Prolonged Piezo1 Activation Induces Cardiac Arrhythmia
Abstract
1. Introduction
2. Results
2.1. piezo1 is Expressed in Zebrafish Cardiomyocytes
2.2. Yoda1 Increases Zebrafish Piezo1 Activation Kinetics
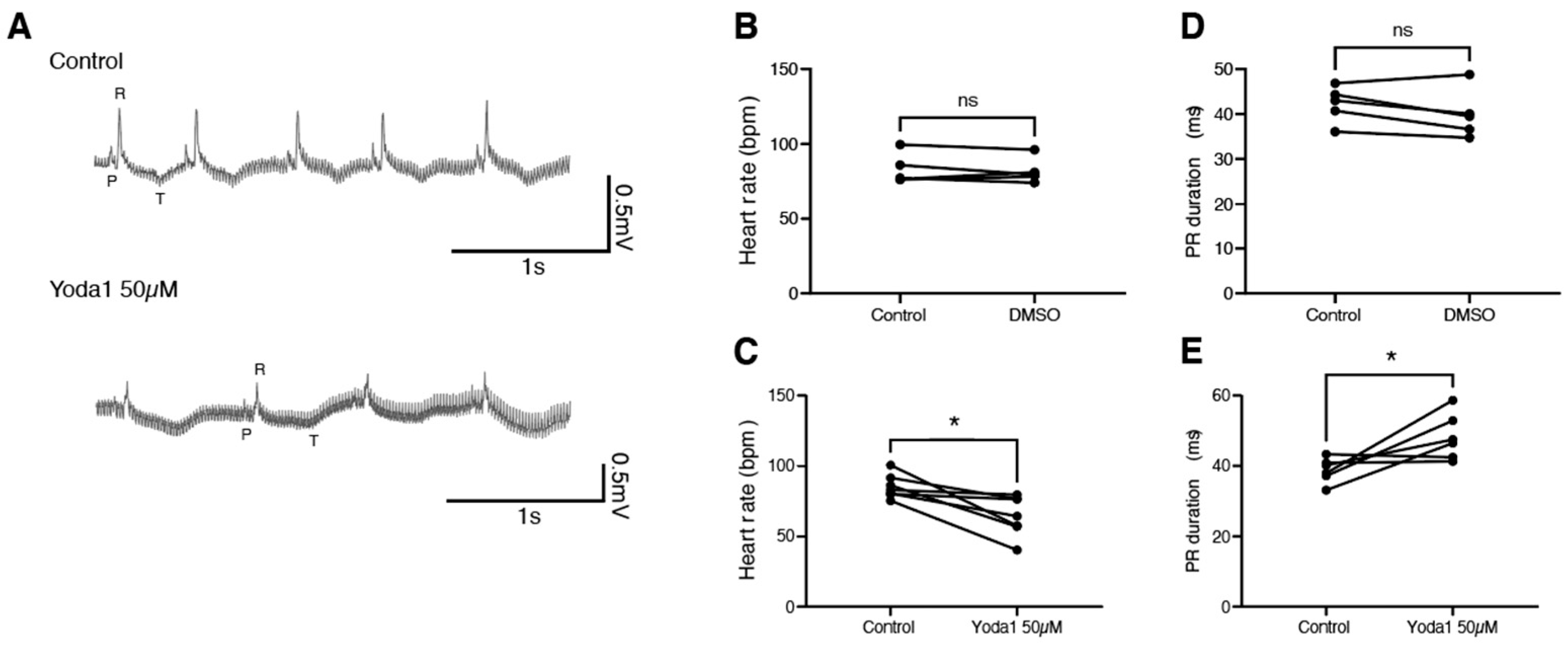
2.3. Prolonged Piezo1 Activation In Vivo Causes Cardiac Arrhythmia
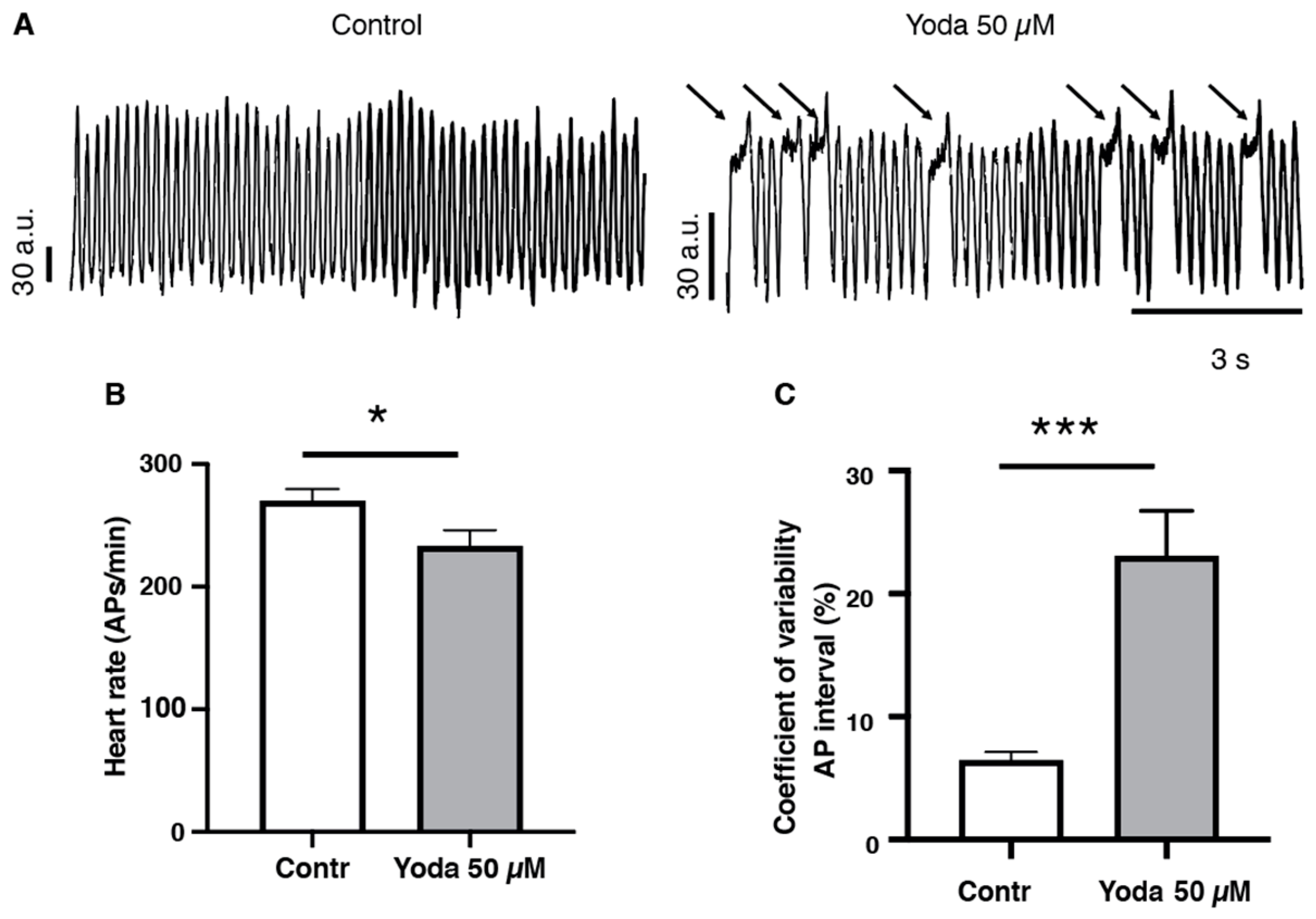
2.4. Prolonged Piezo1 Activation In Vivo Affects the Dynamics of the Cardiac Action Potential
3. Discussion
4. Materials and Methods
4.1. Zebrafish Husbandry
4.2. Cell Culture and Transfection
4.3. Patch Clamp Recordings
4.4. Single-Nuclei RNA Sequencing
4.5. Cardiac Performance Analysis
4.6. Optical Mapping
4.7. ECG Measurement
4.8. Real-Time Quantitative PCR (RT-qPCR)
4.9. Data Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Desai, D.S.; Hajouli, S. Arrhythmias. In StatPearls; StatPearls Publishing: Treasure Island, FL, USA, 2022. [Google Scholar]
- Peyronnet, R.; Nerbonne, J.M.; Kohl, P. Cardiac Mechano-Gated Ion Channels and Arrhythmias. Circ. Res. 2016, 118, 311–329. [Google Scholar] [CrossRef]
- Turner, D.; Kang, C.; Mesirca, P.; Hong, J.; Mangoni, M.E.; Glukhov, A.V.; Sah, R. Electrophysiological and Molecular Mechanisms of Sinoatrial Node Mechanosensitivity. Front. Cardiovasc. Med. 2021, 8, 662410. [Google Scholar] [CrossRef] [PubMed]
- Bode, F.; Sachs, F.; Franz, M.R. Tarantula peptide inhibits atrial fibrillation. Nature 2001, 409, 35–36. [Google Scholar] [CrossRef] [PubMed]
- Fang, X.-Z.; Zhou, T.; Xu, J.-Q.; Wang, Y.-X.; Sun, M.-M.; He, Y.-J.; Pan, S.-W.; Xiong, W.; Peng, Z.-K.; Gao, X.-H.; et al. Structure, kinetic properties and biological function of mechanosensitive Piezo channels. Cell Biosci. 2021, 11, 13. [Google Scholar] [CrossRef] [PubMed]
