Collateral Sensitivity to Fosfomycin of Tobramycin-Resistant Mutants of Pseudomonas aeruginosa Is Contingent on Bacterial Genomic Background
Abstract
1. Introduction
2. Results and Discussion
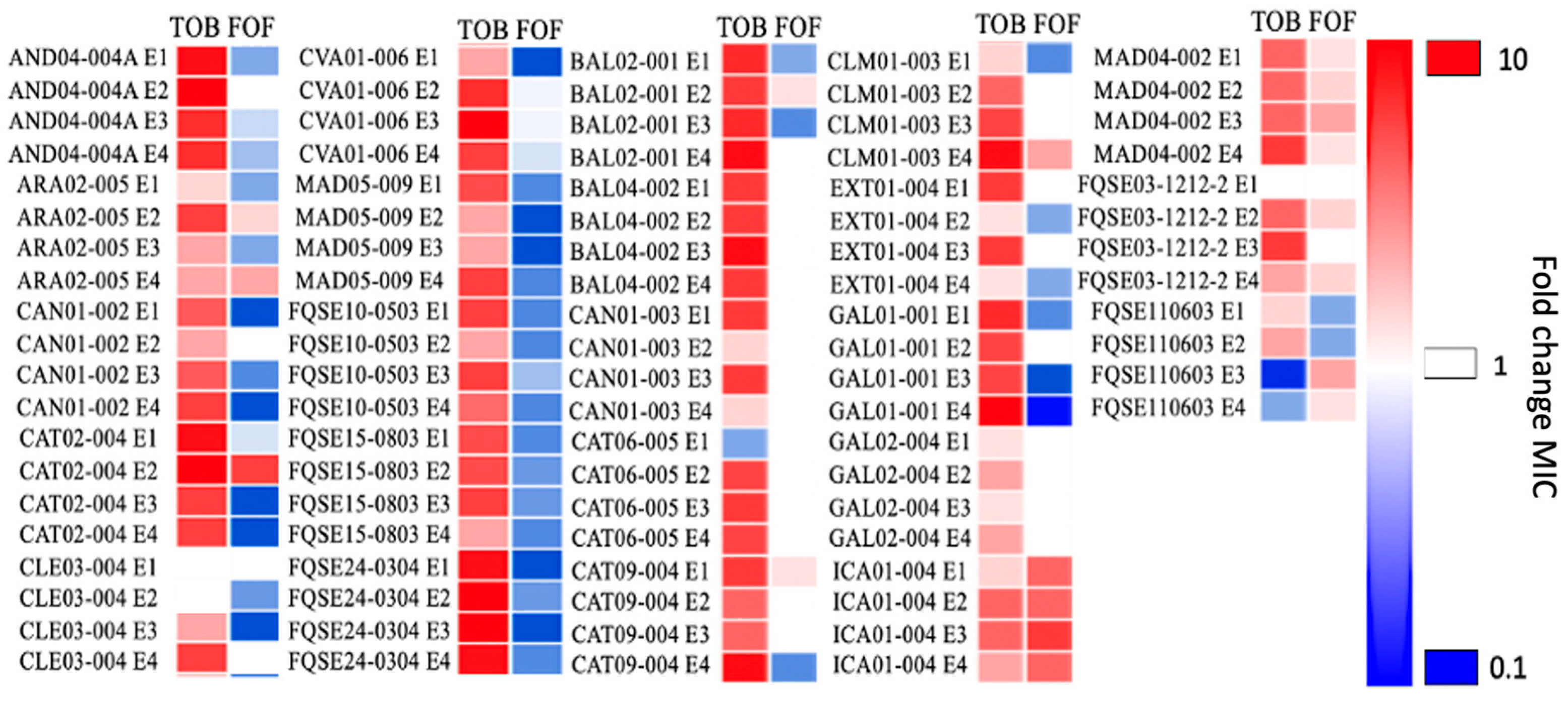
2.1. Collateral Sensitivity to Fosfomycin of Pseudomonas aeruginosa Clinical Isolates Evolved in the Presence of Tobramycin Is Not Robust
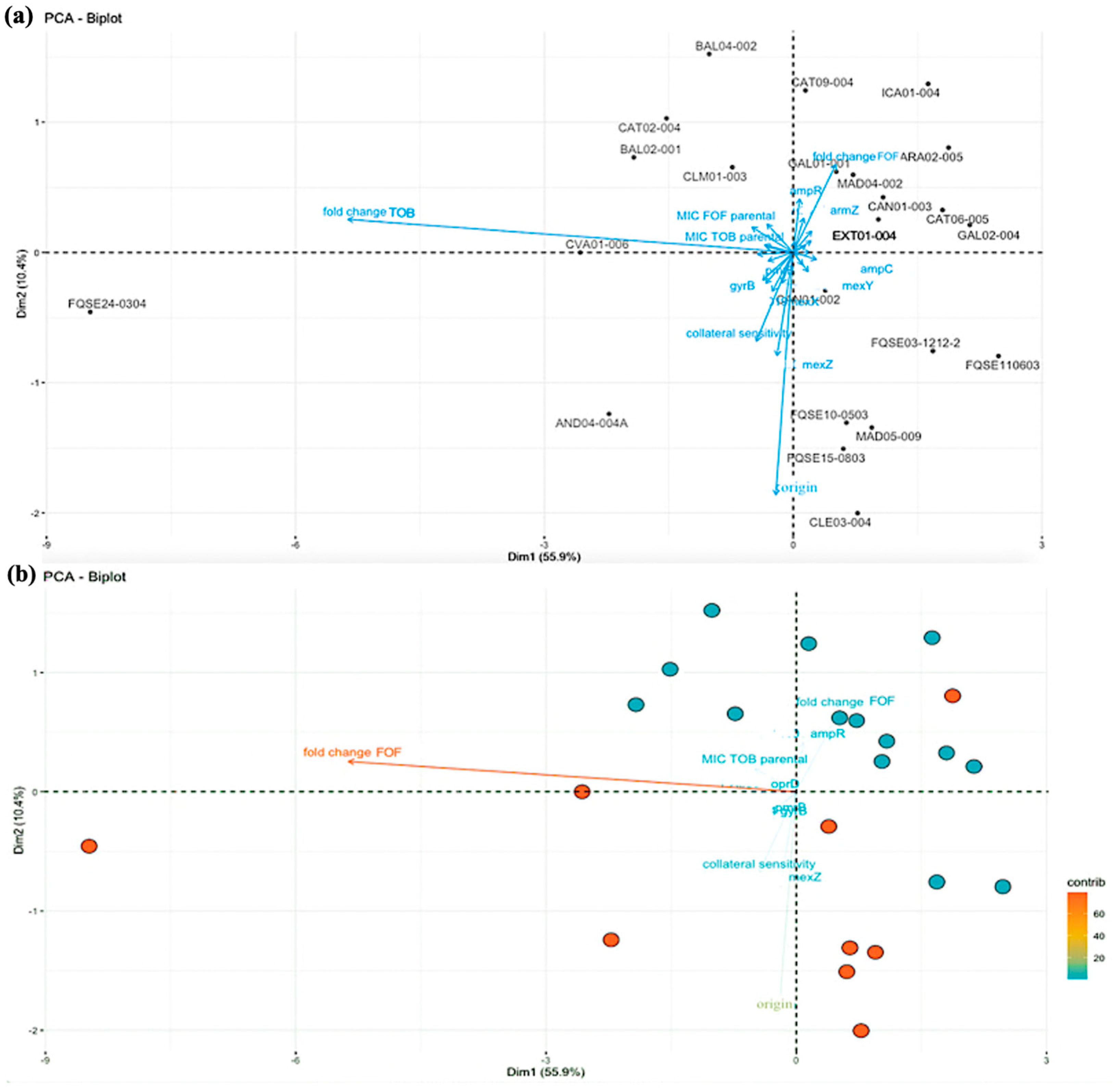
2.2. Principal Component Analysis: The Fold Change in Tobramycin MIC of the Clinical Strains Correlates with the Development of Fosfomycin Collateral Sensitivity
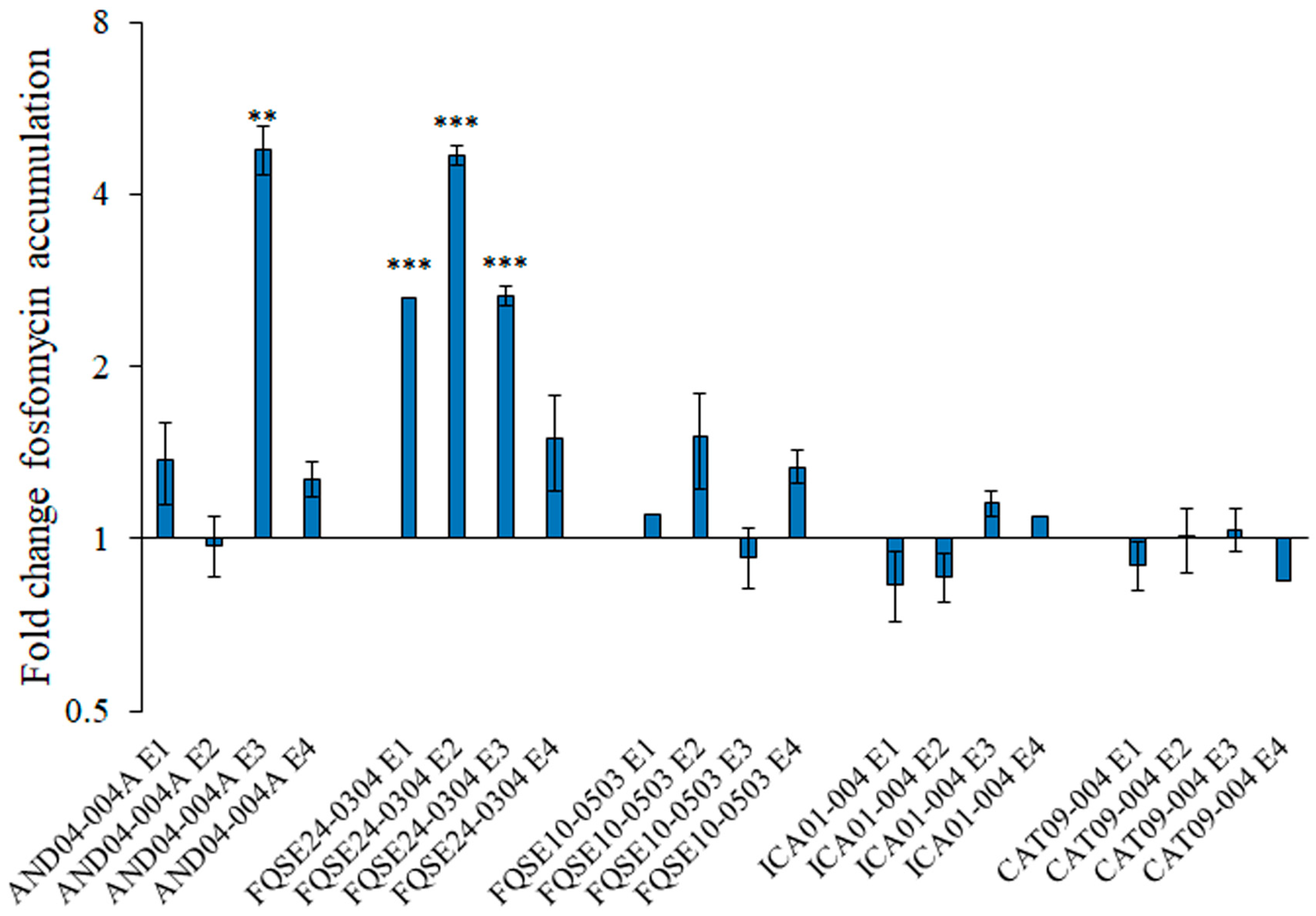
2.3. The Accumulation of Active Fosfomycin Is Increased in Some Tobramycin-Resistant Mutants Presenting Fosfomycin Collateral Sensitivity and Remains the Same in Mutants without Collateral Sensitivity
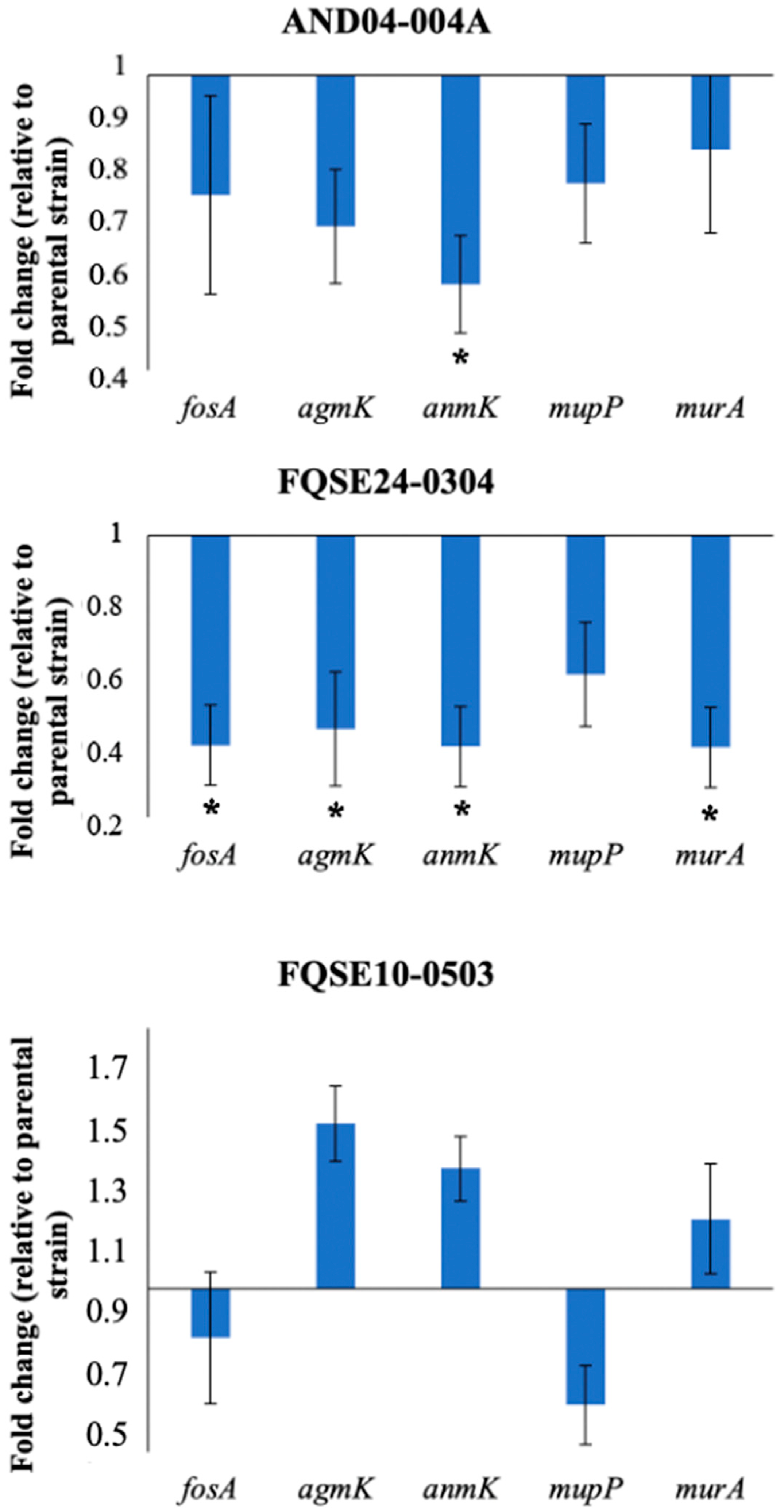
2.4. Reduced Expression of Genes Involved in the Alternative Peptidoglycan-Recycling Pathway May Contribute to Fosfomycin Collateral Sensitivity in P. aeruginosa Clinical Isolates
3. Materials and Methods
3.1. Bacterial Strains and Oligonucleotides
3.2. Adaptive Laboratory Evolution
3.3. Analysis of Susceptibility to Antibiotics: E-Test
3.4. Principal Component Analysis
3.5. RNA Extraction and qRT-PCR
3.6. Measurement of Intracellular Fosfomycin Accumulation
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Rice, L.B. The clinical consequences of antimicrobial resistance. Curr. Opin. Microbiol. 2009, 12, 476–481. [Google Scholar] [CrossRef]
- Hernando-Amado, S.; Coque, T.M.; Baquero, F.; Martinez, J.L. Defining and combating antibiotic resistance from One Health and Global Health perspectives. Nat. Microbiol. 2019, 4, 1432–1442. [Google Scholar] [CrossRef] [PubMed]
- Imamovic, L.; Sommer, M.O. Use of collateral sensitivity networks to design drug cycling protocols that avoid resistance development. Sci. Transl. Med. 2013, 5, 204ra132. [Google Scholar] [CrossRef] [PubMed]
- Pal, C.; Papp, B.; Lazar, V. Collateral sensitivity of antibiotic-resistant microbes. Trends Microbiol. 2015, 23, 401–407. [Google Scholar] [CrossRef] [PubMed]
