The Yeast Permease Agp2 Senses Cycloheximide and Undergoes Degradation That Requires the Small Protein Brp1-Cellular Fate of Agp2 in Response to Cycloheximide
Abstract
1. Introduction
2. Results
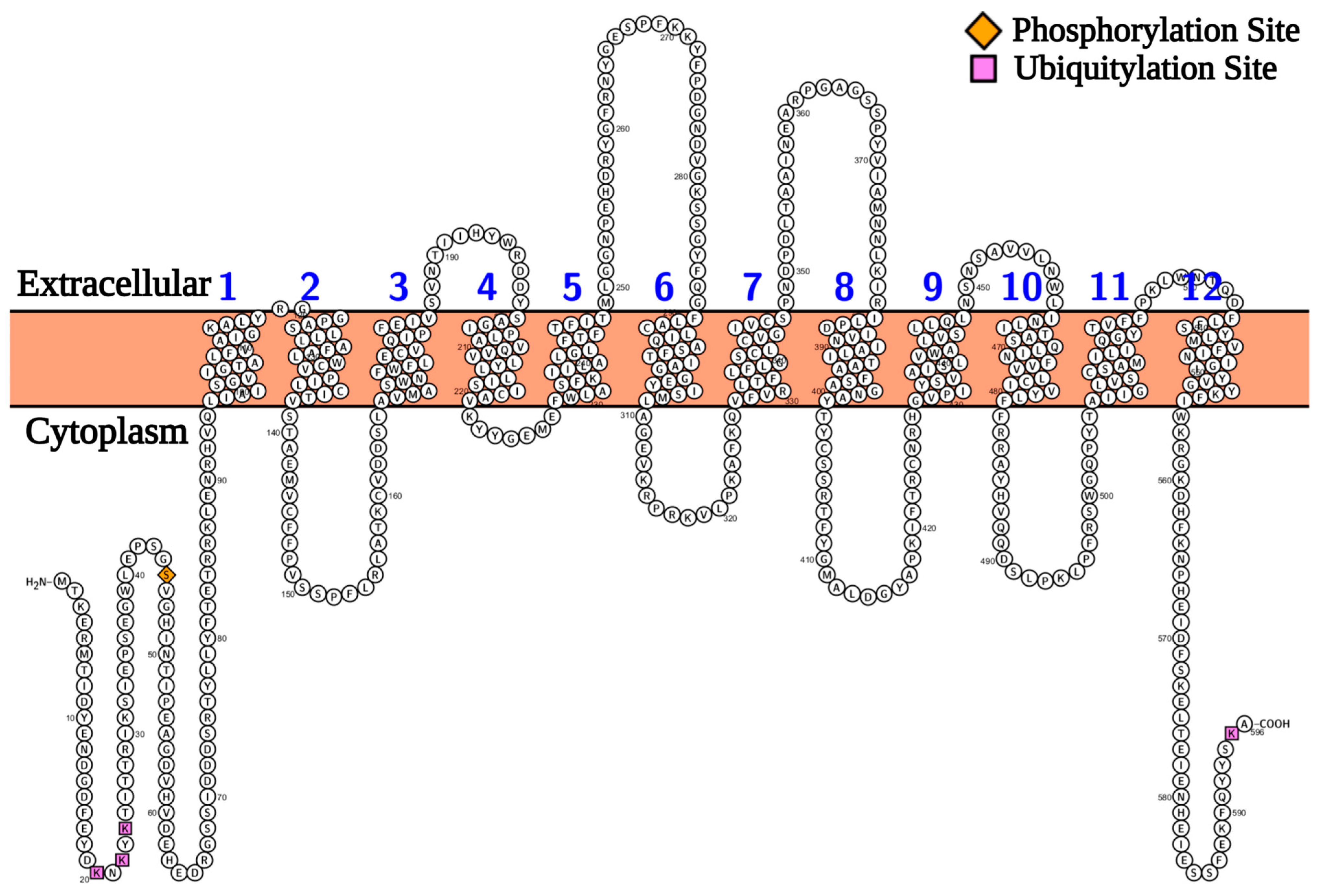
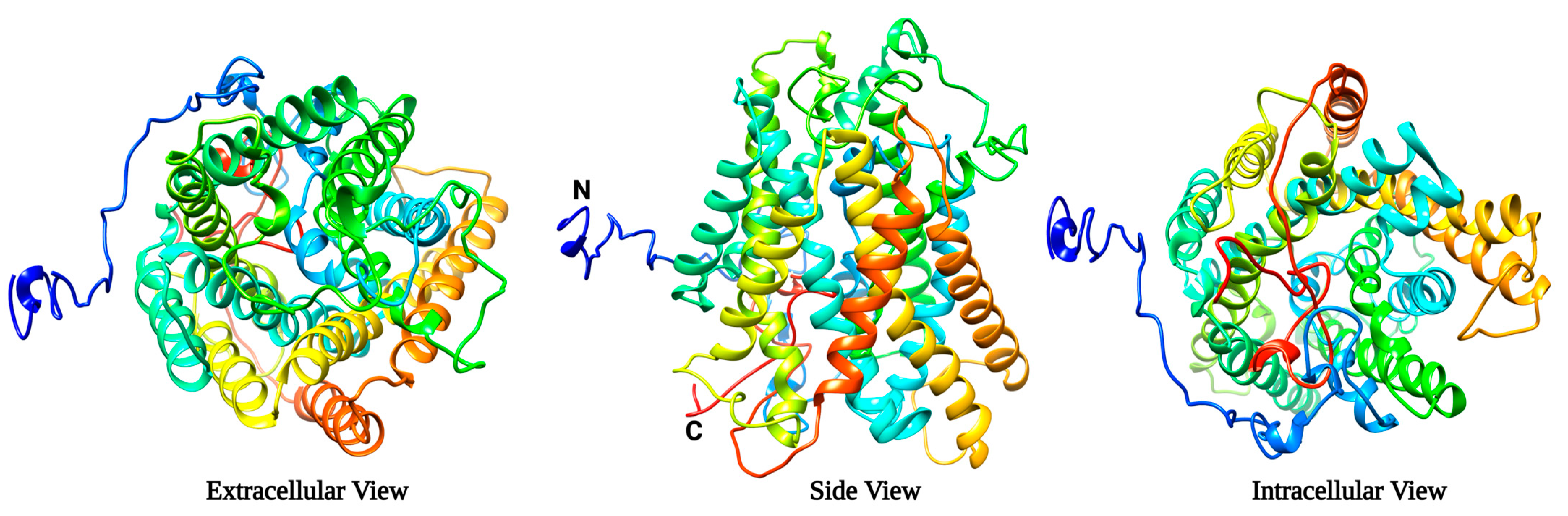
2.1. Protein Modelling Studies Reveal Several Structural Features of Agp2
2.2. Protein-Ligand Docking Reveals That CHX Binds to Agp2
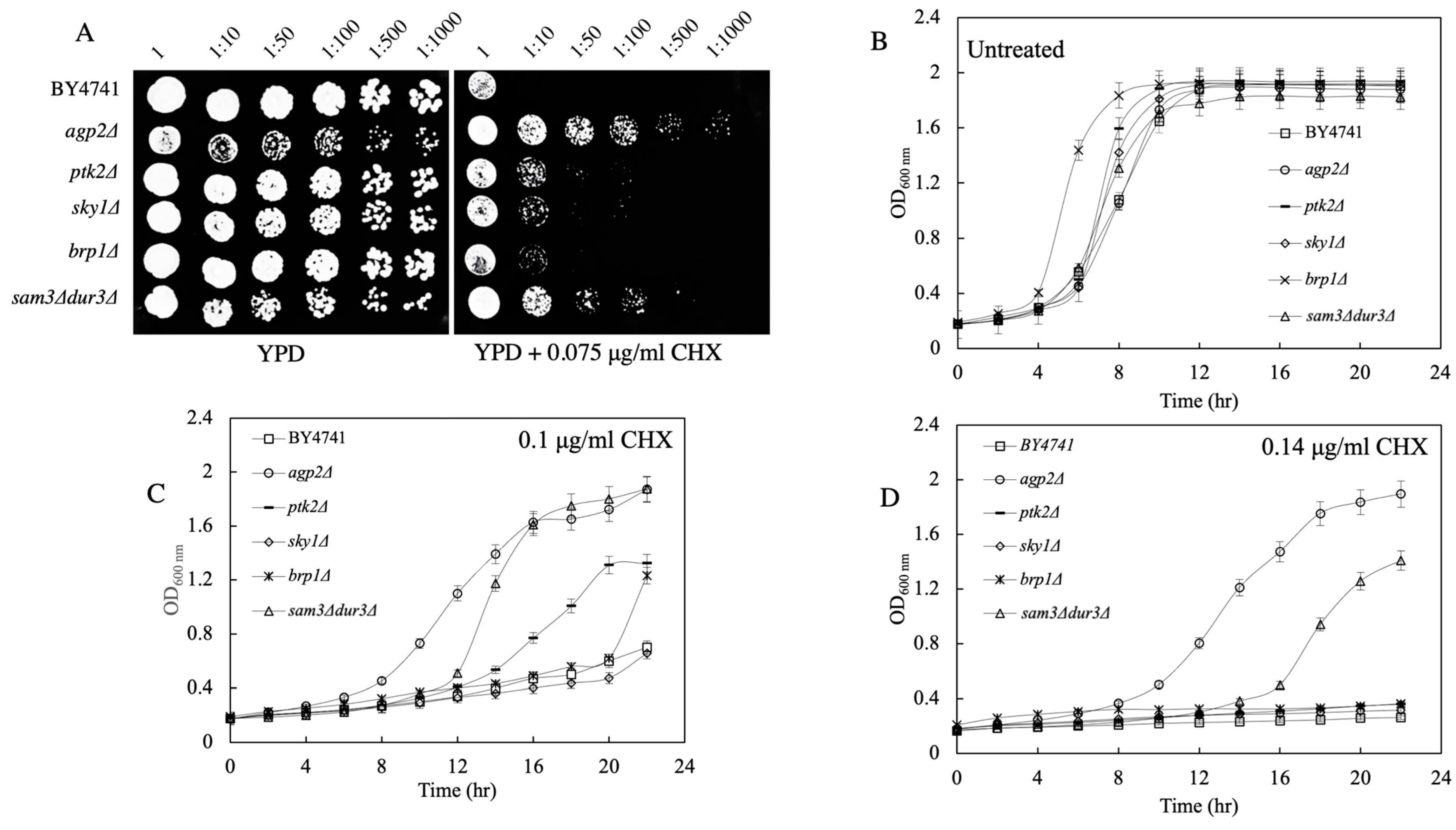
2.3. Spot Test and Growth Curve Analyses Reveal That agp2Δ Mutants Are Resistant to CHX
2.4. agp2Δ Mutant Displays WT Resistance to Rapamycin
2.5. CHX Triggers Agp2 Disappearance in a Time-and Concentration-Dependent Manner
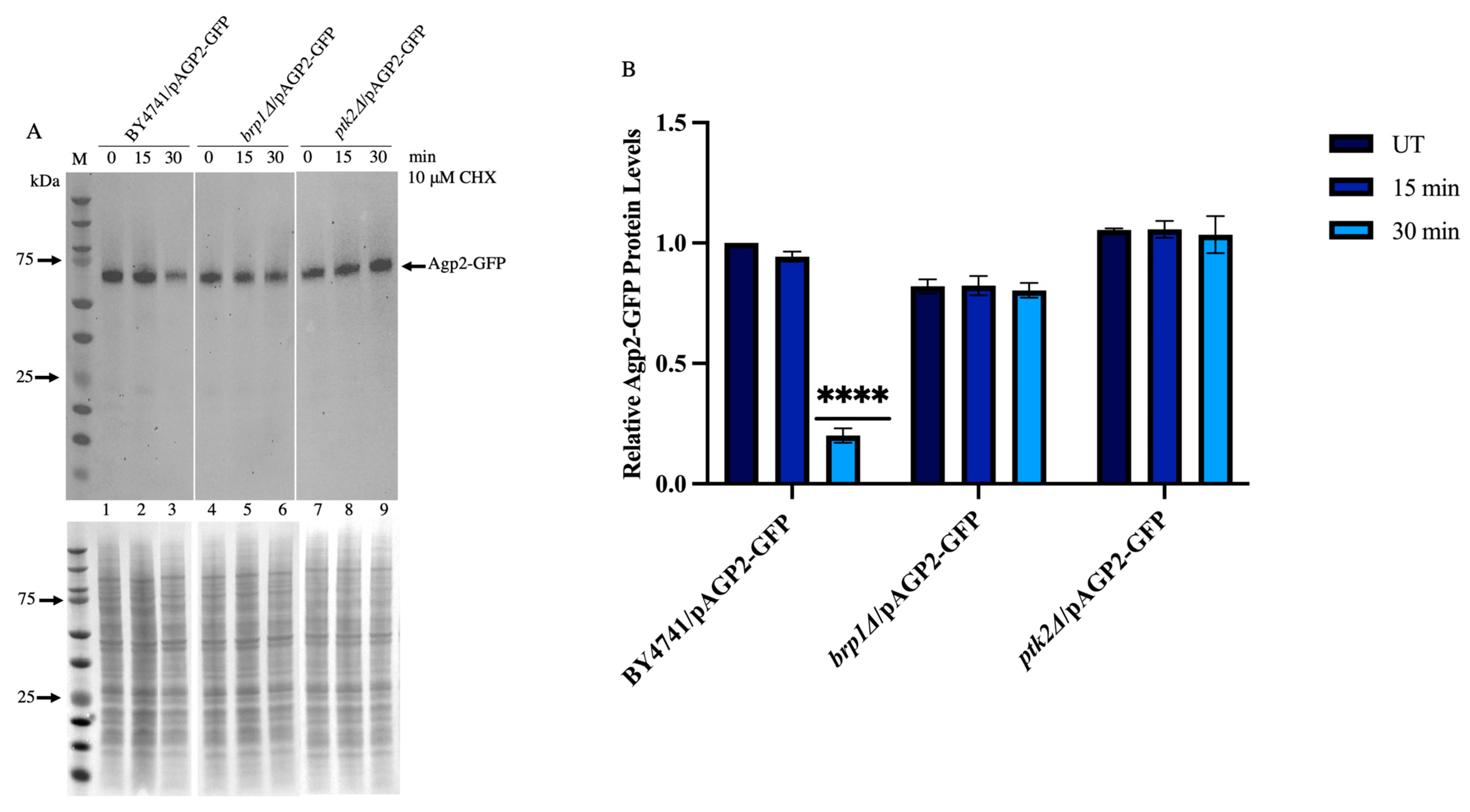
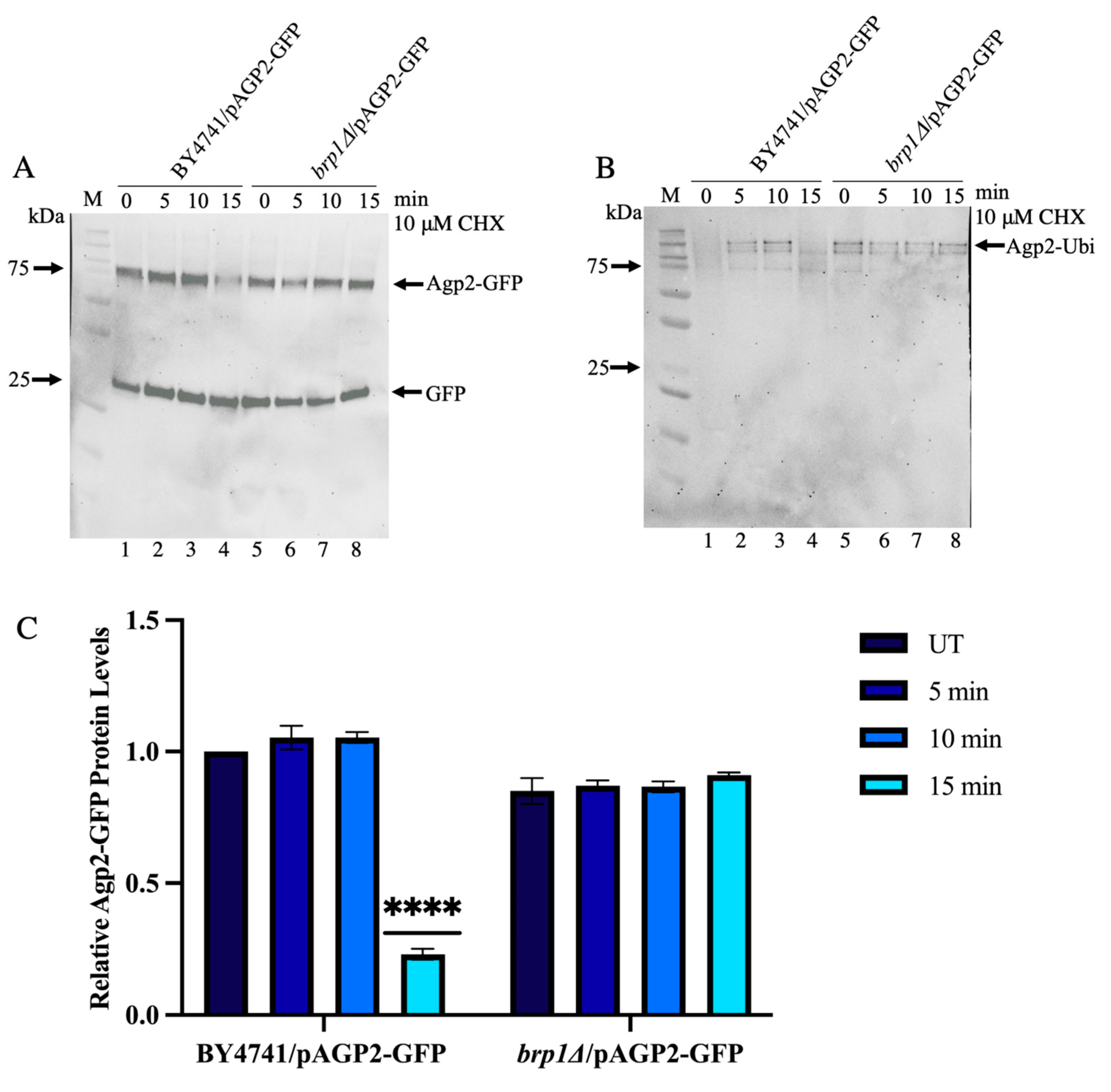
2.6. CHX-Induced Disappearance of Agp2-GFP Is Defective in the brp1Δ and ptk2Δ Mutants
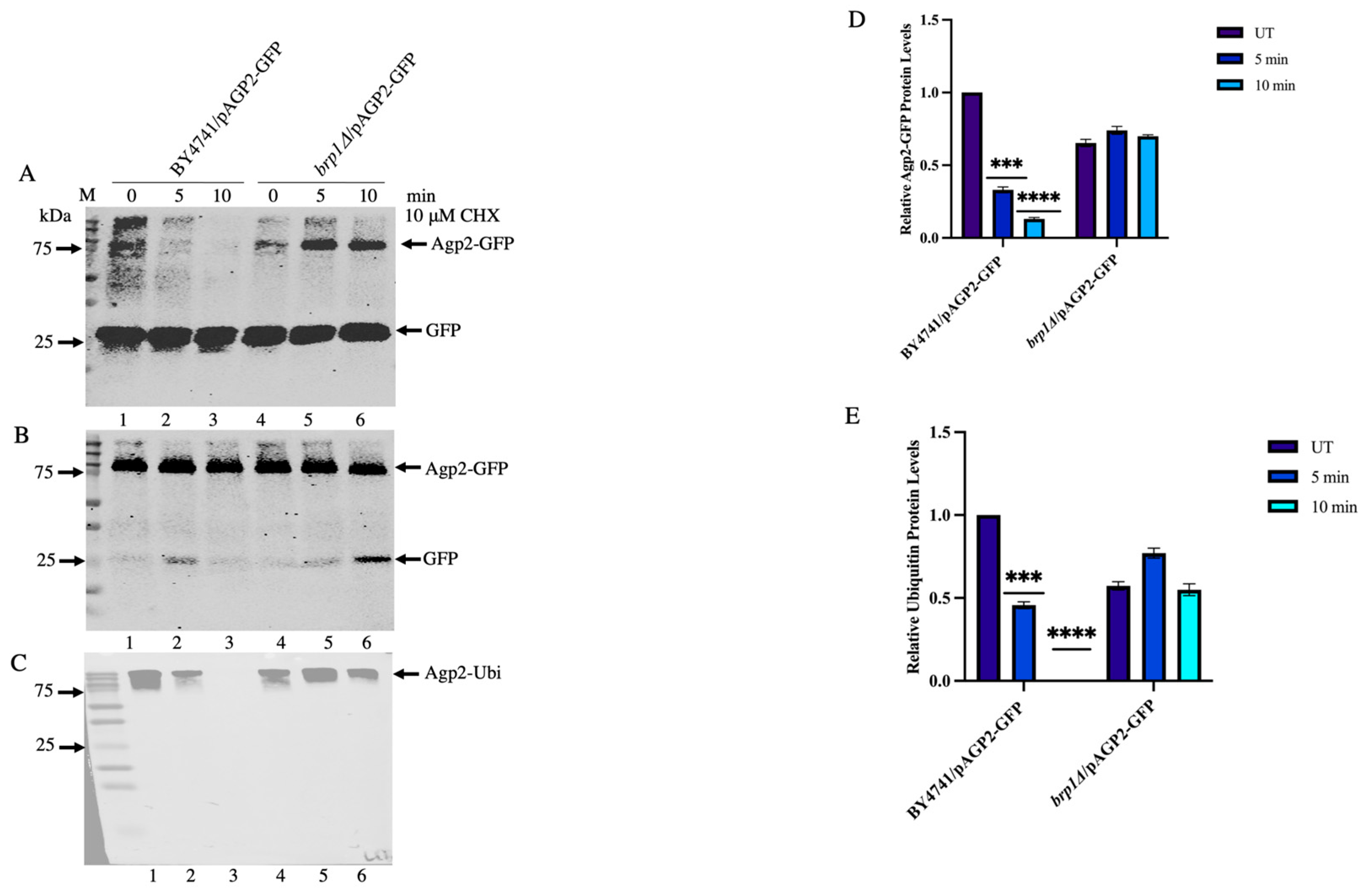
2.7. Immunoprecipitated Agp2-GFP Contains High Molecular Weight Species That Are Ubiquitinylated and Disappear upon CHX Treatment in WT, Not in the brp1Δ Mutant
2.8. Saturated Cultures Contain Significantly High Levels of Ubiquitinylated Agp2-GFP Species, Which Are Susceptible to CHX-Induced Loss in the WT and Not in the brp1Δ Mutant
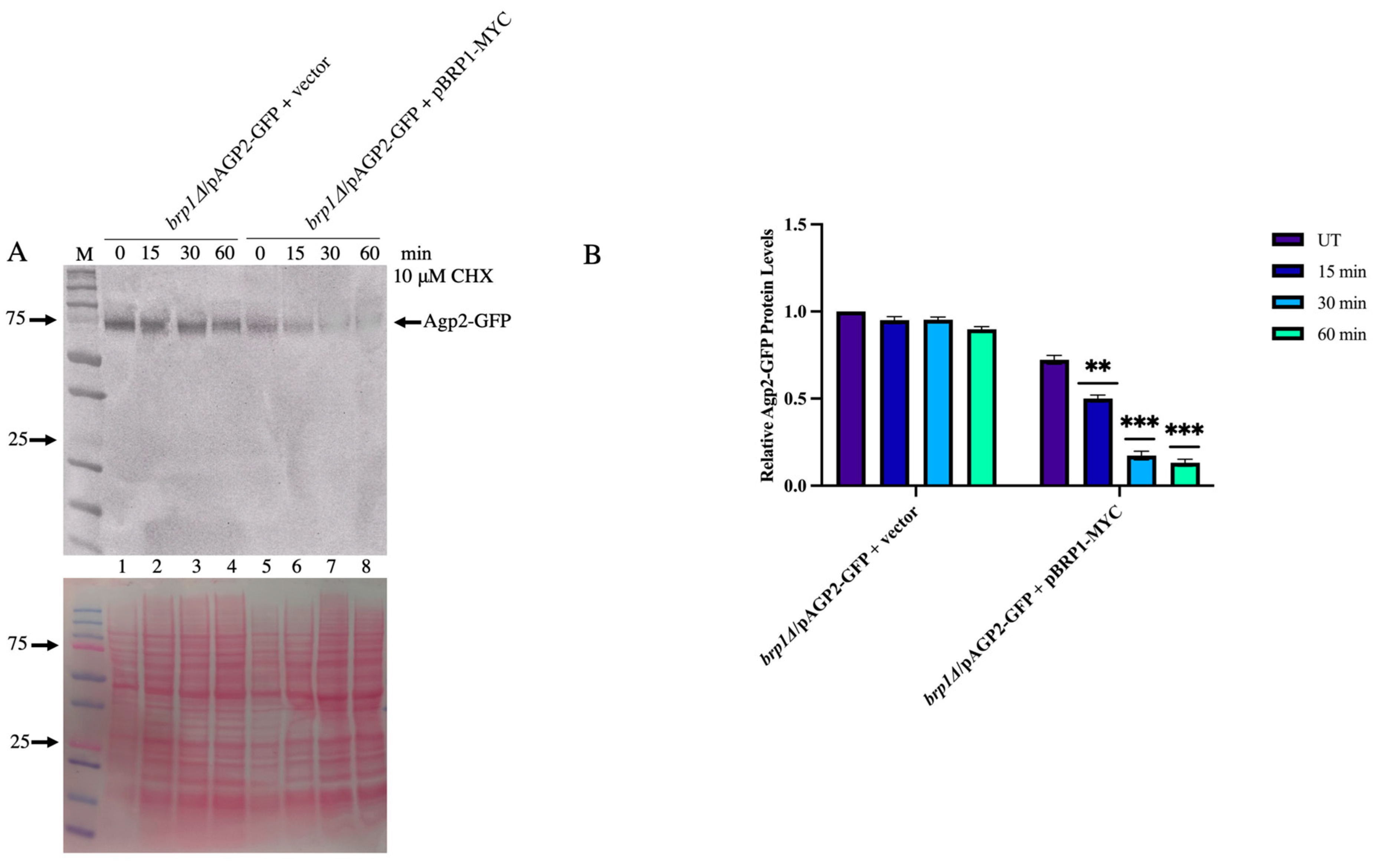
2.9. Expression of Brp1 in the brp1Δ Mutant Re-Instates the Degradation of Agp2-GFP
2.10. Intracellular Distribution of Agp2-GFP Is Dependent upon Brp1
3. Discussion
4. Materials and Methods
4.1. Strains, Media, Transformation and Reagents—The S. cerevisiae Strains Used in the Present Study Are Listed in Table 1