- Jiang, F.; Yin, K.; Wu, K.; Zhang, M.; Wang, S.; Cheng, H.; Zhou, Z.; Xiao, B. The mechanosensitive Piezo1 channel mediates heart mechano-chemo transduction. Nat. Commun. 2021, 12, 869. [Google Scholar] [CrossRef] [PubMed]
- Zarychanski, R.; Schulz, V.P.; Houston, B.L.; Maksimova, Y.; Houston, D.S.; Smith, B.; Rinehart, J.; Gallagher, P.G. Mutations in the mechanotransduction protein PIEZO1 are associated with hereditary xerocytosis. Blood 2012, 120, 1908–1915. [Google Scholar] [CrossRef]
- Bartoli, F.; Evans, E.L.; Blythe, N.M.; Stewart, L.; Chuntharpursat-Bon, E.; Debant, M.; Musialowski, K.E.; Lichtenstein, L.; Parsonage, G.; Futers, T.S.; et al. Global PIEZO1 Gain-of-Function Mutation Causes Cardiac Hypertrophy and Fibrosis in Mice. Cells 2022, 11, 1199. [Google Scholar] [CrossRef]
- Blackwell, D.J.; Schmeckpeper, J.; Knollmann, B.C. Animal Models to Study Cardiac Arrhythmias. Circ. Res. 2022, 130, 1926–1964. [Google Scholar] [CrossRef]
- Fu, F.; Pietropaolo, M.; Cui, L.; Pandit, S.; Li, W.; Tarnavski, O.; Shetty, S.S.; Liu, J.; Lussier, J.M.; Murakami, Y.; et al. Lack of authentic atrial fibrillation in commonly used murine atrial fibrillation models. PLoS ONE 2022, 17, e0256512. [Google Scholar] [CrossRef]
- Narumanchi, S.; Wang, H.; Perttunen, S.; Tikkanen, I.; Lakkisto, P.; Paavola, J. Zebrafish Heart Failure Models. Front. Cell Dev. Biol. 2021, 9, 662583. [Google Scholar] [CrossRef]
- Echeazarra, L.; Hortigón-Vinagre, M.P.; Casis, O.; Gallego, M. Adult and Developing Zebrafish as Suitable Models for Cardiac Electrophysiology and Pathology in Research and Industry. Front. Physiol. 2021, 11, 607860. [Google Scholar] [CrossRef]
- Rolland, L.; Harrington, A.; Faucherre, A.; Abaroa, J.M.; Gangatharan, G.; Gamba, L.; Severac, D.; Pratlong, M.; Moore-Morris, T.; Jopling, C. The regenerative response of cardiac interstitial cells. J. Mol. Cell Biol. 2022, mjac059. [Google Scholar] [CrossRef]