- Pinheiro, F.; Warsi, O.; Andersson, D.I.; Lässig, M. Metabolic fitness landscapes predict the evolution of antibiotic resistance. Nat. Ecol. Evol. 2021, 5, 677–687. [Google Scholar] [CrossRef] [PubMed]
- Roemhild, R.; Andersson, D.I. Mechanisms and therapeutic potential of collateral sensitivity to antibiotics. PLoS Pathog. 2021, 17, e1009172. [Google Scholar] [CrossRef] [PubMed]
- Szybalski, W.; Bryson, V. Genetic studies on microbial cross resistance to toxic agents. I. Cross resistance of Escherichia coli to fifteen antibiotics. J. Bacteriol. 1952, 64, 489–499. [Google Scholar] [CrossRef]
- Roemhild, R.; Bollenbach, T.; Andersson, D.I. The physiology and genetics of bacterial responses to antibiotic combinations. Nat. Rev. Microbiol. 2022, 20, 478–490. [Google Scholar] [CrossRef]
- Baym, M.; Stone, L.K.; Kishony, R. Multidrug evolutionary strategies to reverse antibiotic resistance. Science 2016, 351, aad3292. [Google Scholar] [CrossRef]
- Barbosa, C.; Beardmore, R.; Schulenburg, H.; Jansen, G. Antibiotic combination efficacy (ACE) networks for a Pseudomonas aeruginosa model. PLoS Biol. 2018, 16, e2004356. [Google Scholar] [CrossRef]
- Jahn, L.J.; Simon, D.; Jensen, M.; Bradshaw, C.; Ellabaan, M.M.H.; Sommer, M.O.A. Compatibility of Evolutionary Responses to Constituent Antibiotics Drive Resistance Evolution to Drug Pairs. Mol. Biol. Evol. 2021, 38, 2057–2069. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.; Lieberman, T.D.; Kishony, R. Alternating antibiotic treatments constrain evolutionary paths to multidrug resistance. Proc. Natl. Acad. Sci. USA 2014, 111, 14494–14499. [Google Scholar] [CrossRef] [PubMed]
- Lopez-Causape, C.; Cabot, G.; Del Barrio-Tofino, E.; Oliver, A. The Versatile Mutational Resistome of Pseudomonas aeruginosa. Front. Microbiol. 2018, 9, 685. [Google Scholar] [CrossRef] [PubMed]
- Barbosa, C.; Trebosc, V.; Kemmer, C.; Rosenstiel, P.; Beardmore, R.; Schulenburg, H.; Jansen, G. Alternative Evolutionary Paths to Bacterial Antibiotic Resistance Cause Distinct Collateral Effects. Mol. Biol. Evol. 2017, 34, 2229–2244. [Google Scholar] [CrossRef] [PubMed]
