Identification and Characterization of PRE Genes in Moso Bamboo (Phyllostachys edulis)
Abstract
:1. Introduction
2. Results
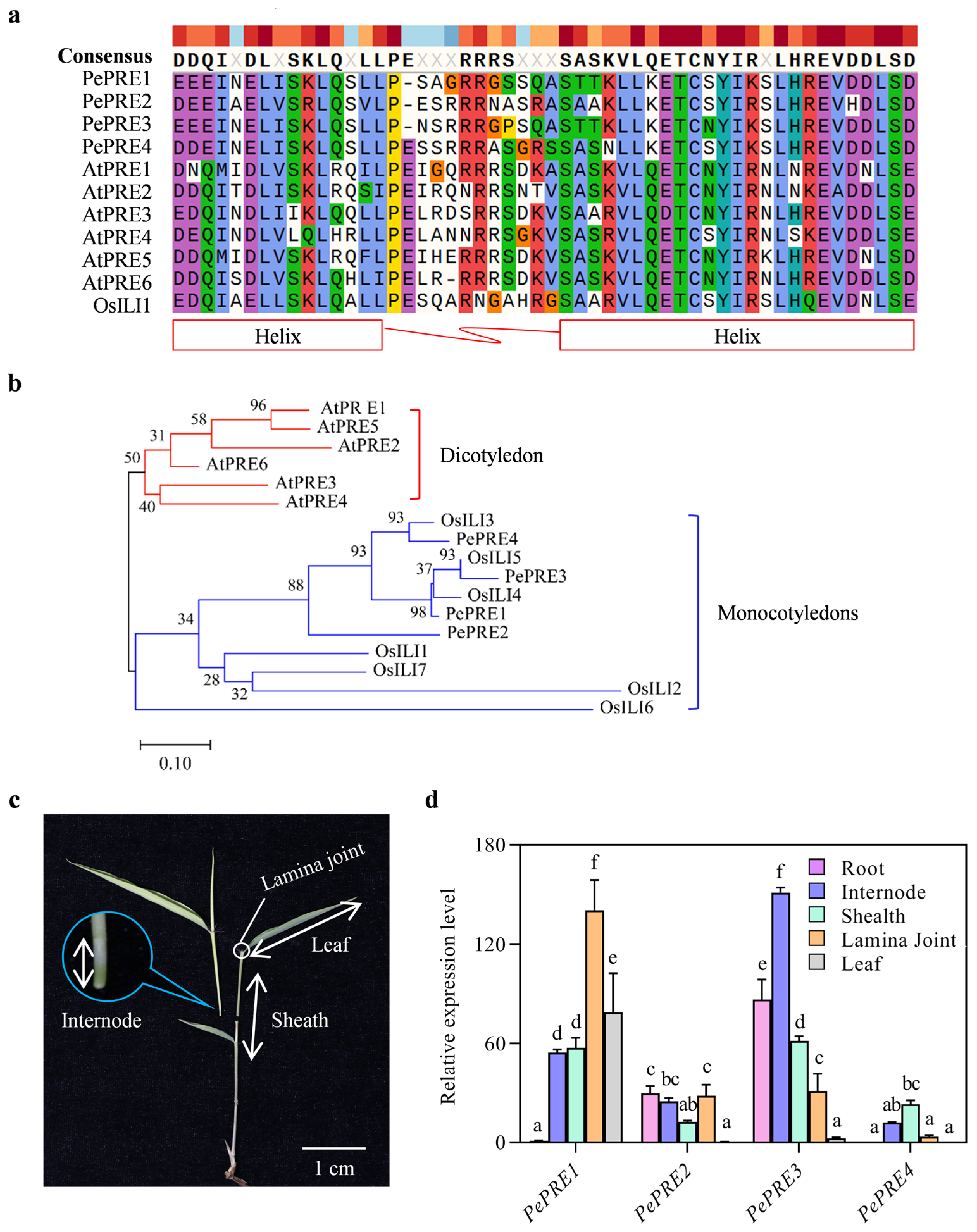
2.1. Identification of PePREs in Moso Bamboo
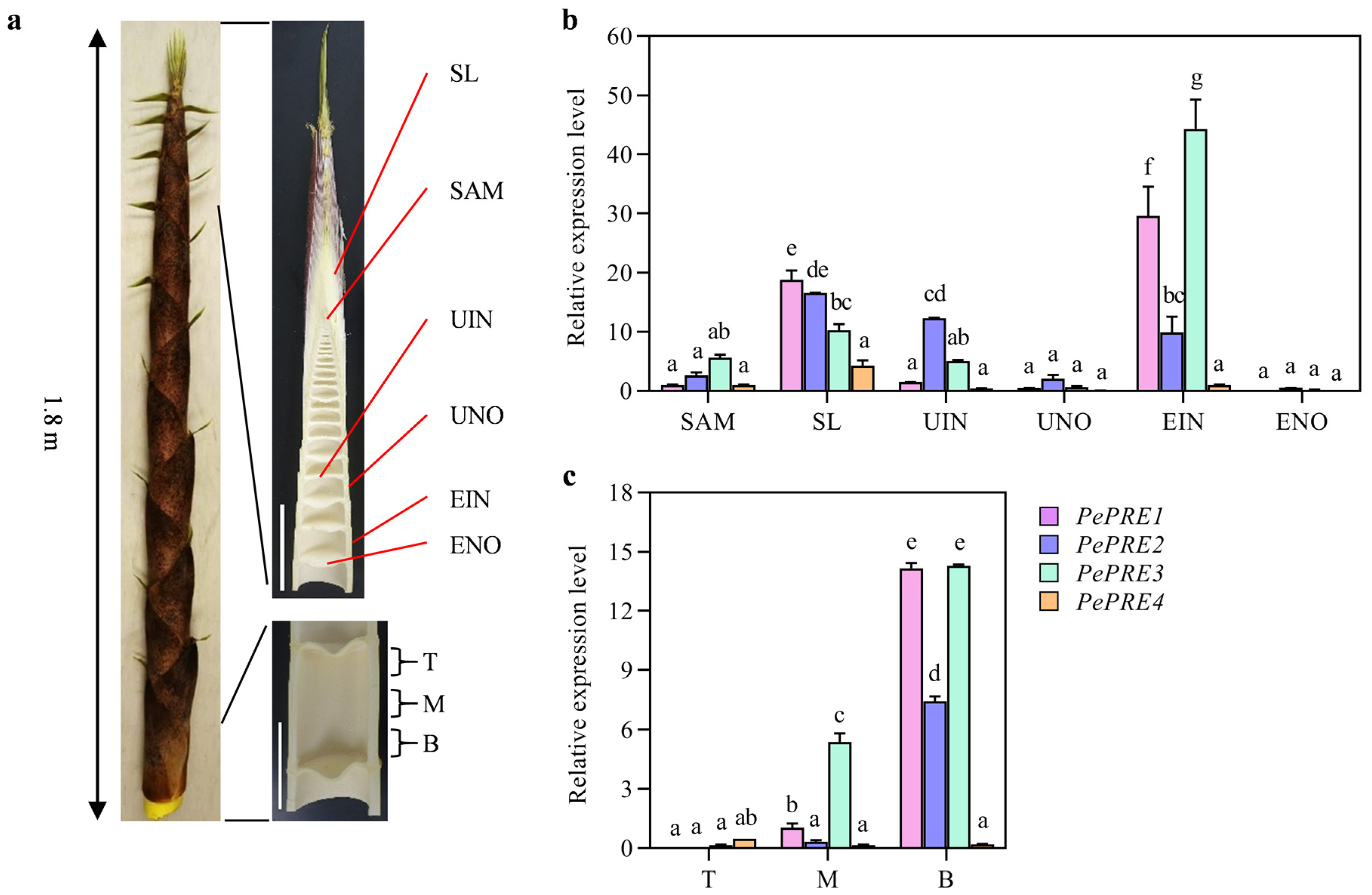
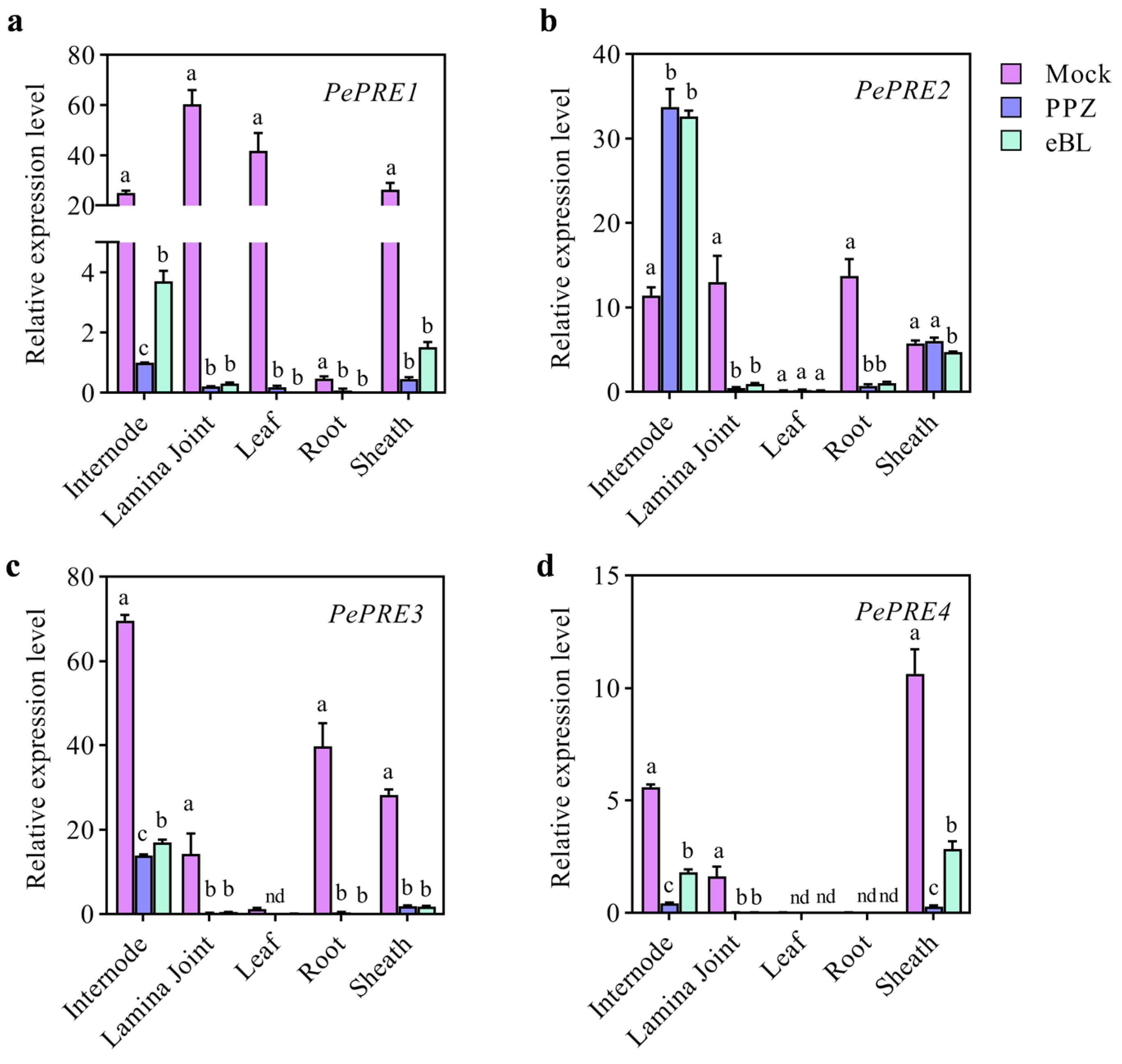
2.2. Expression of PePRE Genes in Elongating Bamboo Shoots
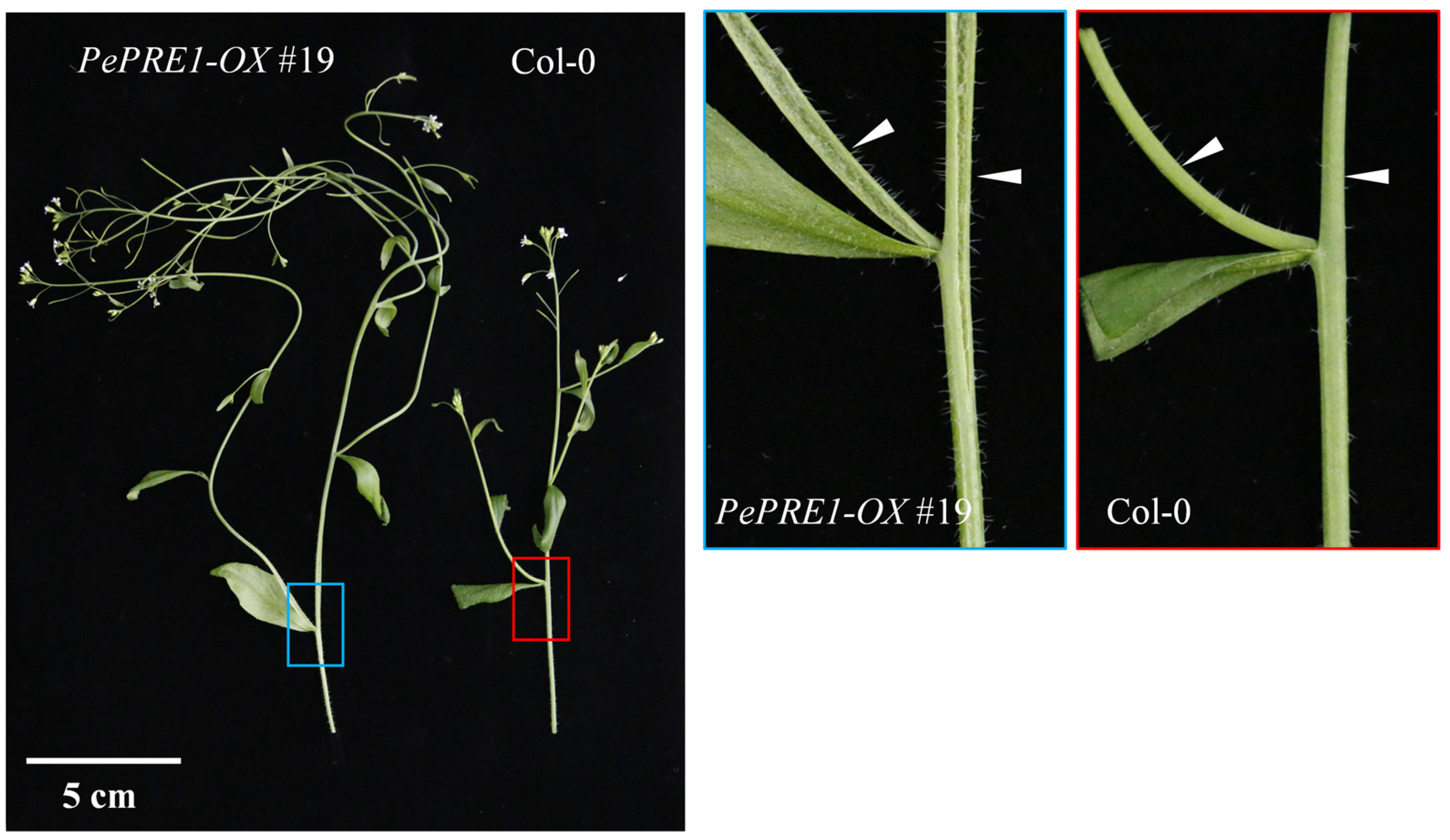
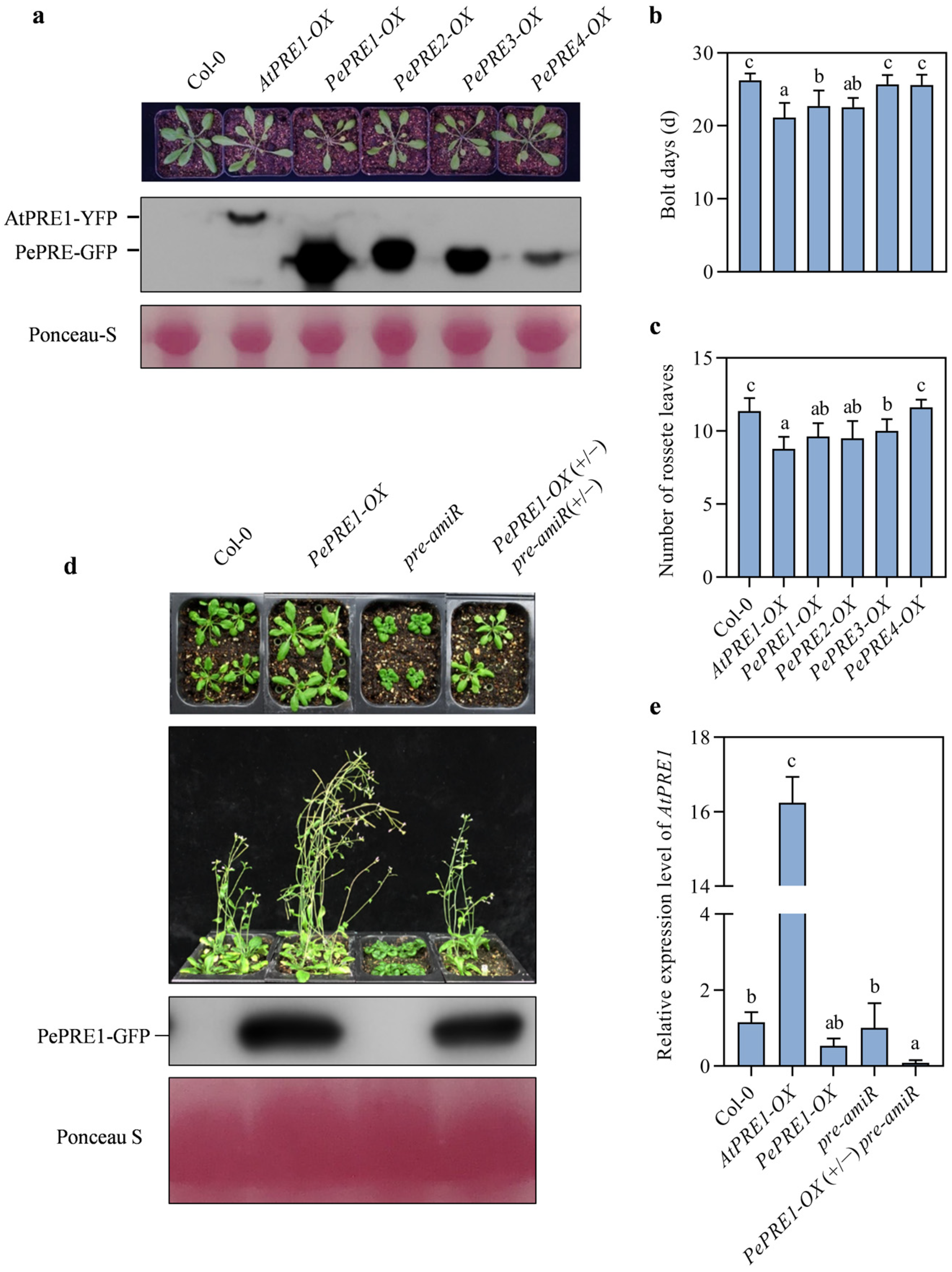
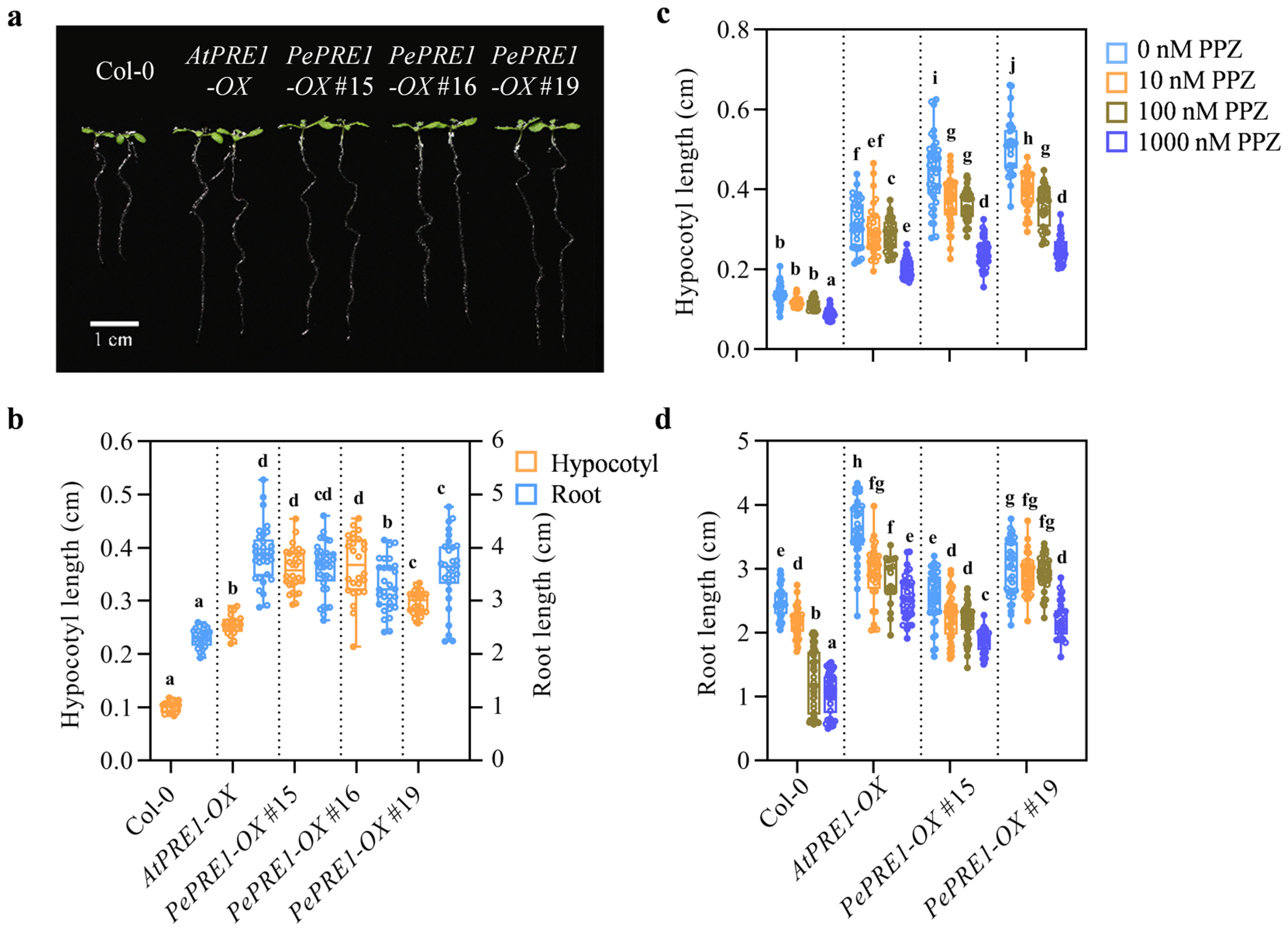
2.3. Overexpressing PePREs in Arabidopsis
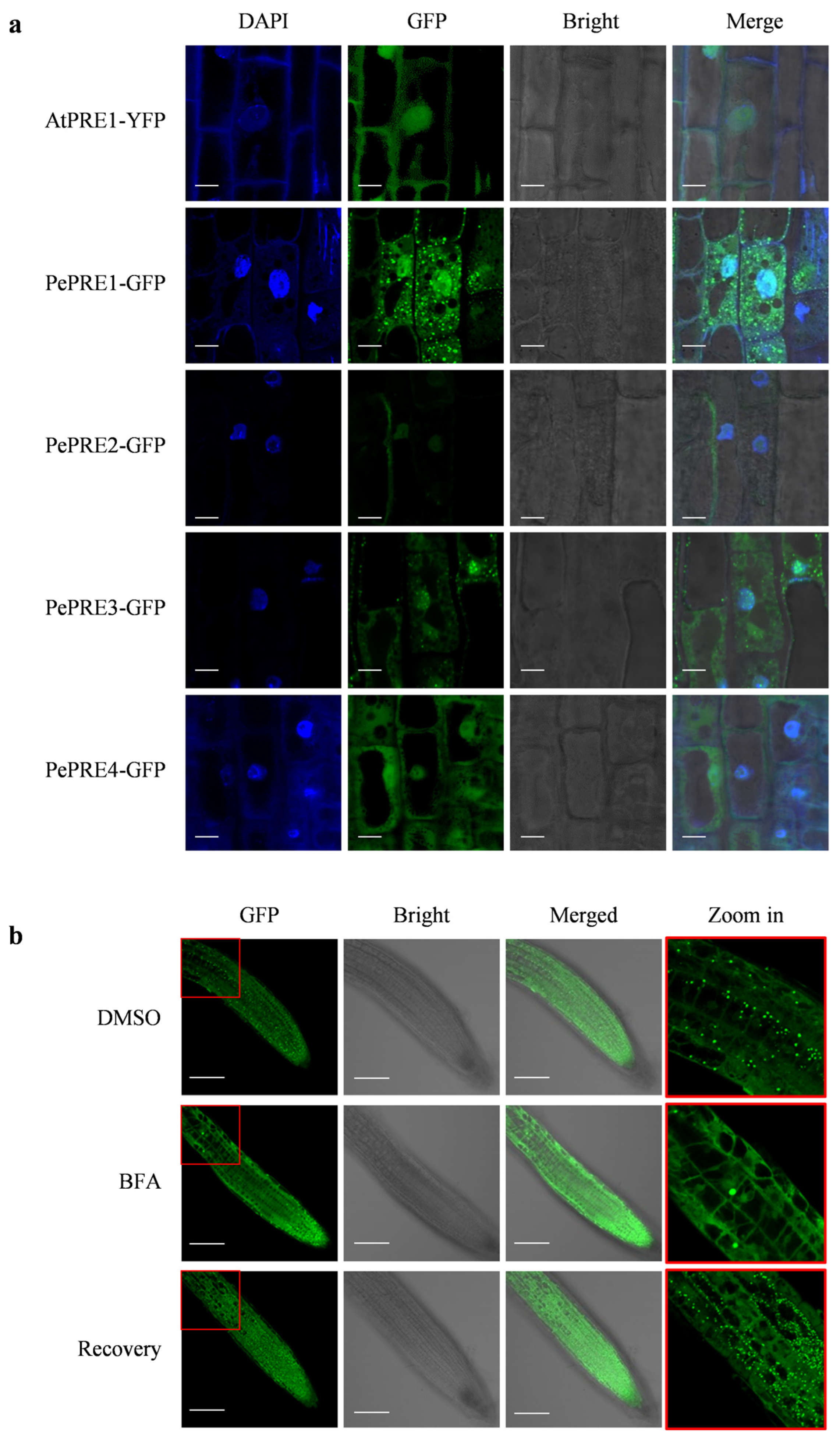
2.4. Subcellular Localization of PePREs in Arabidopsis
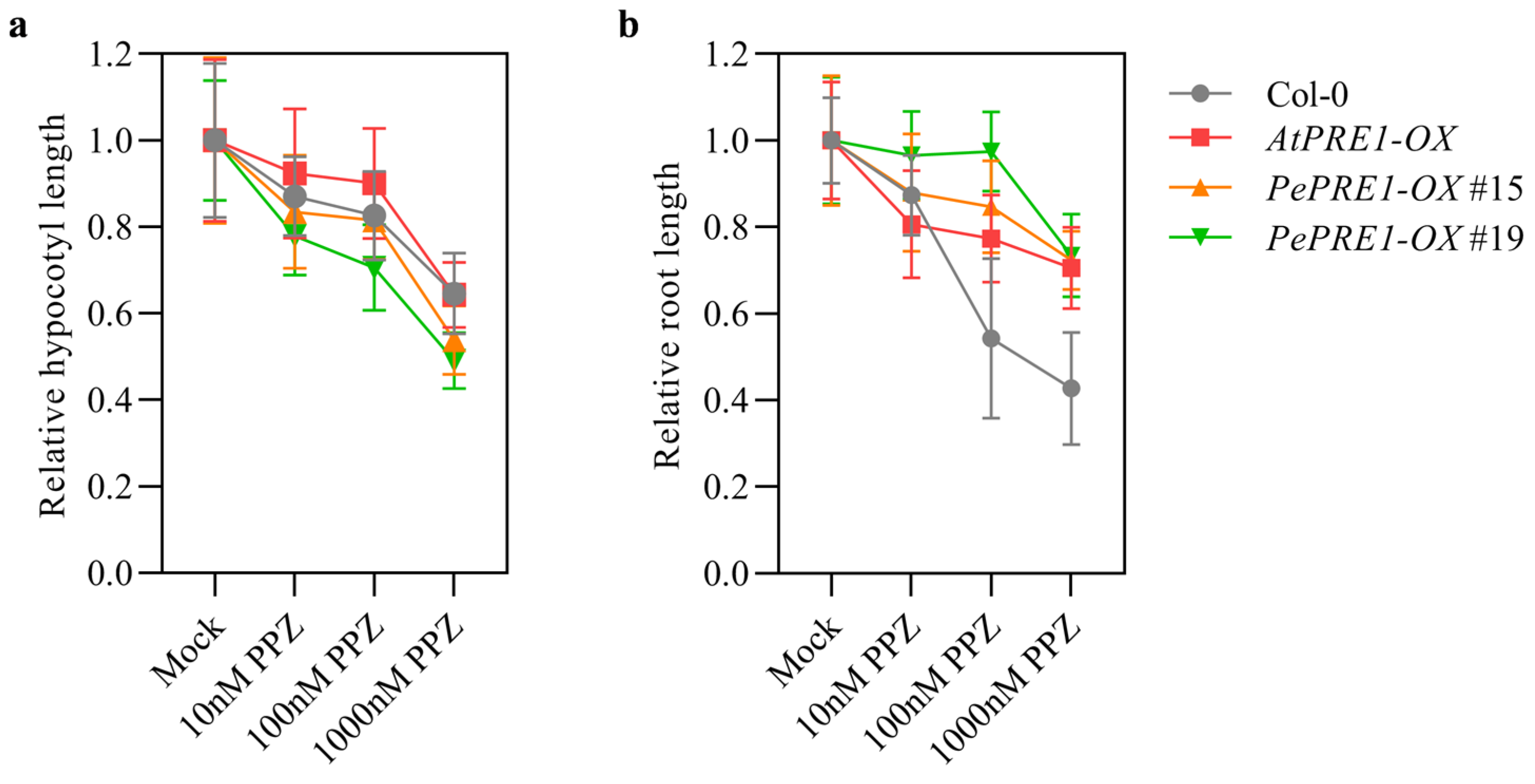
2.5. BR Regulates the Expression Levels of PePREs
3. Discussion
3.1. PePREs Have Tissue-Specific Expression Patterns in Moso Bamboo
3.2. Phytohormones Regulates PRE Function in Bamboo Elongation
3.3. Various Functions of PRE in Regulating Plant Growth through Multiple Signaling Pathways
4. Materials and Methods
4.1. Plant Material and Growth Conditions
4.2. Protein Sequence Alignment and Phylogenic Tree Construction
4.3. Gene Expression Analysis
4.4. Vector Construction and Transformation
4.5. Hypocotyl Measurements and Statistical Analysis
4.6. BFA Treatment
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Appendix A
| Primers | Sequences (5′-3′) | Note |
|---|---|---|
| PePRE1 qPCR F1 | ATTATGTCCATGGATCTCTG | Gene expression level detection of PePRE1 |
| PePRE1 qPCR R1 | ACTTAGTCCTTAGGTATCGA | Gene expression level detection of PePRE1 |
| PePRE2 qPCR F1 | CCCGCAGGCAGACATCAT | Gene expression level detection of PePRE2 |
| PePRE2 qPCR R1 | GTGCTTCGAACTGAGTAAAT | Gene expression level detection of PePRE2 |
| PePRE3 qPCR F1 | CAAGACCTCACCCAGCTTAG | Gene expression level detection of PePRE3 |
| PePRE3 qPCR R1 | TGCCGCGCGACGACCTTC | Gene expression level detection of PePRE3 |
| PePRE4 qPCR F1 | TGAGTTTAAGCTCACACTACTC | Gene expression level detection of PePRE4 |
| PePRE4 qPCR R1 | AGAGGTCGCGATCTCCTAGC | Gene expression level detection of PePRE4 |
| qPeTIP41-F1 | AAAATCATTGTAGGCCATTGTCG | Internal control gene in Ph. edulis |
| qPeTIP41-R1 | ACTAAATTAAGCCAGCGGGAGTG | Internal control gene in Ph. edulis |