| Strains | Genotypes | Sources |
|---|---|---|
| BY4741 Wild type | MATα his3Δ leu2Δ met15Δ ura3Δ | This laboratory collection |
| BY4741(sky1Δ) | Isogenic to BY4741, except sky1Δ::KAN | This laboratory collection |
| BY4741(brp1Δ) | Isogenic to BY4741, except brp1Δ::KAN | This laboratory collection |
| BY4741(ptk2Δ) | Isogenic to BY4741, except ptk2Δ::KAN | This laboratory collection |
| BY4741(agp2Δ) | Isogenic to BY4741, except agp2Δ::KAN | This laboratory collection |
| BY4741 Wild type | carrying the single copy plasmid pAGP2-GFP on URA3 selection. | This study |
| BY4741(agp2Δ) | carrying the single copy plasmid pAGP2-GFP on URA3 selection. | This study |
| BY4741 (brp1Δ) | carrying the single copy plasmid pAGP2-GFP on URA3 selection. | This study |
| BY4741 (ptk2Δ) | carrying the single copy plasmid pAGP2-GFP on URA3 selection. | This study |
| BY4741 (sam3Δ dur3Δ) | Isogenic to BY4741, except sam3Δ::HIS3 dur3Δ::KAN | This laboratory collection |
| BY4741 (brp1Δ)/pAGP2-GFP + vector. | carrying the single copy plasmid pAGP2-GFP on URA3 selection. | This study |
| BY4741 (brp1Δ)/pAGP2-GFP + pBRP1-MYC. | carrying the single copy plasmids pAGP2-GFP on URA3 selection and pBRP-MYC on LEU2 selection | This study |
4.2. Protein Modelling and Docking Studies
4.3. Spot Test Analysis of Cell Growth
4.4. Construction of Plasmid pBRP1-MYC
4.5. CHX Treatment
4.6. Plasma Membrane and Total Cell Extraction
4.7. Western Blotting
4.8. Immunoprecipitation Assay