- Lazar, V.; Pal Singh, G.; Spohn, R.; Nagy, I.; Horvath, B.; Hrtyan, M.; Busa-Fekete, R.; Bogos, B.; Mehi, O.; Csorgo, B.; et al. Bacterial evolution of antibiotic hypersensitivity. Mol. Syst. Biol. 2013, 9, 700. [Google Scholar] [CrossRef]
- Podnecky, N.L.; Fredheim, E.G.A.; Kloos, J.; Sorum, V.; Primicerio, R.; Roberts, A.P.; Rozen, D.E.; Samuelsen, O.; Johnsen, P.J. Conserved collateral antibiotic susceptibility networks in diverse clinical strains of Escherichia coli. Nat. Commun. 2018, 9, 3673. [Google Scholar] [CrossRef]
- Martinez-Solano, L.; Macia, M.D.; Fajardo, A.; Oliver, A.; Martinez, J.L. Chronic Pseudomonas aeruginosa infection in chronic obstructive pulmonary disease. Clin. Infect. Dis. 2008, 47, 1526–1533. [Google Scholar] [CrossRef]
- Rossi, E.; La Rosa, R.; Bartell, J.A.; Marvig, R.L.; Haagensen, J.A.J.; Sommer, L.M.; Molin, S.; Johansen, H.K. Pseudomonas aeruginosa adaptation and evolution in patients with cystic fibrosis. Nat. Rev. Microbiol. 2021, 19, 331–342. [Google Scholar] [CrossRef]
- Laborda, P.; Sanz-García, F.; Hernando-Amado, S.; Martínez, J.L. Pseudomonas aeruginosa: An antibiotic resilient pathogen with environmental origin. Curr. Opin. Microbiol. 2021, 64, 125–132. [Google Scholar] [CrossRef]
- Kahan, F.M.; Kahan, J.S.; Cassidy, P.J.; Kropp, H. The mechanism of action of fosfomycin (phosphonomycin). Ann. N. Y. Acad. Sci. 1974, 235, 364–386. [Google Scholar] [CrossRef]
- Castaneda-Garcia, A.; Rodriguez-Rojas, A.; Guelfo, J.R.; Blazquez, J. The glycerol-3-phosphate permease GlpT is the only fosfomycin transporter in Pseudomonas aeruginosa. J. Bacteriol. 2009, 191, 6968–6974. [Google Scholar] [CrossRef] [PubMed]
- De Groote, V.N.; Fauvart, M.; Kint, C.I.; Verstraeten, N.; Jans, A.; Cornelis, P.; Michiels, J. Pseudomonas aeruginosa fosfomycin resistance mechanisms affect non-inherited fluoroquinolone tolerance. J. Med. Microbiol. 2011, 60 Pt 3, 329–336. [Google Scholar] [CrossRef]
- Silver, L.L. Fosfomycin: Mechanism and Resistance. Cold Spring Harb. Perspect. Med. 2017, 7, a025262. [Google Scholar] [CrossRef]