| AtPRE1 qPCR F | GTTCTGATAAGGCATCAGCCTCG | Gene expression level detection of AtPRE1 |
| AtPRE1 qPCR R | CATGAGTAGGCTTCTAATAACGG | Gene expression level detection of AtPRE1 |
| UBQ10-F | ATCACCCTTGAAGTGGA | Internal control gene in Arabidopsis |
| UBQ10-F | ATCACCCTTGAAGTGGA | Internal control gene in Arabidopsis |
| PePRE1 MoClo F | tttgaagacaaAATGTCGAGCCGGAGGTCGCG | For amplifying CDS of PePRE1 |
| PePRE1 MoClo R | tttgaagacaacgaaccGCGGAGGAGGCTGCGGATGA | For amplifying CDS of PePRE1 |
| PePRE2 MoClo F1 | tttgaagacaaAATGTCGAGGCGGCGGGGG | For amplifying CDS of PePRE2 |
| PePRE2 MoClo R | tttgaagacaacgaaccGCTAGGAGGACCGGAGCTGA | For amplifying CDS of PePRE2 |
| PePRE3 MoClo F | tttgaagacaaAATGTCGGGCCGAAGGTCGTC | For amplifying CDS of PePRE3 |
| PePRE3 MoClo R | tttgaagacaacgaaccGGAGCGGAGGATGTTGCGGA | For amplifying CDS of PePRE3 |
| PePRE4 MoClo F | tttgaagacaaAATGTCGGGGCGCAGAGCCGG | For amplifying CDS of PePRE4 |
| PePRE4 MoClo R | tttgaagacaacgaaccGGATGAGCGGAGGAGACTGC | For amplifying CDS of PePRE4 |
| PePRE4 MoClo F(-R)1 | tttgaagacaaGTCaTCGGCGTCGAACCTGCTG | For amplifying CDS of PePRE4 |
| PePRE4 MoClo (F-)R | tttgaagacaatGACTGCAACAAAAACATTA | For amplifying CDS of PePRE4 |
| level 0 F | CGTTATCCCCTGATTCTGTGGATAAC | MoClo cloning general primer |
| level 0 R | GTCTCATGAGCGGATACATATTTGAATG | MoClo cloning general primer |
| level 1 F | GAACCCTGTGGTTGGCATGCACATAC | MoClo cloning general primer |
| level 1 R | CTGGTGGCAGGATATATTGTGGTG | MoClo cloning general primer |
| level 2 F | GTGGTGTAAACAAATTGACGC | MoClo cloning general primer |
| level 2 R | GGATAAACCTTTTCACGCCC | MoClo cloning general primer |
Appendix B

References
- Kelchner, S.A.; Bamboo Phylogeny, G. Higher level phylogenetic relationships within the bamboos (Poaceae: Bambusoideae) based on five plastid markers. Mol. Phylogenet. Evol. 2013, 67, 404–413. [Google Scholar] [CrossRef] [PubMed]
- Wysocki, W.P.; Clark, L.G.; Attigala, L.; Ruiz-Sanchez, E.; Duvall, M.R. Evolution of the bamboos (Bambusoideae; Poaceae): A full plastome phylogenomic analysis. BMC Evol. Biol. 2015, 15, 50. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, T.; Wang, H.; Zhang, Y.; Wang, H.; Zhang, Z.; Liu, X.; Zhang, Z.; Liu, K.; Yang, D.; Zhang, H.; et al. Comprehensive profiling of epigenetic modifications in fast-growing Moso bamboo shoots. Plant Physiol. 2023, 191, 1017–1035. [Google Scholar] [CrossRef] [PubMed]
