A.; Chaturvedi, A.; Lerman, J.B.; Aberra, T.M.; Rodante, J.A.; Teague, H.L.; Harrington, C.L.; Rivers, J.P.; Chung, J.H.; et al. Association Between Skin and Aortic Vascular Inflammation in Patients with Psoriasis: A Case-Cohort Study Using Positron Emission Tomography/Computed Tomography. JAMA Cardiol. 2017, 2, 1013–1018. [Google Scholar] [CrossRef] [PubMed]
- Mansouri, B.; Kivelevitch, D.; Natarajan, B.; Joshi, A.A.; Ryan, C.; Benjegerdes, K.; Schussler, J.M.; Rader, D.J.; Reilly, M.P.; Menter, A.; et al. Comparison of Coronary Artery Calcium Scores Between Patients with Psoriasis and Type 2 Diabetes. JAMA Dermatol. 2016, 152, 1244–1253. [Google Scholar] [CrossRef]
- Lerman, J.B.; Joshi, A.A.; Chaturvedi, A.; Aberra, T.M.; Dey, A.K.; Rodante, J.A.; Salahuddin, T.; Chung, J.H.; Rana, A.; Teague, H.L.; et al. Coronary Plaque Characterization in Psoriasis Reveals High-Risk Features That Improve after Treatment in a Prospective Observational Study. Circulation 2017, 136, 263–276. [Google Scholar] [CrossRef]
- Armstrong, A.W.; Harskamp, C.T.; Armstrong, E.J. Psoriasis and the Risk of Diabetes Mellitus: A Systematic Review and Meta-Analysis. JAMA Dermatol. 2013, 149, 84–91. [Google Scholar] [CrossRef] [PubMed]
- Armstrong, A.W.; Guérin, A.; Sundaram, M.; Wu, E.Q.; Faust, E.S.; Ionescu-Ittu, R.; Mulani, P. Psoriasis and Risk of Diabetes-Associated Microvascular and Macrovascular Complications. J. Am. Acad. Dermatol. 2015, 72, 968–977.e2. [Google Scholar] [CrossRef]
- Gelfand, J.M.; Dommasch, E.D.; Shin, D.B.; Azfar, R.S.; Kurd, S.K.; Wang, X.; Troxel, A.B. The Risk of Stroke in Patients with Psoriasis. J. Investig. Dermatol. 2009, 129, 2411–2418. [Google Scholar] [CrossRef]
- Gelfand, J.M.; Neimann, A.L.; Shin, D.B.; Wang, X.; Margolis, D.J.; Troxel, A.B. Risk of Myocardial Infarction in Patients with Psoriasis. JAMA 2006, 296, 1735–1741. [Google Scholar] [CrossRef] [PubMed]
- Lin, H.W.; Wang, K.H.; Lin, H.C.; Lin, H.C. Increased Risk of Acute Myocardial Infarction in Patients with Psoriasis: A 5-Year Population-Based Study in Taiwan. J. Am. Acad. Dermatol. 2011, 64, 495–501. [Google Scholar] [CrossRef] [PubMed]
- Hedin, C.R.H.; Sonkoly, E.; Eberhardson, M.; Ståhle, M. Inflammatory Bowel Disease and Psoriasis: Modernizing the Multidisciplinary Approach. J. Intern. Med. 2021, 290, 257–278. [Google Scholar] [CrossRef]
- Fu, Y.; Lee, C.H.; Chi, C.C. Association of Psoriasis with Inflammatory Bowel Disease: A Systematic Review and Meta-Analysis. JAMA Dermatol. 2018, 154, 1417–1427. [Google Scholar] [CrossRef]
- Reynolds, G.; Vegh, P.; Fletcher, J.; Poyner, E.F.M.; Stephenson, E.; Goh, I.; Botting, R.A.; Huang, N.; Olabi, B.; Dubois, A.; et al. Developmental Cell Programs Are Co-Opted in Inflammatory Skin Disease. Science 2021, 371, 730–735. [Google Scholar] [CrossRef]
- Grjibovski, A.M.; Olsen, A.O.; Magnus, P.; Harris, J.R. Psoriasis in Norwegian Twins: Contribution of Genetic and Environmental Effects. J. Eur. Acad. Dermatol. Venereol. 2007, 21, 1337–1343. [Google Scholar] [CrossRef]
- Singh, S.; Pradhan, D.; Puri, P.; Ramesh, V.; Aggarwal, S.; Nayek, A.; Jain, A.K. Genomic Alterations Driving Psoriasis Pathogenesis. Gene 2019, 683, 61–71. [Google Scholar] [CrossRef]
- Jenisch, S.; Westphal, E.; Nair, R.P.; Stuart, P.; Voorhees, J.J.; Christophers, E.; Krönke, M.; Elder, J.T.; Henseler, T. Linkage Disequilibrium Analysis of Familial Psoriasis: Identification of Multiple Disease-Associated MHC Haplotypes. Tissue Antigens 1999, 53, 135–146. [Google Scholar] [CrossRef] [PubMed]
- Ozawa, A.; Ohkido, M.; Inoko, H.; Ando, A.; Tsuji, K. Specific Restriction Fragment Length Polymorphism on the HLA-C Region and Susceptibility to Psoriasis Vulgaris. J. Investig. Dermatol. 1988, 90, 402–405. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.; Tsai, T.F. HLA-Cw6 and Psoriasis. Br. J. Dermatol. 2018, 178, 854–862. [Google Scholar] [CrossRef]
- Scher, J.U.; Ubeda, C.; Artacho, A.; Attur, M.; Isaac, S.; Reddy, S.M.; Marmon, S.; Neimann, A.; Brusca, S.; Patel, T.; et al. Decreased Bacterial Diversity Characterizes the Altered Gut Microbiota in Patients with Psoriatic Arthritis, Resembling Dysbiosis in Inflammatory Bowel Disease. Arthritis Rheumatol. 2015, 67, 128–139. [Google Scholar] [CrossRef]