- Gisin, J.; Schneider, A.; Nagele, B.; Borisova, M.; Mayer, C. A cell wall recycling shortcut that bypasses peptidoglycan de novo biosynthesis. Nat. Chem. Biol. 2013, 9, 491–493. [Google Scholar] [CrossRef]
- Fumeaux, C.; Bernhardt, T.G. Identification of MupP as a New Peptidoglycan Recycling Factor and Antibiotic Resistance Determinant in Pseudomonas aeruginosa. mBio 2017, 8, e00102-17. [Google Scholar] [CrossRef] [PubMed]
- Borisova, M.; Gisin, J.; Mayer, C. Blocking peptidoglycan recycling in Pseudomonas aeruginosa attenuates intrinsic resistance to fosfomycin. Microb. Drug Resist. 2014, 20, 231–237. [Google Scholar] [CrossRef] [PubMed]
- Sanz-Garcia, F.; Hernando-Amado, S.; Martinez, J.L. Mutational Evolution of Pseudomonas aeruginosa Resistance to Ribosome-Targeting Antibiotics. Front. Genet. 2018, 9, 451. [Google Scholar] [CrossRef]
- Sanz-García, F.; Hernando-Amado, S.; Martínez, J.L. Mutation-Driven Evolution of Pseudomonas aeruginosa in the Presence of either Ceftazidime or Ceftazidime-Avibactam. Antimicrob. Agents Chemother. 2018, 62, e01379-18. [Google Scholar] [CrossRef]
- Bassetti, M.; Vena, A.; Croxatto, A.; Righi, E.; Guery, B. How to manage Pseudomonas aeruginosa infections. Drugs Context 2018, 7, 212527. [Google Scholar] [CrossRef]
- Laborda, P.; Martínez, J.L.; Hernando-Amado, S. Evolution of Habitat-Dependent Antibiotic Resistance in Pseudomonas aeruginosa. Microbiol. Spectr. 2022, 10, e0024722. [Google Scholar] [CrossRef]
- Laborda, P.; Martinez, J.L.; Hernando-Amado, S. Convergent phenotypic evolution towards fosfomycin collateral sensitivity of Pseudomonas aeruginosa antibiotic-resistant mutants. Microb. Biotechnol. 2022, 15, 613–629. [Google Scholar] [CrossRef] [PubMed]
- Hernando-Amado, S.; López-Causapé, C.; Laborda, P.; Sanz-García, F.; Oliver, A.; Martínez, J.L. Rapid Phenotypic Convergence towards Collateral Sensitivity in Clinical Isolates of Pseudomonas aeruginosa Presenting Different Genomic Backgrounds. Microbiol. Spectr. 2022, 11, e0227622. [Google Scholar] [CrossRef] [PubMed]