- Faucherre, A.; Kissa, K.; Nargeot, J.; Mangoni, M.E.; Jopling, C. Piezo1 plays a role in erythrocyte volume homeostasis. Haematologica 2014, 99, 70–75. [Google Scholar] [CrossRef]
- Faucherre, A.; Moha ou Maati, H.; Nasr, N.; Pinard, A.; Theron, A.; Odelin, G.; Desvignes, J.-P.; Salgado, D.; Collod-Béroud, G.; Avierinos, J.-F.; et al. Piezo1 is required for outflow tract and aortic valve development. J. Mol. Cell Cardiol. 2020, 143, 51–62. [Google Scholar] [CrossRef]
- Syeda, R.; Xu, J.; Dubin, A.E.; Coste, B.; Mathur, J.; Huynh, T.; Matzen, J.; Lao, J.; Tully, D.C.; Engels, I.H.; et al. Chemical activation of the mechanotransduction channel Piezo1. eLife 2015, 4, e07369. [Google Scholar] [CrossRef]
- Wang, Y.; Chi, S.; Guo, H.; Li, G.; Wang, L.; Zhao, Q.; Rao, Y.; Zu, L.; He, W.; Xiao, B. A lever-like transduction pathway for long-distance chemical- and mechano-gating of the mechanosensitive Piezo1 channel. Nat. Commun. 2018, 9, 1300. [Google Scholar] [CrossRef]
- Coste, B.; Mathur, J.; Schmidt, M.; Earley, T.J.; Ranade, S.; Petrus, M.J.; Dubin, A.E.; Patapoutian, A. Piezo1 and Piezo2 are essential components of distinct mechanically activated cation channels. Science 2010, 330, 55–60. [Google Scholar] [CrossRef]
- Arel, E.; Rolland, L.; Thireau, J.; Torrente, A.G.; Bechard, E.; Bride, J.; Jopling, C.; Demion, M.; Le Guennec, J.-Y. The Effect of Hypothermia and Osmotic Shock on the Electrocardiogram of Adult Zebrafish. Biology 2022, 11, 603. [Google Scholar] [CrossRef]
- Torrente, A.G.; Zhang, R.; Zaini, A.; Giani, J.F.; Kang, J.; Lamp, S.T.; Philipson, K.D.; Goldhaber, J.I. Burst pacemaker activity of the sinoatrial node in sodium–calcium exchanger knockout mice. Proc. Natl. Acad. Sci. USA 2015, 112, 9769–9774. [Google Scholar] [CrossRef]