- Chen, Y.J.; Ho, H.J.; Tseng, C.H.; Lai, Z.L.; Shieh, J.J.; Wu, C.Y. Intestinal Microbiota Profiling and Predicted Metabolic Dysregulation in Psoriasis Patients. Exp. Dermatol. 2018, 27, 1336–1343. [Google Scholar] [CrossRef]
- Hidalgo-Cantabrana, C.; Gómez, J.; Delgado, S.; Requena-López, S.; Queiro-Silva, R.; Margolles, A.; Coto, E.; Sánchez, B.; Coto-Segura, P. Gut Microbiota Dysbiosis in a Cohort of Patients with Psoriasis. Br. J. Dermatol. 2019, 181, 1287–1295. [Google Scholar] [CrossRef]
- Myers, B.; Brownstone, N.; Reddy, V.; Chan, S.; Thibodeaux, Q.; Truong, A.; Bhutani, T.; Chang, H.W.; Liao, W. The Gut Microbiome in Psoriasis and Psoriatic Arthritis. Best Pract. Res. Clin. Rheumatol. 2019, 33, 101494. [Google Scholar] [CrossRef]
- Shapiro, J.; Cohen, N.A.; Shalev, V.; Uzan, A.; Koren, O.; Maharshak, N. Psoriatic Patients Have a Distinct Structural and Functional Fecal Microbiota Compared with Controls. J. Dermatol. 2019, 46, 595–603. [Google Scholar] [CrossRef] [PubMed]
- Widhiati, S.; Purnomosari, D.; Wibawa, T.; Soebono, H. The Role of Gut Microbiome in Inflammatory Skin Disorders: A Systematic Review. Dermatol. Rep. 2022, 14, 9188. [Google Scholar] [CrossRef] [PubMed]
- Polak, K.; Bergler-czop, B.; Szczepanek, M.; Wojciechowska, K.; Frątczak, A.; Kiss, N. Psoriasis and Gut Microbiome—Current State of Art. Int. J. Mol. Sci. 2021, 22, 4529. [Google Scholar] [CrossRef]
- Thye, A.Y.-K.; Bah, Y.-R.; Law, J.W.-F.; Tan, L.T.-H.; He, Y.-W.; Wong, S.-H.; Thurairajasingam, S.; Chan, K.-G.; Lee, L.-H.; Letchumanan, V. Gut-Skin Axis: Unravelling the Connection between the Gut Microbiome and Psoriasis. Biomedicines 2022, 10, 1037. [Google Scholar] [CrossRef] [PubMed]
- Zeng, M.Y.; Inohara, N.; Nuñez, G. Mechanisms of Inflammation-Driven Bacterial Dysbiosis in the Gut. Mucosal Immunol. 2016, 10, 18–26. [Google Scholar] [CrossRef] [PubMed]
- Navarro-lópez, V.; Núñez-delegido, E.; Ruzafa-costas, B.; Sánchez-pellicer, P.; Agüera-santos, J.; Navarro-moratalla, L. Probiotics in the Therapeutic Arsenal of Dermatologists. Microorganisms 2021, 9, 1513. [Google Scholar] [CrossRef]
- Notay, M.; Foolad, N.; Vaughn, A.R.; Sivamani, R.K. Probiotics, Prebiotics, and Synbiotics for the Treatment and Prevention of Adult Dermatological Diseases. Am. J. Clin. Dermatol. 2017, 18, 721–732. [Google Scholar] [CrossRef]
- Atabati, H.; Esmaeili, S.A.; Saburi, E.; Akhlaghi, M.; Raoofi, A.; Rezaei, N.; Momtazi-Borojeni, A.A. Probiotics with Ameliorating Effects on the Severity of Skin Inflammation in Psoriasis: Evidence from Experimental and Clinical Studies. J. Cell. Physiol. 2020, 235, 8925–8937. [Google Scholar] [CrossRef] [PubMed]
- Kaur, M.; Conde, J.; Willard, J.D.; Taylor, S.; Camacho, F.; Fleischer, A.B., Jr.; Feldman, S.R. A Randomized, Double-Blind Clinical Trial of a Probiotic Nutritional Intervention in the Treatment of Mild to Moderate Non-Scalp Psoriasis. Psoriasis Forum 2018, 13, 12–15. [Google Scholar] [CrossRef]
- Groeger, D.; O’Mahony, L.; Murphy, E.F.; Bourke, J.F.; Dinan, T.G.; Kiely, B.; Shanahan, F.; Quigley, E.M.M. Bifidobacterium Infantis 35624 Modulates Host Inflammatory Processes beyond the Gut. Gut Microbes 2013, 4, 325–339. [Google Scholar] [CrossRef] [PubMed]
- Navarro-López, V.; Martínez-Andrés, A.; Ramírez-Boscà, A.; Ruzafa-Costas, B.; Núñez-Delegido, E.; Carrión-Gutiérrez, M.A.; Prieto-Merino, D.; Codoñer-Cortés, F.; Ramón-Vidal, D.; Genovés-Martínez, S.; et al. Efficacy and Safety of Oral Administration of a Mixture of Probiotic Strains in Patients with Psoriasis: A Randomized Controlled Clinical Trial. Acta Derm. Venereol. 2019, 99, 1078–1084. [Google Scholar] [CrossRef]
- Zeng, L.; Yu, G.; Wu, Y.; Hao, W.; Zeng, L. The Effectiveness and Safety of Probiotic Supplements for Psoriasis: A Systematic Review and Meta-Analysis of Randomized Controlled Trials and Preclinical Trials. J. Immunol. Res. 2021, 2021, 7552546. [Google Scholar] [CrossRef]