- Cabot, G.; Ocampo-Sosa, A.A.; Dominguez, M.A.; Gago, J.F.; Juan, C.; Tubau, F.; Rodriguez, C.; Moya, B.; Pena, C.; Martinez-Martinez, L.; et al. Genetic markers of widespread extensively drug-resistant Pseudomonas aeruginosa high-risk clones. Antimicrob. Agents Chemother. 2012, 56, 6349–6357. [Google Scholar] [CrossRef] [PubMed]
- Del Barrio-Tofino, E.; Lopez-Causape, C.; Oliver, A. Pseudomonas aeruginosa epidemic high-risk clones and their association with horizontally-acquired beta-lactamases: 2020 update. Int. J. Antimicrob. Agents 2020, 56, 106196. [Google Scholar] [CrossRef] [PubMed]
- Hernando-Amado, S.; Laborda, P.; Valverde, J.R.; Martínez, J.L. Rapid Decline of Ceftazidime Resistance in Antibiotic-Free and Sublethal Environments Is Contingent on Genetic Background. Mol. Biol. Evol. 2022, 39, msac049. [Google Scholar] [CrossRef]
- Roemhild, R.; Gokhale, C.S.; Dirksen, P.; Blake, C.; Rosenstiel, P.; Traulsen, A.; Andersson, D.I.; Schulenburg, H. Cellular hysteresis as a principle to maximize the efficacy of antibiotic therapy. Proc. Natl. Acad. Sci. USA 2018, 115, 9767–9772. [Google Scholar] [CrossRef]
- Knoppel, A.; Nasvall, J.; Andersson, D.I. Evolution of antibiotic resistance without antibiotic exposure. Antimicrob. Agents Chemother. 2017, 61, e01495-17. [Google Scholar] [CrossRef]
- Diez-Aguilar, M.; Morosini, M.I.; Tedim, A.P.; Rodriguez, I.; Aktas, Z.; Canton, R. Antimicrobial Activity of Fosfomycin-Tobramycin Combination against Pseudomonas aeruginosa Isolates Assessed by Time-Kill Assays and Mutant Prevention Concentrations. Antimicrob. Agents Chemother. 2015, 59, 6039–6045. [Google Scholar] [CrossRef]
- Hernando-Amado, S.; Sanz-García, F.; Martínez, J.L. Rapid and robust evolution of collateral sensitivity in Pseudomonas aeruginosa antibiotic-resistant mutants. Sci. Adv. 2020, 6, eaba5493. [Google Scholar] [CrossRef]
- Sarbu, C.; Pop, H.F. Principal component analysis versus fuzzy principal component analysis A case study: The quality of danube water (1985–1996). Talanta 2005, 65, 1215–1220. [Google Scholar] [CrossRef]