4.9. Confocal Imaging
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- van Roermund, C.W.; Hettema, E.H.; van den Berg, M.; Tabak, H.F.; Wanders, R.J. Molecular characterization of carnitine-dependent transport of acetyl-CoA from peroxisomes to mitochondria in Saccharomyces cerevisiae and identification of a plasma membrane carnitine transporter, Agp2p. Embo J. 1999, 18, 5843–5852. [Google Scholar] [CrossRef] [PubMed]
- Schreve, J.L.; Garrett, J.M. Yeast Agp2p and Agp3p function as amino acid permeases in poor nutrient conditions. Biochem. Biophys. Res. Commun. 2004, 313, 745–751. [Google Scholar] [CrossRef]
- Iraqui, I.; Vissers, S.; Bernard, F.; de Craene, J.O.; Boles, E.; Urrestarazu, A.; Andre, B. Amino acid signaling in Saccharomyces cerevisiae: A permease-like sensor of external amino acids and F-Box protein Grr1p are required for transcriptional induction of the AGP1 gene, which encodes a broad-specificity amino acid permease. Mol. Cell. Biol. 1999, 19, 989–1001. [Google Scholar] [CrossRef]
- Lee, J.; Lee, B.; Shin, N.; Kwak, S.-S.; Bahk, J.D.; Lim, C.O.; Yun, D.-J. Carnitine uptake by AGP2 in yeast Saccharomyces cerevisiae is dependent on Hog1 MAP kinase pathway. Mol. Cells 2002, 13, 407–412. [Google Scholar]
- Aouida, M.; Pagé, N.; Leduc, A.; Peter, M.; Ramotar, D. A Genome-Wide Screen in Saccharomyces cerevisiae Reveals Altered Transport As a Mechanism of Resistance to the Anticancer Drug Bleomycin. Cancer Res. 2004, 64, 1102–1109. [Google Scholar] [CrossRef] [PubMed]