- Svendsen, M.T.; Andersen, F.; Andersen, K.H.; Pottegård, A.; Johannessen, H.; Möller, S.; August, B.; Feldman, S.R.; Andersen, K.E. A Smartphone Application Supporting Patients with Psoriasis Improves Adherence to Topical Treatment: A Randomized Controlled Trial. Br. J. Dermatol. 2018, 179, 1062–1071. [Google Scholar] [CrossRef]
- Iskandar, I.Y.K.; Ashcroft, D.M.; Warren, R.B.; Yiu, Z.Z.N.; McElhone, K.; Lunt, M.; Barker, J.N.W.N.; Burden, A.D.; Ormerod, A.D.; Reynolds, N.J.; et al. Demographics and Disease Characteristics of Patients with Psoriasis Enrolled in the British Association of Dermatologists Biologic Interventions Register. Br. J. Dermatol. 2015, 173, 510–518. [Google Scholar] [CrossRef]
- Budu-Aggrey, A.; Brumpton, B.; Tyrrell, J.; Watkins, S.; Modalsli, E.H.; Celis-Morales, C.; Ferguson, L.D.; Vie, G.Å.; Palmer, T.; Fritsche, L.G.; et al. Evidence of a Causal Relationship between Body Mass Index and Psoriasis: A Mendelian Randomization Study. PLoS Med. 2019, 16, e1002739. [Google Scholar] [CrossRef] [PubMed]
- Norden, A.; Rekhtman, S.; Strunk, A.; Garg, A. Risk of Psoriasis According to Body Mass Index: A Retrospective Cohort Analysis. J. Am. Acad. Dermatol. 2022, 86, 1020–1026. [Google Scholar] [CrossRef] [PubMed]
- Gupta, V.K.; Kim, M.; Bakshi, U.; Cunningham, K.Y.; Davis, J.M.; Lazaridis, K.N.; Nelson, H.; Chia, N.; Sung, J. A Predictive Index for Health Status Using Species-Level Gut Microbiome Profiling. Nat. Commun. 2020, 11, 4635. [Google Scholar] [CrossRef]
- Marcos-Zambrano, L.J.; Karaduzovic-Hadziabdic, K.; Loncar Turukalo, T.; Przymus, P.; Trajkovik, V.; Aasmets, O.; Berland, M.; Gruca, A.; Hasic, J.; Hron, K.; et al. Applications of Machine Learning in Human Microbiome Studies: A Review on Feature Selection, Biomarker Identification, Disease Prediction and Treatment. Front. Microbiol. 2021, 12, 313. [Google Scholar] [CrossRef] [PubMed]
- Miyauchi, E.; Shimokawa, C.; Steimle, A.; Desai, M.S.; Ohno, H. The Impact of the Gut Microbiome on Extra-Intestinal Autoimmune Diseases. Nat. Rev. Immunol. 2022, 23, 9–23. [Google Scholar] [CrossRef] [PubMed]
- Singh, R.K.; Chang, H.W.; Yan, D.; Lee, K.M.; Ucmak, D.; Wong, K.; Abrouk, M.; Farahnik, B.; Nakamura, M.; Zhu, T.H.; et al. Influence of Diet on the Gut Microbiome and Implications for Human Health. J. Transl. Med. 2017, 15, 73. [Google Scholar] [CrossRef]
- Wu, G.D.; Chen, J.; Hoffmann, C.; Bittinger, K.; Chen, Y.Y.; Keilbaugh, S.A.; Bewtra, M.; Knights, D.; Walters, W.A.; Knight, R.; et al. Linking Long-Term Dietary Patterns with Gut Microbial Enterotypes. Science 2011, 334, 105. [Google Scholar] [CrossRef]
- David, L.A.; Materna, A.C.; Friedman, J.; Campos-Baptista, M.I.; Blackburn, M.C.; Perrotta, A.; Erdman, S.E.; Alm, E.J. Host Lifestyle Affects Human Microbiota on Daily Timescales. Genome Biol. 2014, 15, R89. [Google Scholar] [CrossRef]
- Rothschild, D.; Weissbrod, O.; Barkan, E.; Kurilshikov, A.; Korem, T.; Zeevi, D.; Costea, P.I.; Godneva, A.; Kalka, I.N.; Bar, N.; et al. Environment Dominates over Host Genetics in Shaping Human Gut Microbiota. Nature 2018, 555, 210–215. [Google Scholar] [CrossRef]
- Liu, X.; Mao, B.; Gu, J.; Wu, J.; Cui, S.; Wang, G.; Zhao, J.; Zhang, H.; Chen, W. Blautia—A New Functional Genus with Potential Probiotic Properties? Gut Microbes 2021, 13, 1–21. [Google Scholar] [CrossRef] [PubMed]
- Sorbara, M.T.; Littmann, E.R.; Fontana, E.; Moody, T.U.; Kohout, C.E.; Gjonbalaj, M.; Eaton, V.; Seok, R.; Leiner, I.M.; Pamer, E.G. Functional and Genomic Variation between Human-Derived Isolates of Lachnospiraceae Reveals Inter- and Intra-Species Diversity. Cell Host Microbe 2020, 28, 134–146.e4. [Google Scholar] [CrossRef]
- Dei-Cas, I.; Giliberto, F.; Luce, L.; Dopazo, H.; Penas-Steinhardt, A. Metagenomic Analysis of Gut Microbiota in Non-Treated Plaque Psoriasis Patients Stratified by Disease Severity: Development of a New Psoriasis-Microbiome Index. Sci. Rep. 2020, 10, 12754. [Google Scholar] [CrossRef]
- Jenq, R.R.; Taur, Y.; Devlin, S.M.; Ponce, D.M.; Goldberg, J.D.; Ahr, K.F.; Littmann, E.R.; Ling, L.; Gobourne, A.C.; Miller, L.C.; et al. Intestinal Blautia Is Associated with Reduced Death from Graft-versus-Host Disease. Biol. Blood Marrow Transplant. 2015, 21, 1373–1383. [Google Scholar] [CrossRef]