- Gamuyao, R.; Nagai, K.; Ayano, M.; Mori, Y.; Minami, A.; Kojima, M.; Suzuki, T.; Sakakibara, H.; Higashiyama, T.; Ashikari, M.; et al. Hormone distribution and transcriptome profiles in bamboo shoots provide insights on bamboo stem emergence and growth. Plant Cell Physiol. 2017, 58, 702–716. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chaiwanon, J.; Wang, W.; Zhu, J.Y.; Oh, E.; Wang, Z.Y. Information integration and communication in plant growth regulation. Cell 2016, 164, 1257–1268. [Google Scholar] [CrossRef] [Green Version]
- Oh, E.; Zhu, J.Y.; Bai, M.Y.; Arenhart, R.A.; Sun, Y.; Wang, Z.Y. Cell elongation is regulated through a central circuit of interacting transcription factors in the Arabidopsis hypocotyl. Elife 2014, 3, e03031. [Google Scholar] [CrossRef]
- Zhiponova, M.K.; Morohashi, K.; Vanhoutte, I.; Machemer-Noonan, K.; Revalska, M.; Van Montagu, M.; Grotewold, E.; Russinova, E. Helix-loop-helix/basic helix-loop-helix transcription factor network represses cell elongation in Arabidopsis through an apparent incoherent feed-forward loop. Proc. Natl. Acad. Sci. USA 2014, 111, 2824–2829. [Google Scholar] [CrossRef] [Green Version]
- Ruzinova, M.B.; Benezra, R. Id proteins in development, cell cycle and cancer. Trends Cell Biol. 2003, 13, 410–418. [Google Scholar] [CrossRef]
- Castelain, M.; Le Hir, R.; Bellini, C. The non-DNA-binding bHLH transcription factor PRE3/bHLH135/ATBS1/TMO7 is involved in the regulation of light signaling pathway in Arabidopsis. Physiol. Plant 2012, 145, 450–460. [Google Scholar] [CrossRef]
- Bai, M.Y.; Fan, M.; Oh, E.; Wang, Z.Y. A triple Helix-Loop-Helix/basic Helix-Loop-Helix cascade controls cell elongation downstream of multiple hormonal and environmental signaling pathways in Arabidopsis. Plant Cell 2012, 24, 4917–4929. [Google Scholar] [CrossRef] [Green Version]