- Aouida, M.; Leduc, A.; Wang, H.; Ramotar, D. Characterization of a transport and detoxification pathway for the antitumour drug bleomycin in Saccharomyces cerevisiae. Biochem. J. 2004, 384, 47–58. [Google Scholar] [CrossRef] [PubMed]
- Aouida, M.; Leduc, A.; Poulin, R.; Ramotar, D. AGP2 Encodes the Major Permease for High Affinity Polyamine Import in Saccharomyces cerevisiae. J. Biol. Chem. 2005, 280, 24267–24276. [Google Scholar] [CrossRef]
- Uemura, T.; Kashiwagi, K.; Igarashi, K. Polyamine Uptake by DUR3 and SAM3 in Saccharomyces cerevisiae. J. Biol. Chem. 2007, 282, 7733–7741. [Google Scholar] [CrossRef]
- Gaber, R.F.; Ottow, K.; Andersen, H.A.; Kielland-Brandt, M.C. Constitutive and Hyperresponsive Signaling by Mutant Forms of Saccharomyces cerevisiae Amino Acid Sensor Ssy1. Eukaryot. Cell 2003, 2, 922–929. [Google Scholar] [CrossRef]
- Van Nuland, A.; Vandormael, P.; Donaton, M.; Alenquer, M.; Lourenço, A.; Quintino, E.; Versele, M.; Thevelein, J.M. Ammonium permease-based sensing mechanism for rapid ammonium activation of the protein kinase A pathway in yeast. Mol. Microbiol. 2006, 59, 1485–1505. [Google Scholar] [CrossRef]
- Van Zeebroeck, G.; Kimpe, M.; Vandormael, P.; Thevelein, J.M. A Split-Ubiquitin Two-Hybrid Screen for Proteins Physically Interacting with the Yeast Amino Acid Transceptor Gap1 and Ammonium Transceptor Mep2. PLoS ONE 2011, 6, e24275. [Google Scholar] [CrossRef]