- Bajaj, J.S.; Hylemon, P.B.; Ridlon, J.M.; Heuman, D.M.; Daita, K.; White, M.B.; Monteith, P.; Noble, N.A.; Sikaroodi, M.; Gillevet, P.M. Colonic Mucosal Microbiome Differs from Stool Microbiome in Cirrhosis and Hepatic Encephalopathy and Is Linked to Cognition and Inflammation. Am. J. Physiol. Gastrointest. Liver Physiol. 2012, 303, G675–G685. [Google Scholar] [CrossRef] [PubMed]
- Chen, W.; Liu, F.; Ling, Z.; Tong, X.; Xiang, C. Human Intestinal Lumen and Mucosa-Associated Microbiota in Patients with Colorectal Cancer. PLoS ONE 2012, 7, e39743. [Google Scholar] [CrossRef]
- Hao, Y.; Zhou, P.; Zhu, Y.J.; Zou, S.; Zhao, Q.; Yu, J.; Hu, Y.; Li, J. Gut Microbiota Dysbiosis and Altered Bile Acid Catabolism Lead to Metabolic Disorder in Psoriasis Mice. Front. Microbiol. 2022, 13, 853566. [Google Scholar] [CrossRef] [PubMed]
- Stehlikova, Z.; Kostovcikova, K.; Kverka, M.; Rossmann, P.; Dvorak, J.; Novosadova, I.; Kostovcik, M.; Coufal, S.; Srutkova, D.; Prochazkova, P.; et al. Crucial Role of Microbiota in Experimental Psoriasis Revealed by a Gnotobiotic Mouse Model. Front. Microbiol. 2019, 10, 236. [Google Scholar] [CrossRef] [PubMed]
- Shinno-Hashimoto, H.; Hashimoto, Y.; Wei, Y.; Chang, L.; Fujita, Y.; Ishima, T.; Matsue, H.; Hashimoto, K. Abnormal Composition of Microbiota in the Gut and Skin of Imiquimod-Treated Mice. Sci. Rep. 2021, 11, 11265. [Google Scholar] [CrossRef]
- Koh, G.Y.; Kane, A.v.; Wu, X.; Crott, J.W. Parabacteroides Distasonis Attenuates Tumorigenesis, Modulates Inflammatory Markers and Promotes Intestinal Barrier Integrity in Azoxymethane-Treated A/J Mice. Carcinogenesis 2020, 41, 909–917. [Google Scholar] [CrossRef] [PubMed]
- Ezeji, J.C.; Sarikonda, D.K.; Hopperton, A.; Erkkila, H.L.; Cohen, D.E.; Martinez, S.P.; Cominelli, F.; Kuwahara, T.; Dichosa, A.E.K.; Good, C.E.; et al. Parabacteroides Distasonis: Intriguing Aerotolerant Gut Anaerobe with Emerging Antimicrobial Resistance and Pathogenic and Probiotic Roles in Human Health. Gut Microbes 2021, 13, 1922241. [Google Scholar] [CrossRef]
- Koh, G.Y.; Kane, A.V.; Lee, K.; Xu, Q.; Wu, X.; Mason, J.B.; Crott, J.W. Parabacteroides Distasonis Attenuates Colonic Inflammation and Prevents Tumor Formation in Azoxymethane-Treated High-Fat Diet-Fed Mice. FASEB J. 2017, 31, 435.2. [Google Scholar] [CrossRef]
- Koh, G.Y.; Kane, A.; Lee, K.; Xu, Q.; Wu, X.; Roper, J.; Mason, J.B.; Crott, J.W. Parabacteroides Distasonis Attenuates Toll-like Receptor 4 Signaling and Akt Activation and Blocks Colon Tumor Formation in High-Fat Diet-Fed Azoxymethane-Treated Mice. Int. J. Cancer 2018, 143, 1797–1805. [Google Scholar] [CrossRef] [PubMed]
- Sun, H.; Guo, Y.; Wang, H.; Yin, A.; Hu, J.; Yuan, T.; Zhou, S.; Xu, W.; Wei, P.; Yin, S.; et al. Gut Commensal Parabacteroides Distasonis Alleviates Inflammatory Arthritis. Gut 2023, gutjnl-2022-327756. [Google Scholar] [CrossRef]
- Vinolo, M.A.R.; Rodrigues, H.G.; Nachbar, R.T.; Curi, R. Regulation of Inflammation by Short Chain Fatty Acids. Nutrients 2011, 3, 858. [Google Scholar] [CrossRef]
- Akhtar, M.; Chen, Y.; Ma, Z.; Zhang, X.; Shi, D.; Khan, J.A.; Liu, H. Gut Microbiota-Derived Short Chain Fatty Acids Are Potential Mediators in Gut Inflammation. Anim. Nutr. 2022, 8, 350–360. [Google Scholar] [CrossRef]
- Liu, P.; Wang, Y.; Yang, G.; Zhang, Q.; Meng, L.; Xin, Y.; Jiang, X. The Role of Short-Chain Fatty Acids in Intestinal Barrier Function, Inflammation, Oxidative Stress, and Colonic Carcinogenesis. Pharmacol. Res. 2021, 165, 105420. [Google Scholar] [CrossRef]
- Park, J.; Wang, Q.; Wu, Q.; Mao-Draayer, Y.; Kim, C.H. Bidirectional Regulatory Potentials of Short-Chain Fatty Acids and Their G-Protein-Coupled Receptors in Autoimmune Neuroinflammation. Sci. Rep. 2019, 9, 8837. [Google Scholar] [CrossRef]
- Van der Hee, B.; Wells, J.M. Microbial Regulation of Host Physiology by Short-Chain Fatty Acids. Trends Microbiol. 2021, 29, 700–712. [Google Scholar] [CrossRef]