- Gower, J.C.; Hand, D.J. Biplots; Chapman Hall: London, UK, 1996; p. 71630. [Google Scholar]
- Wang, D.; Dorosky, R.J.; Han, C.S.; Lo, C.C.; Dichosa, A.E.; Chain, P.S.; Yu, J.M.; Pierson, L.S., 3rd; Pierson, E.A. Adaptation genomics of a small-colony variant in a Pseudomonas chlororaphis 30-84 biofilm. Appl. Environ. Microbiol. 2015, 81, 890–899. [Google Scholar] [CrossRef] [PubMed]
- Feng, Y.; Jonker, M.J.; Moustakas, I.; Brul, S.; Ter Kuile, B.H. Dynamics of Mutations during Development of Resistance by Pseudomonas aeruginosa against Five Antibiotics. Antimicrob. Agents Chemother. 2016, 60, 4229–4236. [Google Scholar] [CrossRef]
- McCoy, A.J.; Sandlin, R.C.; Maurelli, A.T. In vitro and in vivo functional activity of Chlamydia MurA, a UDP-N-acetylglucosamine enolpyruvyl transferase involved in peptidoglycan synthesis and fosfomycin resistance. J. Bacteriol. 2003, 185, 1218–1228. [Google Scholar] [CrossRef]
- Borisova, M.; Gisin, J.; Mayer, C. The N-Acetylmuramic Acid 6-Phosphate Phosphatase MupP Completes the Pseudomonas Peptidoglycan Recycling Pathway Leading to Intrinsic Fosfomycin Resistance. mBio 2017, 8, e00092-17. [Google Scholar] [CrossRef]
- R Core Team. R Package Stats: A Language and Environment for Statistical Computing; The R Project for Statistical Computing: Vienna, Austria, 2015. [Google Scholar]
- Lê, S.; Josse, J.; Husson, F. FactoMineR: An R Package for Multivariate Analysis. J. Stat. Softw. 2008, 25, 1–18. [Google Scholar]
- Kassambara, A. Practical Guide to Principal Component Methods in R: PCA, M (CA), FAMD, MFA, HCPC, Factoextra; Sthda: Marseille, France, 2016; p. 2. [Google Scholar]
- Pfaffl, M.W. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 2001, 29, e45. [Google Scholar] [CrossRef] [PubMed]
| P. aeruginosa Clinical Isolates | |||
|---|---|---|---|
| Isolate ID | Origin | ST | Mutations in Genes/Proteins Associated with Resistance |
| AND04-004A | Tracheal aspirate | 244 | mexT (nt234∆8). |
| ARA02-005 | Blood | 267 | mexT (nt234∆8); OprN (R151L); ArmZ (R320C). |
| BAL02-001 | Sputum | 1619 | mexT (nt234∆8). |