- Baudot, M.; Torre, E.; Bidaud, I.; Louradour, J.; Torrente, A.G.; Fossier, L.; Talssi, L.; Nargeot, J.; Barrère-Lemaire, S.; Mesirca, P.; et al. Concomitant genetic ablation of L-type Cav1.3 (α1D) and T-type Cav3.1 (α1G) Ca2+ channels disrupts heart automaticity. Sci. Rep. 2020, 10, 18906. [Google Scholar] [CrossRef]
- Liang, J.; Huang, B.; Yuan, G.; Chen, Y.; Liang, F.; Zeng, H.; Zheng, S.; Cao, L.; Geng, D.; Zhou, S. Stretch-activated channel Piezo1 is up-regulated in failure heart and cardiomyocyte stimulated by AngII. Am. J. Transl. Res. 2017, 9, 2945–2955. [Google Scholar] [PubMed]
- Yamaguchi, Y.; Allegrini, B.; Rapetti-Mauss, R.; Picard, V.; Garçon, L.; Kohl, P.; Soriani, O.; Peyronnet, R.; Guizouarn, H. Hereditary Xerocytosis: Differential Behavior of PIEZO1 Mutations in the N-Terminal Extracellular Domain Between Red Blood Cells and HEK Cells. Front. Physiol. 2021, 12, 736585. [Google Scholar] [CrossRef] [PubMed]
- Patterson, S.W.; Piper, H.; Starling, E.H. The regulation of the heart beat. J. Physiol. 1914, 48, 465–513. [Google Scholar] [CrossRef] [PubMed]
- von Anrep, G. On the part played by the suprarenals in the normal vascular reactions of the body. J. Physiol. 1912, 45, 307–317. [Google Scholar] [CrossRef]
- Neves, J.S.; Leite-Moreira, A.M.; Neiva-Sousa, M.; Almeida-Coelho, J.; Castro-Ferreira, R.; Leite-Moreira, A.F. Acute Myocardial Response to Stretch: What We (don’t) Know. Front. Physiol. 2015, 6, 408. [Google Scholar] [CrossRef]
- Yamaguchi, Y.; Iribe, G.; Nishida, M.; Naruse, K. Role of TRPC3 and TRPC6 channels in the myocardial response to stretch: Linking physiology and pathophysiology. Prog. Biophys. Mol. Biol. 2017, 130, 264–272. [Google Scholar] [CrossRef]