- Sikora, M.; Chrabąszcz, M.; Waśkiel-Burnat, A.; Rakowska, A.; Olszewska, M.; Rudnicka, L. Claudin-3–A New Intestinal Integrity Marker in Patients with Psoriasis: Association with Disease Severity. J. Eur. Acad. Dermatol. Venereol. 2019, 33, 1907–1912. [Google Scholar] [CrossRef]
- Humbert, P.; Bidet, A.; Treffel, P.; Drobacheff, C.; Agache, P. Intestinal Permeability in Patients with Psoriasis. J. Dermatol. Sci. 1991, 2, 324–326. [Google Scholar] [CrossRef] [PubMed]
- Sikora, M.; Chrabąszcz, M.; Maciejewski, C.; Zaremba, M.; Waśkiel, A.; Olszewska, M.; Rudnicka, L. Intestinal Barrier Integrity in Patients with Plaque Psoriasis. J. Dermatol. 2018, 45, 1468–1470. [Google Scholar] [CrossRef] [PubMed]
- Sikora, M.; Stec, A.; Chrabaszcz, M.; Giebultowicz, J.; Samborowska, E.; Jazwiec, R.; Dadlez, M.; Olszewska, M.; Rudnicka, L. Clinical Implications of Intestinal Barrier Damage in Psoriasis. J. Inflamm. Res. 2021, 14, 237–243. [Google Scholar] [CrossRef]
- Richetta, A.G.; Grassi, S.; Moliterni, E.; Chello, C.; Calvieri, C.; Carnevale, R.; Peruzzi, M.; Violi, F.; Calvieri, S. Increased Intestinal Barrier Permeability in Patients with Moderate to Severe Plaque-Type Psoriasis. J. Dermatol. 2020, 47, e366–e368. [Google Scholar] [CrossRef]
- Kamleh, M.A.; Snowden, S.G.; Grapov, D.; Blackburn, G.J.; Watson, D.G.; Xu, N.; Ståhle, M.; Wheelock, C.E. LC–MS Metabolomics of Psoriasis Patients Reveals Severity-Dependent Increases in Circulating Amino Acids That Ameliorated by Anti-TNFα Treatment. J. Proteome Res. 2015, 14, 557. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.; Wan, H.; He, Q.; He, S.; Deng, M. Statistical Methods for Microbiome Compositional Data Network Inference: A Survey. J. Comput. Biol. 2022, 29, 704–723. [Google Scholar] [CrossRef]
- Hirano, H.; Takemoto, K. Difficulty in Inferring Microbial Community Structure Based on Co-Occurrence Network Approaches. BMC Bioinform. 2019, 20, 329. [Google Scholar] [CrossRef]
- Matchado, M.S.; Lauber, M.; Reitmeier, S.; Kacprowski, T.; Baumbach, J.; Haller, D.; List, M. Network Analysis Methods for Studying Microbial Communities: A Mini Review. Comput. Struct. Biotechnol. J. 2021, 19, 2687–2698. [Google Scholar] [CrossRef] [PubMed]
- Bullman, S.; Eggermont, A.; Johnston, C.D.; Zitvogel, L. Harnessing the Microbiome to Restore Immunotherapy Response. Nat. Cancer 2021, 2, 1301–1304. [Google Scholar] [CrossRef] [PubMed]
- Barone, M.; Rampelli, S.; Biagi, E.; Bertozzi, S.M.; Falchi, F.; Cavalli, A.; Armirotti, A.; Brigidi, P.; Turroni, S.; Candela, M. Searching for New Microbiome-Targeted Therapeutics through a Drug Repurposing Approach. J. Med. Chem. 2021, 64, 17277–17286. [Google Scholar] [CrossRef]
- Ting, N.L.N.; Lau, H.C.H.; Yu, J. Cancer Pharmacomicrobiomics: Targeting Microbiota to Optimise Cancer Therapy Outcomes. Gut 2022, 71, 1412–1425. [Google Scholar] [CrossRef]
- Liu, L.; Shah, K. The Potential of the Gut Microbiome to Reshape the Cancer Therapy Paradigm: A Review. JAMA Oncol. 2022, 8, 1059–1067. [Google Scholar] [CrossRef]
- Sandler, R.H.; Finegold, S.M.; Bolte, E.R.; Buchanan, C.P.; Maxwell, A.P.; Väisänen, M.L.; Nelson, M.N.; Wexler, H.M. Short-Term Benefit from Oral Vancomycin Treatment of Regressive-Onset Autism. J. Child Neurol. 2000, 15, 429–435. [Google Scholar] [CrossRef]
- Stewart Campbell, A.; Needham, B.D.; Meyer, C.R.; Tan, J.; Conrad, M.; Preston, G.M.; Bolognani, F.; Rao, S.G.; Heussler, H.; Griffith, R.; et al. Safety and Target Engagement of an Oral Small-Molecule Sequestrant in Adolescents with Autism Spectrum Disorder: An Open-Label Phase 1b/2a Trial. Nat. Med. 2022, 28, 528–534. [Google Scholar] [CrossRef]
- Wong, A.C.; Levy, M. New Approaches to Microbiome-Based Therapies. mSystems 2019, 4, e00122-19. [Google Scholar] [CrossRef]
- Strati, F.; Lattanzi, G.; Amoroso, C.; Facciotti, F. Microbiota-Targeted Therapies in Inflammation Resolution. Semin. Immunol. 2022, 59, 101599. [Google Scholar] [CrossRef] [PubMed]
- Caporaso, J.G.; Lauber, C.L.; Walters, W.A.; Berg-Lyons, D.; Huntley, J.; Fierer, N.; Owens, S.M.; Betley, J.; Fraser, L.; Bauer, M.; et al. Ultra-High-Throughput Microbial Community Analysis on the Illumina HiSeq and MiSeq Platforms. ISME J. 2012, 6, 1621–1624. [Google Scholar] [CrossRef]