- Malinovsky, F.G.; Batoux, M.; Schwessinger, B.; Youn, J.H.; Stransfeld, L.; Win, J.; Kim, S.K.; Zipfel, C. Antagonistic regulation of growth and immunity by the Arabidopsis basic helix-loop-helix transcription factor homolog of brassinosteroid enhanced expression2 interacting with increased leaf inclination1 binding bHLH1. Plant Physiol. 2014, 164, 1443–1455. [Google Scholar] [CrossRef] [Green Version]
- Zhang, L.Y.; Bai, M.Y.; Wu, J.; Zhu, J.Y.; Wang, H.; Zhang, Z.; Wang, W.; Sun, Y.; Zhao, J.; Sun, X.; et al. Antagonistic HLH/bHLH transcription factors mediate brassinosteroid regulation of cell elongation and plant development in rice and Arabidopsis. Plant Cell 2009, 21, 3767–3780. [Google Scholar] [CrossRef] [Green Version]
- Choi, H.; Oh, E. PIF4 integrates multiple environmental and hormonal signals for plant growth regulation in Arabidopsis. Mol. Cells 2016, 39, 587–593. [Google Scholar] [CrossRef] [Green Version]
- Oh, E.; Zhu, J.Y.; Wang, Z.Y. Interaction between BZR1 and PIF4 integrates brassinosteroid and environmental responses. Nat. Cell Biol. 2012, 14, 802–809. [Google Scholar] [CrossRef] [Green Version]
- Hao, Y.; Oh, E.; Choi, G.; Liang, Z.; Wang, Z.Y. Interactions between HLH and bHLH factors modulate light-regulated plant development. Mol. Plant 2012, 5, 688–697. [Google Scholar] [CrossRef] [Green Version]
- Hyun, Y.; Lee, I. KIDARI, encoding a non-DNA Binding bHLH protein, represses light signal transduction in Arabidopsis thaliana. Plant Mo.l Biol. 2006, 61, 283–296. [Google Scholar] [CrossRef]
- Buti, S.; Pantazopoulou, C.K.; van Gelderen, K.; Hoogers, V.; Reinen, E.; Pierik, R. A Gas-and-Brake mechanism of bHLH proteins modulates shade avoidance. Plant Physiol. 2020, 184, 2137–2153. [Google Scholar] [CrossRef]
- Ikeda, M.; Mitsuda, N.; Ishizuka, T.; Satoh, M.; Ohme-Takagi, M. The CIB1 transcription factor regulates light- and heat-inducible cell elongation via a two-step HLH/bHLH system. J. Exp. Bot. 2021, 72, 1795–1808. [Google Scholar] [CrossRef]
- Chapman, E.J.; Greenham, K.; Castillejo, C.; Sartor, R.; Bialy, A.; Sun, T.P.; Estelle, M. Hypocotyl transcriptome reveals auxin regulation of growth-promoting genes through GA-dependent and -independent pathways. PLoS ONE 2012, 7, e36210. [Google Scholar] [CrossRef] [Green Version]