- Didion, T.; Regenberg, B.; Jorgensen, M.U.; Kielland-Brandt, M.; Andersen, H.A. The permease homologue Ssy1p controls the expression of amino acid and peptide transporter genes in Saccharomyces cerevisiae. Mol. Microbiol. 1998, 27, 643–650. [Google Scholar] [CrossRef]
- Ljungdahl, P.O. Amino-acid-induced signalling via the SPS-sensing pathway in yeast. Biochem. Soc. Trans. 2009, 37 Pt 1, 242–247. [Google Scholar] [CrossRef] [PubMed]
- Brosseau, N.; Andreev, E.; Ramotar, D. Complementation of the Yeast Model System Reveals that Caenorhabditis elegans OCT-1 Is a Functional Transporter of Anthracyclines. PLoS ONE 2015, 10, e0133182. [Google Scholar] [CrossRef] [PubMed]
- Aouida, M.; Texeira, M.R.; Thevelein, J.M.; Poulin, R.; Ramotar, D. Agp2, a Member of the Yeast Amino Acid Permease Family, Positively Regulates Polyamine Transport at the Transcriptional Level. PLoS ONE 2013, 8, e65717. [Google Scholar] [CrossRef]
- Kaouass, M.; Audette, M.; Ramotar, D.; Verma, S.; De Montigny, D.; Gamache, I.; Torossian, K.; Poulin, R. The STK2 gene, which encodes a putative Ser/Thr protein kinase, is required for high-affinity spermidine transport in Saccharomyces cerevisiae. Mol. Cell. Biol. 1997, 17, 2994–3004. [Google Scholar] [CrossRef]
- Goossens, A.; de la Fuente, N.; Forment, J.; Serrano, R.; Portillo, F. Regulation of Yeast H+-ATPase by Protein Kinases Belonging to a Family Dedicated to Activation of Plasma Membrane Transporters. Mol. Cell. Biol. 2000, 20, 7654–7661. [Google Scholar] [CrossRef]