| BAL04-002 | Blood | 1816 | MexA (K86E); mexT (nt234∆8); AmpR (G295R); AmpC (A278G); ParE (E215Q). |
| CAN01-002 | Sputum | 111 | MexB (Q319X); MexY (G530S); MexZ (Q140K); MexT (G276D, nt234∆8); |
| CAN01-003 | Sputum | 111 | ParS (L137P, nt998∆1); MexY (G530S); mexT (nt234∆8); GyrA (T83I, V671I, G860S); Mpl (aa436InsGGF); ParC (S87L). |
| CAT02-004 | Blood | 244 | mexR (nt266InsG); mexT (nt234∆8); DacB (PBP4) (A340G). |
| CAT06-005 | Sputum | 175 | OprM (T198P); OprD (Q142X); MexZ (G195D); mexT (nt234∆8); GyrA (T83I, D87N); AmpR (G154R); ParC (S87W, L168Q); ArmZ (V266M). |
| CAT09-004 | Blood | 244 | mexT (nt234∆8). |
| CLE03-004 | Tracheal aspirate | 381 | mexZ (nt386∆1); mexT (nt234∆8); AmpC (S173N). |
| CLM01-003 | Sputum | 709 | GyrB (S466Y); oprM (nt501∆5); mexT (nt234∆8); AmpR (D135N); FusA1 (Q678L). |
| CVA01-006 | Sputum | 175 | GyrB (aadB); mexB (nt881InsGG); oprD (nt55InsGCACT); MexZ (G195D); mexT (nt234∆8); GyrA (T83I, D87N); ParC (S87W, L168Q); ArmZ (V266M). |
| EXT01-004 | Sputum | 3353 | GyrB (S466Y); MexY (D724V); MexS (V308I); mexT (nt234∆8); Mpl (P274L); AmpC (D280N, Q311L); FtsI (PBP3) (L219V); AmpD (D83G); OprJ (N121S); MexD (H953N). |
| GAL01-001 | Sputum | 560 | mexT (nt234∆8); GyrA (T83I); mpl (nt111InsC). |
| GAL02-004 | Sputum | 217 | GyrB (R22C); mexB (nt3018∆5); MexS (P225L); mexT (nt234∆8); GyrA (A908T); Mpl (H190Y); AmpC (V239A); FusA1 (T671A). |
| ICA01-004 | Sputum | 698 | mexT (nt234∆8); MexE (V156A); AmpR (E162Q); ArmZ (V266M). |
| MAD04-002 | Sputum | 242 | mexT (nt234∆8); ParC (K726R) |
| MAD05-009 | Sputum (CF) | 27 | GyrB (S466F); MexZ (A88P); mexT (nt234∆8); AmpC (P274L). |
| FQSE03-1212-2 | Sputum (CF) | 274 | MexA (L338P); MexZ (A144V); mexT (nt234∆8); GyrA (D87N). |
| FQSE10-0503 | Sputum (CF) | 274 | MexY (V875M); MexZ (IS); mexT (nt234∆8). |
| FQSE110603 | Sputum (CF) | 701 | MexB (473962delC); MexY (N709H,A586T); MexX (A38P); mexT (nt234∆8); OprN (R363H); AmpDh2 (P116S). |
| FQSE15-0803 | Sputum (CF) | 274 | GyrB (E788A); MexA (L338P); MexX (D346H); MexZ (A144V); mexT (nt234∆8); PmrB (E213D); |