- Bolyen, E.; Rideout, J.R.; Dillon, M.R.; Bokulich, N.A.; Abnet, C.C.; Al-Ghalith, G.A.; Alexander, H.; Alm, E.J.; Arumugam, M.; Asnicar, F.; et al. Reproducible, Interactive, Scalable and Extensible Microbiome Data Science Using QIIME 2. Nat. Biotechnol. 2019, 37, 852–857. [Google Scholar] [CrossRef] [PubMed]
- Callahan, B.J.; McMurdie, P.J.; Rosen, M.J.; Han, A.W.; Johnson, A.J.A.; Holmes, S.P. DADA2: High-Resolution Sample Inference from Illumina Amplicon Data. Nat. Methods 2016, 13, 581–583. [Google Scholar] [CrossRef] [PubMed]
- Callahan, B.J.; McMurdie, P.J.; Holmes, S.P. Exact Sequence Variants Should Replace Operational Taxonomic Units in Marker-Gene Data Analysis. ISME J. 2017, 11, 2639–2643. [Google Scholar] [CrossRef]
- Katoh, K.; Misawa, K.; Kuma, K.; Miyata, T. MAFFT: A Novel Method for Rapid Multiple Sequence Alignment Based on Fast Fourier Transform. Nucleic Acids Res. 2002, 30, 3059–3066. [Google Scholar] [CrossRef] [PubMed]
- Price, M.N.; Dehal, P.S.; Arkin, A.P. FastTree 2–Approximately Maximum-Likelihood Trees for Large Alignments. PLoS ONE 2010, 5, e9490. [Google Scholar] [CrossRef] [PubMed]
- Bokulich, N.A.; Kaehler, B.D.; Rideout, J.R.; Dillon, M.; Bolyen, E.; Knight, R.; Huttley, G.A.; Gregory Caporaso, J. Optimizing Taxonomic Classification of Marker-Gene Amplicon Sequences with QIIME 2’s Q2-Feature-Classifier Plugin. Microbiome 2018, 6, 90. [Google Scholar] [CrossRef] [PubMed]
- Quast, C.; Pruesse, E.; Yilmaz, P.; Gerken, J.; Schweer, T.; Yarza, P.; Peplies, J.; Glöckner, F.O. The SILVA Ribosomal RNA Gene Database Project: Improved Data Processing and Web-Based Tools. Nucleic Acids Res. 2012, 41, D590–D596. [Google Scholar] [CrossRef]
- Yilmaz, P.; Parfrey, L.W.; Yarza, P.; Gerken, J.; Pruesse, E.; Quast, C.; Schweer, T.; Peplies, J.; Ludwig, W.; Glöckner, F.O. The SILVA and “All-Species Living Tree Project (LTP)” Taxonomic Frameworks. Nucleic Acids Res. 2013, 42, D643–D648. [Google Scholar] [CrossRef] [PubMed]
- Anderson, M.J. Permutational Multivariate Analysis of Variance (PERMANOVA). In Wiley StatsRef: Statistics Reference Online; John and Wiley and Sons: Hoboken, NJ, USA, 2017; pp. 1–15. ISBN 9781118445112. [Google Scholar]
- Kurtz, Z.D.; Müller, C.L.; Miraldi, E.R.; Littman, D.R.; Blaser, M.J.; Bonneau, R.A. Sparse and Compositionally Robust Inference of Microbial Ecological Networks. PLoS Comput. Biol. 2015, 11, e1004226. [Google Scholar] [CrossRef] [PubMed]







| Psoriasis (n = 58 ^) | Control (n = 49) | p Value | ||
|---|---|---|---|---|
| Sex, No. (%) | 0.0122 | |||
| Male | 38 (65.5) | 20 (40.8%) | ||
| Female | 20 (34.5) | 29 (59.2%) | ||
| Age, mean (SD) (range), y | 44.4 (12.1) (18–65) | 45.5 (13.2) (21–64) | 0.4624 | |
| Weight, mean (SD), kg | 71.8 (16.3) | 63.1 (13.4) | 0.0026 | |
| BMI, mean (SD) † | 25.8 (4.8) | 23.4 (4.0) | 0.0049 | |
| BSA, mean (SD) | 13.1 (16.2) | N/A | ||
| PASI, mean (SD) | 7.6 (7.1) | N/A | ||
| DLQI, mean (SD) | 7.6 (7.1) | N/A | ||
| Psoriatic arthritis, No. (%) | 20 (34.5) | N/A | ||
| Feature ID | Taxon | W |
|---|---|---|
| 01f2a9a0f64a0c8e9f380e4759f68e42 | f_Bacteroidaceae;g_Bacteroides;s_Bacteroides_vulgatus | 4972 |
| ccc00ff50c15bc9d115d53589fd1db6f | f_Lachnospiraceae;g_Fusicatenibacter;s_uncultured_organism | 4945 |
| 725ae09bd27ee835f1e2afaee85aafe0 | f_Lachnospiraceae;g_Blautia;s_Blautia_wexlerae | 4940 |
| 16feabdbf1087e04e1eef52210ed93ad | f_Erysipelatoclostridiaceae;g_Erysipelotrichaceae_UCG-003;s_uncultured_bacterium | 4918 |
| 7523ad91056301e66e3a8f299c0878fc | f_Ruminococcaceae;g_Faecalibacterium | 4905 |
| f678835c929c59443b1e5e736048e209 | f_Ruminococcaceae;g_Faecalibacterium;s_human_gut | 4903 |
| fd947e8f706a04316b167e2935696c96 | f_Lachnospiraceae;g_Dorea | 4896 |
| f86c8767307c0f2ab691b9892c1e99c6 | f_Lachnospiraceae;g_Agathobacter;s_[Eubacterium]_rectale | 4890 |
| d37e5aa22497be27a11bce9eda0c8cf7 | f_Lachnospiraceae;g_Blautia;s_Blautia_wexlerae | 4875 |
| da5fdabeacf9b6cafc8d2dcbe81db05d | f_Bacteroidaceae;g_Bacteroides;s_Bacteroides_thetaiotaomicron | 4850 |