- Erez, O.; Kahana, C. Screening for Modulators of Spermine Tolerance Identifies Sky1, the SR Protein Kinase of Saccharomyces cerevisiae, as a Regulator of Polyamine Transport and Ion Homeostasis. Mol. Cell. Biol. 2001, 21, 175–184. [Google Scholar] [CrossRef]
- Erez, O.; Kahana, C. Deletions of SKY1 or PTK2 in the Saccharomyces cerevisiae trk1Deltatrk2Delta mutant cells exert dual effect on ion homeostasis. Biochem. Biophys. Res. Commun. 2002, 295, 1142–1149. [Google Scholar] [CrossRef]
- Forment, J.; Mulet, J.M.; Vicente, O.; Serrano, R. The yeast SR protein kinase Sky1p modulates salt tolerance, membrane potential and the Trk1,2 potassium transporter. Biochim. Biophys. Acta (BBA)-Biomembr. 2002, 1565, 36–40. [Google Scholar] [CrossRef]
- The UniProt Consortium; Bateman, A.; Martin, M.-J.; Orchard, S.; Magrane, M.; Ahmad, S.; Alpi, E.; Bowler-Barnett, E.H.; Britto, R.; Bye-A-Jee, H.; et al. UniProt: The Universal Protein Knowledgebase in 2023. Nucleic Acids Res. 2023, 51, D523–D531. [Google Scholar] [CrossRef]
- Omasits, U.; Ahrens, C.H.; Müller, S.; Wollscheid, B. Protter: Interactive protein feature visualization and integration with experimental proteomic data. Bioinformatics 2013, 30, 884–886. [Google Scholar] [CrossRef] [PubMed]
- Beck, T.D.I.; Schmidt, A.; Hall, M.N. Starvation Induces Vacuolar Targeting and Degradation of the Tryptophan Permease in Yeast. J. Cell Biol. 1999, 146, 1227–1238. [Google Scholar] [CrossRef] [PubMed]
- Yang, J.; Yan, R.; Roy, A.; Xu, D.; Poisson, J.; Zhang, Y. The I-TASSER Suite: Protein structure and function prediction. Nat. Methods 2015, 12, 7–8. [Google Scholar] [CrossRef] [PubMed]