- Reed, J.W.; Wu, M.F.; Reeves, P.H.; Hodgens, C.; Yadav, V.; Hayes, S.; Pierik, R. Three auxin response factors promote hypocotyl elongation. Plant Physiol. 2018, 178, 864–875. [Google Scholar] [CrossRef] [Green Version]
- Lee, S.; Lee, S.; Yang, K.Y.; Kim, Y.M.; Park, S.Y.; Kim, S.Y.; Soh, M.S. Overexpression of PRE1 and its homologous genes activates Gibberellin-dependent responses in Arabidopsis thaliana. Plant Cell Physiol. 2006, 47, 591–600. [Google Scholar] [CrossRef] [PubMed]
- Dai, X.; Lu, Q.; Wang, J.; Wang, L.; Xiang, F.; Liu, Z. MiR160 and its target genes ARF10, ARF16 and ARF17 modulate hypocotyl elongation in a light, BRZ, or PAC-dependent manner in Arabidopsis: miR160 promotes hypocotyl elongation. Plant Sci. 2021, 303, 110686. [Google Scholar] [CrossRef] [PubMed]
- Medina-Puche, L.; Martinez-Rivas, F.J.; Molina-Hidalgo, F.J.; Garcia-Gago, J.A.; Mercado, J.A.; Caballero, J.L.; Munoz-Blanco, J.; Blanco-Portales, R. Ectopic expression of the atypical HLH FaPRE1 gene determines changes in cell size and morphology. Plant Sci. 2021, 305, 110830. [Google Scholar] [CrossRef] [PubMed]
- Zhao, B.; Cao, J.F.; Hu, G.J.; Chen, Z.W.; Wang, L.Y.; Shangguan, X.X.; Wang, L.J.; Mao, Y.B.; Zhang, T.Z.; Wendel, J.F.; et al. Core cis-element variation confers subgenome-biased expression of a transcription factor that functions in cotton fiber elongation. New Phytol. 2018, 218, 1061–1075. [Google Scholar] [CrossRef] [Green Version]
- Zhang, Z.; Yang, X.; Cheng, L.; Guo, Z.; Wang, H.; Wu, W.; Shin, K.; Zhu, J.; Zheng, X.; Bian, J.; et al. Physiological and transcriptomic analyses of brassinosteroid function in moso bamboo (Phyllostachys edulis) seedlings. Planta 2020, 252, 27. [Google Scholar] [CrossRef]
- Zhao, H.; Peng, Z.; Fei, B.; Li, L.; Hu, T.; Gao, Z.; Jiang, Z. BambooGDB: A bamboo genome database with functional annotation and an analysis platform. Database 2014, 2014, bau006. [Google Scholar] [CrossRef] [Green Version]
- Mara, C.D.; Huang, T.; Irish, V.F. The Arabidopsis floral homeotic proteins APETALA3 and PISTILLATA negatively regulate the BANQUO genes implicated in light signaling. Plant Cell 2010, 22, 690–702. [Google Scholar] [CrossRef] [Green Version]