| 18a8fe3a9227e73e6f15d50980cb5c75 | f_Lachnospiraceae;g_Anaerostipes;s_human_gut | 4826 |
| 428f4904e0dfb8dfb40952a57526dbb2 | f_Lachnospiraceae;g_Lachnospiraceae_ND3007_group;s_metagenome | 4817 |
| f4465d8e3fddd8dd39702387f21ffb4e | f_Bacteroidaceae;g_Bacteroides;s_Bacteroides_vulgatus | 4791 |
| f444b6959a6760ca409a51e8107f729f | f_Ruminococcaceae;g_Subdoligranulum | 4772 |
| 3866fc184b629f22b6b29f848dd90694 | f_Lachnospiraceae;g_Blautia | 4765 |
| bf5fc5be757c244017374074589e41d7 | f_Monoglobaceae;g_Monoglobus;s_uncultured_organism | 4745 |
| 7c73dc21566ae02ef3e98859d879468c | f_Lachnospiraceae;g_[Ruminococcus]_torques_group;s_uncultured_Firmicutes | 4728 |
| a0836bd4a80786ce759ffb26d563eb54 | f_Lachnospiraceae;g_Blautia;s_uncultured_Blautia | 4686 |
| 928a2600b168d35b17d8c7578352c596 | f_Streptococcaceae;g_Streptococcus;s_Streptococcus_salivarius | 4647 |
| 9755d76a1a899b96090b30fe9582c058 | f_Butyricicoccaceae;g_Butyricicoccus | 4634 |
| 60d6ecdf998b630c36e1277da586e204 | f_Ruminococcaceae;g_Faecalibacterium | 4621 |
| 434c8851e06baea123e89217030e6e23 | f_Bacteroidaceae;g_Bacteroides;s_Bacteroides_uniformis | 4620 |
| 93198e1fc1b71b9e7d3abf522a926278 | f_Lachnospiraceae;g_Blautia;s_Blautia_faecis | 4588 |
| 65b9f8997ba78cf0dae03e7146efc23b | f_Lachnospiraceae;g_Blautia;s_Blautia_wexlerae | 4543 |
| 4655a784c774d4acb1bb902a65ed9612 | f_Lachnospiraceae;g_Lachnoclostridium | 4483 |
| Mild & (n = 20) | Moderate to Severe & (n = 32) | p Value | ||
|---|---|---|---|---|
| Sex, No. (%) | 0.2237 | |||
| Male | 11 (55.0) | 24 (75.0) | ||
| Female | 9 (45.0) | 8 (25.0) | ||
| Age, mean (SD), y | 44.1 (13.5) | 44.2 (10.8) | 0.9795 | |
| Weight, mean (SD), kg | 68.9 (16.6) | 75.3 (15.8) | 0.1722 | |
| BMI, mean (SD) † | 25.4 (5.8) | 26.5 (4.4) | 0.4200 | |
| Psoriatic arthritis, No. (%) | 6 (30.0) | 10 (31.3) | 1.0000 | |
| ∆BSA, mean (SD) | −2.1 (2.1) | −5.8 (12.1) | 0.1925 | |
| ∆%PASI, mean (SD) | −21.7 (15.9) | −26.5 (31.4) | 0.7692 | |
| ∆DLQI, mean (SD) | −2.6 (6.5) | −2.1 (6.2) | 0.5443 | |
| Responder, No. (%) ^ | 8 (40.0) | 14 (43.8) | 1.0000 | |
| Feature_ID | Taxon | W |
|---|---|---|
| 721a741bef4a5db0211de1a5b84a8b5b | f_Tannerellaceae;g_Parabacteroides;s_Parabacteroides_distasonis | 4139 |
| 72af122f180225b2b0c90e487b25af6c | f_Lactobacillaceae;g_Lactobacillus;s_Lactobacillus_plantarum | 4139 |
| 8cec479da287209d3c7d464f14242794 | f_Ruminococcaceae;g_CAG-352;s_uncultured_bacterium | 4139 |
| bd4606ad663e209e745c8c51b4deeee8 | f_Bacteroidaceae;g_Bacteroides;s_Bacteroides_vulgatus | 4142 |
| f678835c929c59443b1e5e736048e209 | f_Ruminococcaceae;g_Faecalibacterium;s_human_gut | 4141 |
| BioCyc ID | MetaCyc Pathway Name | Group | Log LDA | p |
|---|---|---|---|---|
| PWY_5989 | stearate biosynthesis II (bacteria and plants) | control | 2.7092 | 0.0128 |
| PWYG_321 | mycolate biosynthesis | control | 2.6975 | 0.0113 |
| PWY_7664 | oleate biosynthesis IV (anaerobic) | control | 2.6970 | 0.0140 |
| PWY_6282 | palmitoleate biosynthesis I (from (5Z)-dodec-5-enoate) | control | 2.6923 | 0.0138 |
| PWY0_862 | (5Z)-dodecenoate biosynthesis I | control | 2.6826 | 0.0142 |
| PWY_6519 | 8-amino-7-oxononanoate biosynthesis I | control | 2.6193 | 0.0037 |
| FASYN_ INITIAL_PWY | super pathway of fatty acid biosynthesis initiation | control | 2.5984 | 0.0140 |
| P42_PWY | incomplete reductive TCA cycle | control | 2.5914 | 0.0024 |
| BIOTIN_ BIOSYNTHESIS_PWY | biotin biosynthesis I | control | 2.5560 | 0.0096 |
| PWY_6703 | preQ0 biosynthesis | control | 2.3734 | 0.0252 |
| THISYN_PWY | super pathway of thiamine diphosphate biosynthesis I | control | 2.3712 | 0.0059 |
| HISDEG_PWY | l-histidine degradation I | control | 2.3484 | 0.0070 |
| PWY_7323 | super pathway of GDP-mannose-derived o-antigen building blocks biosynthesis | control | 2.3441 | 0.0074 |
| PWY_5695 | inosine 5′-phosphate degradation | control | 2.3388 | 0.0282 |
| NAGLIPASYN_PWY | lipid IVA biosynthesis (E. coli) | control | 2.3144 | 0.0244 |
| GALACTUROCAT_PWY | d-galacturonate degradation I | control | 2.3052 | 0.0175 |
| POLYISOPRENSYN_PWY | polyisoprenoid biosynthesis (E. coli) | control | 2.2352 | 0.0305 |