- Laskowski, R.A.; MacArthur, M.W.; Moss, D.S.; Thornton, J.M. PROCHECK: A program to check the stereochemical quality of protein structures. J. Appl. Crystallogr. 1993, 26, 283–291. [Google Scholar] [CrossRef]
- Laskowski, R.A.; Jabłońska, J.; Pravda, L.; Vařeková, R.S.; Thornton, J. PDBsum: Structural summaries of PDB entries. Protein Sci. 2017, 27, 129–134. [Google Scholar] [CrossRef]
- Laskowski, R.A.; Swindells, M.B. LigPlot+: Multiple ligand–protein interaction diagrams for drug discovery. J. Chem. Inf. Model. 2011, 51, 2778–2786. [Google Scholar] [CrossRef]
- Paysan-Lafosse, T.; Blum, M.; Chuguransky, S.; Grego, T.; Pinto, B.L.; Salazar, G.A.; Bileschi, M.L.; Bork, P.; Bridge, A.; Colwell, L.; et al. InterPro in 2022. Nucleic Acids Res. 2023, 51, D418–D427. [Google Scholar] [CrossRef]
- Raught, B.; Gingras, A.C.; Sonenberg, N. The Target of Rapamycin (TOR) Proteins. Proc. Natl. Acad. Sci. USA 2001, 98, 7037–7044. [Google Scholar] [CrossRef]
- Sulaiman, A.A.; Ali, R.; Aouida, M.; Moovarkumudalvan, B.; Ramotar, D. The histone H2B Arg95 residue links the pheromone response pathway to rapamycin-induced G1 arrest in yeast. Sci. Rep. 2022, 12, 10023. [Google Scholar] [CrossRef]
- Law, B.K. Rapamycin: An anti-cancer immunosuppressant? Crit. Rev. Oncol. 2005, 56, 47–60. [Google Scholar] [CrossRef]
- Yokoe, H.; Meyer, T. Spatial dynamics of GFP-tagged proteins investigated by local fluorescence enhancement. Nat. Biotechnol. 1996, 14, 1252–1256. [Google Scholar] [CrossRef]