- Klausner, R.D.; Donaldson, J.G.; Lippincott-Schwartz, J. Brefeldin A: Insights into the control of membrane traffic and organelle structure. J. Cell Biol. 1992, 116, 1071–1080. [Google Scholar] [CrossRef]
- Heang, D.; Sassa, H. Antagonistic actions of HLH/bHLH proteins are involved in grain length and weight in rice. PLoS ONE 2012, 7, e31325. [Google Scholar] [CrossRef] [Green Version]
- Tanaka, A.; Nakagawa, H.; Tomita, C.; Shimatani, Z.; Ohtake, M.; Nomura, T.; Jiang, C.J.; Dubouzet, J.G.; Kikuchi, S.; Sekimoto, H.; et al. BRASSINOSTEROID UPREGULATED1, encoding a Helix-Loop-Helix protein, is a novel gene involved in brassinosteroid signaling and controls bending of the lamina joint in rice. Plant Physiol. 2009, 151, 669–680. [Google Scholar] [CrossRef] [Green Version]
- Jang, S.; Cho, J.Y.; Do, G.R.; Kang, Y.; Li, H.Y.; Song, J.; Kim, H.Y.; Kim, B.G.; Hsing, Y.I. Modulation of rice leaf angle and grain size by expressing OsBCL1 and OsBCL2 under the control of OsBUL1 promoter. Int. J. Mol. Sci. 2021, 22, 7792. [Google Scholar] [CrossRef]
- Jang, S.; An, G.; Li, H.Y. Rice leaf angle and grain size are affected by the OsBUL1 transcriptional activator complex. Plant Physiol. 2017, 173, 688–702. [Google Scholar] [CrossRef] [Green Version]
- Medina-Puche, L.; Martinez-Rivas, F.J.; Molina-Hidalgo, F.J.; Mercado, J.A.; Moyano, E.; Rodriguez-Franco, A.; Caballero, J.L.; Munoz-Blanco, J.; Blanco-Portales, R. An atypical HLH transcriptional regulator plays a novel and important role in strawberry ripened receptacle. BMC Plant Biol. 2019, 19, 586. [Google Scholar] [CrossRef]
- McKim, S.M. Moving on up—Controlling internode growth. New Phytol. 2020, 226, 672–678. [Google Scholar] [CrossRef]
- Wang, K.L.; Zhang, Y.; Zhang, H.M.; Lin, X.C.; Xia, R.; Song, L.; Wu, A.M. MicroRNAs play important roles in regulating the rapid growth of the Phyllostachys edulis culm internode. New Phytol. 2021, 231, 2215–2230. [Google Scholar] [CrossRef]
- Li, J.; Liu, Z.; Gao, C.; Miao, Y.; Cui, K. Overexpression of DsEXLA2 gene from Dendrocalamus sinicus accelerates the plant growth rate of Arabidopsis. Phytochemistry 2022, 199, 113178. [Google Scholar] [CrossRef]