- Ravelli, F. Mechano-electric feedback and atrial fibrillation. Prog. Biophys. Mol. Biol. 2003, 82, 137–149. [Google Scholar] [CrossRef]
- Tse, G. Mechanisms of cardiac arrhythmias. J. Arrhythmia 2016, 32, 75–81. [Google Scholar] [CrossRef]
- Zeng, W.-Z.; Marshall, K.L.; Min, S.; Daou, I.; Chapleau, M.W.; Abboud, F.M.; Liberles, S.D.; Patapoutian, A. PIEZOs mediate neuronal sensing of blood pressure and the baroreceptor reflex. Science 2018, 362, 464–467. [Google Scholar] [CrossRef]
- Aleström, P.; D’Angelo, L.; Midtlyng, P.J.; Schorderet, D.F.; Schulte-Merker, S.; Sohm, F.; Warner, S. Zebrafish: Housing and husbandry recommendations. Lab. Anim. 2020, 54, 213–224. [Google Scholar] [CrossRef]



Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Rolland, L.; Torrente, A.G.; Bourinet, E.; Maskini, D.; Drouard, A.; Chevalier, P.; Jopling, C.; Faucherre, A. Prolonged Piezo1 Activation Induces Cardiac Arrhythmia. Int. J. Mol. Sci. 2023, 24, 6720. https://doi.org/10.3390/ijms24076720
Rolland L, Torrente AG, Bourinet E, Maskini D, Drouard A, Chevalier P, Jopling C, Faucherre A. Prolonged Piezo1 Activation Induces Cardiac Arrhythmia. International Journal of Molecular Sciences. 2023; 24(7):6720. https://doi.org/10.3390/ijms24076720
Chicago/Turabian StyleRolland, Laura, Angelo Giovanni Torrente, Emmanuel Bourinet, Dounia Maskini, Aurélien Drouard, Philippe Chevalier, Chris Jopling, and Adèle Faucherre. 2023. "Prolonged Piezo1 Activation Induces Cardiac Arrhythmia" International Journal of Molecular Sciences 24, no. 7: 6720. https://doi.org/10.3390/ijms24076720
APA StyleRolland, L., Torrente, A. G., Bourinet, E., Maskini, D., Drouard, A., Chevalier, P., Jopling, C., & Faucherre, A. (2023). Prolonged Piezo1 Activation Induces Cardiac Arrhythmia. International Journal of Molecular Sciences, 24(7), 6720. https://doi.org/10.3390/ijms24076720