| FQSE24-0304 | Sputum (CF) | 1089 | GyrB (S466F); MexA (L338P); OprM (E456G); OprD (V67*); MexY (Y355H); MexX (D346H); MexZ (A194P); GalU (P123L); mexT (nt234∆8); mexF (Nt1051ins9); FusA1 (K430E); PmrB (R287Q). |
| Oligonucleotides | |
|---|---|
| Name | Sequence 5′-3′ |
| murA_fw | CATTTCCGGCGCGAAGAACT |
| murA_rv | ATGCTGCTGGCGTCGACTTCGA |
| agmK_fw | AGCTGAATCGCTGGTTGGAC |
| agmK_rv | AACGGTCGGCAGTCTTCCTG |
| nagZ_fw | AGGTGGGCGGGCTGATCATCTT |
| nagZ_rv | ATTGGGGTTGTCGGCGATCG |
| mupP_fw | GCCGGACTTCATCGCCATCA |
| mupP_rv | AATGCTCCTGGTAGCGGTCGAG |
| fosA_fw | ACCAGGGCGCCTATCTCGAA |
| fosA_rv | CGCTGCGGTTCTGCTTCCAT |
| anmK_fw | CAACGTGCTGATGGACGCCT |
| anmK_rv | AGCCAGGACAGGTTGAAGCG |
| rplU_fw | CGCAGTGATTGTTACCGGTG |
| rplU_rv | AGGCCTGAATGCCGGTGATC |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Genova, R.; Laborda, P.; Cuesta, T.; Martínez, J.L.; Sanz-García, F. Collateral Sensitivity to Fosfomycin of Tobramycin-Resistant Mutants of Pseudomonas aeruginosa Is Contingent on Bacterial Genomic Background. Int. J. Mol. Sci. 2023, 24, 6892. https://doi.org/10.3390/ijms24086892
Genova R, Laborda P, Cuesta T, Martínez JL, Sanz-García F. Collateral Sensitivity to Fosfomycin of Tobramycin-Resistant Mutants of Pseudomonas aeruginosa Is Contingent on Bacterial Genomic Background. International Journal of Molecular Sciences. 2023; 24(8):6892. https://doi.org/10.3390/ijms24086892
Chicago/Turabian StyleGenova, Roberta, Pablo Laborda, Trinidad Cuesta, José Luis Martínez, and Fernando Sanz-García. 2023. "Collateral Sensitivity to Fosfomycin of Tobramycin-Resistant Mutants of Pseudomonas aeruginosa Is Contingent on Bacterial Genomic Background" International Journal of Molecular Sciences 24, no. 8: 6892. https://doi.org/10.3390/ijms24086892
APA StyleGenova, R., Laborda, P., Cuesta, T., Martínez, J. L., & Sanz-García, F. (2023). Collateral Sensitivity to Fosfomycin of Tobramycin-Resistant Mutants of Pseudomonas aeruginosa Is Contingent on Bacterial Genomic Background. International Journal of Molecular Sciences, 24(8), 6892. https://doi.org/10.3390/ijms24086892