- McKim, S.M. How plants grow up. J. Integr. Plant Biol. 2019, 61, 257–277. [Google Scholar] [CrossRef] [Green Version]
- Yeh, S.H.; Lee, B.H.; Liao, S.C.; Tsai, M.H.; Tseng, Y.H.; Chang, H.C.; Yang, C.C.; Jan, H.C.; Chiu, Y.C.; Wang, A.Y. Identification of genes differentially expressed during the growth of Bambusa oldhamii. Plant Physiol. Biochem. 2013, 63, 217–226. [Google Scholar] [CrossRef]
- Nagai, K.; Mori, Y.; Ishikawa, S.; Furuta, T.; Gamuyao, R.; Niimi, Y.; Hobo, T.; Fukuda, M.; Kojima, M.; Takebayashi, Y.; et al. Antagonistic regulation of the gibberellic acid response during stem growth in rice. Nature 2020, 584, 109–114. [Google Scholar] [CrossRef]
- Lee, J.; Moon, S.; Jang, S.; Lee, S.; An, G.; Jung, K.-H.; Park, S.K. OsbHLH073 negatively regulates internode elongation and plant height by modulating ga homeostasis in rice. Plants 2020, 9, 547. [Google Scholar] [CrossRef] [Green Version]
- Wang, Y.; Qiao, G.; Xu, J.; Jin, K.; Fan, M.; Ding, Y.; Wei, Q.; Zhuo, R. Anatomical characteristics and variation mechanisms on the thick-walled and dwarfed culm of shidu bamboo (Phyllostachys nidularia f. farcta). Front. Plant Sci. 2022, 13, 876658. [Google Scholar] [CrossRef] [PubMed]
- Chen, M.; Guo, L.; Ramakrishnan, M.; Fei, Z.; Vinod, K.K.; Ding, Y.; Jiao, C.; Gao, Z.; Zha, R.; Wang, C.; et al. Rapid growth of Moso bamboo (Phyllostachys edulis): Cellular roadmaps, transcriptome dynamics, and environmental factors. Plant Cell 2022, 34, 3577–3610. [Google Scholar] [CrossRef] [PubMed]
- Hofmann, N.R. A tripartite growth regulatory cascade of basic Helix-Loop-Helix transcription factors. Plant Cell 2012, 24, 4774. [Google Scholar] [CrossRef] [PubMed]
- Ikeda, M.; Fujiwara, S.; Mitsuda, N.; Ohme-Takagi, M. A triantagonistic basic helix-loop-helix system regulates cell elongation in Arabidopsis. Plant Cell 2012, 24, 4483–4497. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shin, K.; Lee, I.; Kim, E.; Park, S.K.; Soh, M.S.; Lee, S. PACLOBUTRAZOL-RESISTANCE gene family regulates floral organ growth with unequal genetic redundancy in Arabidopsis thaliana. Int. J. Mol. Sci. 2019, 20, 869. [Google Scholar] [CrossRef] [Green Version]