| PWY_6467 | Kdo transfer to lipid IVA (Chlamydia) | control | 2.1935 | 0.0228 |
| PWY_7328 | super pathway of UDP-glucose-derived o-antigen building blocks biosynthesis | control | 2.1563 | 0.0184 |
| PWY_4984 | urea cycle | control | 2.1505 | 0.0204 |
| COLANSYN_PWY | colanic acid building blocks biosynthesis | control | 2.1109 | 0.0166 |
| ASPASN_PWY | super pathway of l-aspartate and l-asparagine biosynthesis | control | 2.0583 | 0.0305 |
| P161_PWY | acetylene degradation (anaerobic) | psoriasis | 2.5697 | 0.0074 |
| PWY0_1586 | peptidoglycan maturation (meso-diaminopimelate containing) | psoriasis | 2.5201 | 0.0300 |
| PWY_6471 | peptidoglycan biosynthesis IV (Enterococcus faecium) | psoriasis | 2.4510 | 0.0184 |
| PWY_5505 | l-glutamate and l-glutamine biosynthesis | psoriasis | 2.3867 | 0.0330 |
| NONOXIPENT_PWY | pentose phosphate pathway (non-oxidative branch) I | psoriasis | 2.3355 | 0.0454 |
| PWY_7111 | pyruvate fermentation to isobutanol (engineered) | psoriasis | 2.3156 | 0.0296 |
| PWY0_1297 | super pathway of purine deoxyribonucleosides degradation | psoriasis | 2.3149 | 0.0126 |
| PWY_6151 | S-adenosyl-l-methionine salvage I | psoriasis | 2.3130 | 0.0210 |
| PWY_621 | sucrose degradation III (sucrose invertase) | psoriasis | 2.3016 | 0.0207 |
| PWY_5686 | UMP biosynthesis I | psoriasis | 2.2906 | 0.0296 |
| PWY_6317 | d-galactose degradation I (Leloir pathway) | psoriasis | 2.2897 | 0.0128 |
| DAPLYSINESYN_PWY | l-lysine biosynthesis I | psoriasis | 2.2642 | 0.0320 |
| PWY0_1298 | super pathway of pyrimidine deoxyribonucleosides degradation | psoriasis | 2.2569 | 0.0138 |
| ARGSYNBSUB_PWY | l-arginine biosynthesis II (acetyl cycle) | psoriasis | 2.2303 | 0.0286 |
| ARGSYN_PWY | l-arginine biosynthesis I (via L-ornithine) | psoriasis | 2.2162 | 0.0277 |
| PWY_7400 | l-arginine biosynthesis IV (archaea) | psoriasis | 2.2090 | 0.0277 |
| COMPLETE_ARO_PWY | super pathway of aromatic amino acid biosynthesis | psoriasis | 2.1875 | 0.0391 |
| ARO_PWY | chorismate biosynthesis I | psoriasis | 2.1698 | 0.0482 |
| SER_GLYSYN_PWY | super pathway of l-serine and glycine biosynthesis I | psoriasis | 2.1284 | 0.0273 |
| PWY_6163 | chorismate biosynthesis from 3-dehydroquinate | psoriasis | 2.1129 | 0.0403 |
| PWY_6122 | 5-aminoimidazole ribonucleotide biosynthesis II | psoriasis | 2.0896 | 0.0373 |
| PWY_6277 | super pathway of 5-aminoimidazole ribonucleotide biosynthesis | psoriasis | 2.0896 | 0.0373 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Choy, C.T.; Chan, U.K.; Siu, P.L.K.; Zhou, J.; Wong, C.H.; Lee, Y.W.; Chan, H.W.; Tsui, J.C.C.; Loo, S.K.F.; Tsui, S.K.W. A Novel E3 Probiotics Formula Restored Gut Dysbiosis and Remodelled Gut Microbial Network and Microbiome Dysbiosis Index (MDI) in Southern Chinese Adult Psoriasis Patients. Int. J. Mol. Sci. 2023, 24, 6571. https://doi.org/10.3390/ijms24076571
Choy CT, Chan UK, Siu PLK, Zhou J, Wong CH, Lee YW, Chan HW, Tsui JCC, Loo SKF, Tsui SKW. A Novel E3 Probiotics Formula Restored Gut Dysbiosis and Remodelled Gut Microbial Network and Microbiome Dysbiosis Index (MDI) in Southern Chinese Adult Psoriasis Patients. International Journal of Molecular Sciences. 2023; 24(7):6571. https://doi.org/10.3390/ijms24076571
Chicago/Turabian StyleChoy, Chi Tung, Un Kei Chan, Pui Ling Kella Siu, Junwei Zhou, Chi Ho Wong, Yuk Wai Lee, Ho Wang Chan, Joseph Chi Ching Tsui, Steven King Fan Loo, and Stephen Kwok Wing Tsui. 2023. "A Novel E3 Probiotics Formula Restored Gut Dysbiosis and Remodelled Gut Microbial Network and Microbiome Dysbiosis Index (MDI) in Southern Chinese Adult Psoriasis Patients" International Journal of Molecular Sciences 24, no. 7: 6571. https://doi.org/10.3390/ijms24076571
APA StyleChoy, C. T., Chan, U. K., Siu, P. L. K., Zhou, J., Wong, C. H., Lee, Y. W., Chan, H. W., Tsui, J. C. C., Loo, S. K. F., & Tsui, S. K. W. (2023). A Novel E3 Probiotics Formula Restored Gut Dysbiosis and Remodelled Gut Microbial Network and Microbiome Dysbiosis Index (MDI) in Southern Chinese Adult Psoriasis Patients. International Journal of Molecular Sciences, 24(7), 6571. https://doi.org/10.3390/ijms24076571