- Zhu, Z.; Liang, H.; Chen, G.; Li, F.; Wang, Y.; Liao, C.; Hu, Z. The bHLH transcription factor SlPRE2 regulates tomato fruit development and modulates plant response to gibberellin. Plant Cell Rep. 2019, 38, 1053–1064. [Google Scholar] [CrossRef]
- Casal, J.J.; Fankhauser, C. Shade avoidance in the context of climate change. Plant Physiol. 2023, 191, 1475–1491. [Google Scholar] [CrossRef]
- Galstyan, A.; Cifuentes-Esquivel, N.; Bou-Torrent, J.; Martinez-Garcia, J.F. The shade avoidance syndrome in Arabidopsis: A fundamental role for atypical basic helix-loop-helix proteins as transcriptional cofactors. Plant J. 2011, 66, 258–267. [Google Scholar] [CrossRef]
- Huang, W.; Ding, Y.; Wang, S.; Song, C.; Wang, F. Growth and development responses of the Rhizome-Root system in Pleioblastus pygmaeus to light intensity. Plants 2022, 11, 2204. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef]
- Fan, C.; Ma, J.; Guo, Q.; Li, X.; Wang, H.; Lu, M. Selection of reference genes for quantitative real-time PCR in bamboo (Phyllostachys edulis). PLoS ONE 2013, 8, e56573. [Google Scholar] [CrossRef] [Green Version]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zheng, S.; Shin, K.; Lin, W.; Wang, W.; Yang, X. Identification and Characterization of PRE Genes in Moso Bamboo (Phyllostachys edulis). Int. J. Mol. Sci. 2023, 24, 6886. https://doi.org/10.3390/ijms24086886
Zheng S, Shin K, Lin W, Wang W, Yang X. Identification and Characterization of PRE Genes in Moso Bamboo (Phyllostachys edulis). International Journal of Molecular Sciences. 2023; 24(8):6886. https://doi.org/10.3390/ijms24086886
Chicago/Turabian StyleZheng, Sujin, Kihye Shin, Wenxiong Lin, Wenfei Wang, and Xuelian Yang. 2023. "Identification and Characterization of PRE Genes in Moso Bamboo (Phyllostachys edulis)" International Journal of Molecular Sciences 24, no. 8: 6886. https://doi.org/10.3390/ijms24086886
APA StyleZheng, S., Shin, K., Lin, W., Wang, W., & Yang, X. (2023). Identification and Characterization of PRE Genes in Moso Bamboo (Phyllostachys edulis). International Journal of Molecular Sciences, 24(8), 6886. https://doi.org/10.3390/ijms24086886