- Rana, M.S.; Wang, X.; Banerjee, A. An Improved Strategy for Fluorescent Tagging of Membrane Proteins for Overexpression and Purification in Mammalian Cells. Biochemistry 2018, 57, 6741–6751. [Google Scholar] [CrossRef] [PubMed]
- Drew, D.; Newstead, S.; Sonoda, Y.; Kim, H.; von Heijne, G.; Iwata, S. GFP-based optimization scheme for the overexpression and purification of eukaryotic membrane proteins in Saccharomyces cerevisiae. Nat. Protoc. 2008, 3, 784–798. [Google Scholar] [CrossRef] [PubMed]
- Porat, Z.; Wender, N.; Erez, O.; Kahana, C. Mechanism of polyamine tolerance in yeast: Novel regulators and insights. Cell. Mol. Life Sci. 2005, 62, 3106–3116. [Google Scholar] [CrossRef]
- Hirschberg, S.; Dvorzhak, A.; Rasooli-Nejad, S.M.A.; Angelov, S.; Kirchner, M.; Mertins, P.; Lättig-Tünnemann, G.; Harms, C.; Schmitz, D.; Grantyn, R. Uncoupling the Excitatory Amino Acid Transporter 2 From Its C-Terminal Interactome Restores Synaptic Glutamate Clearance at Corticostriatal Synapses and Alleviates Mutant Huntingtin-Induced Hypokinesia. Front. Cell. Neurosci. 2022, 15, 792652. [Google Scholar] [CrossRef]
- Jouvet, N.; Poschmann, J.; Douville, J.; Marrakchi, R.; Ramotar, D. RNA polymerase II degradation in response to rapamycin is not mediated through ubiquitylation. Biochem. Biophys. Res. Commun. 2011, 413, 248–253. [Google Scholar] [CrossRef] [PubMed]
- Bhattacharya, S.; Sae-Tia, S.; Fries, B.C. Candidiasis and Mechanisms of Antifungal Resistance. Antibiotics 2020, 9, 312. [Google Scholar] [CrossRef]
- Lauwers, E.; Jacob, C.; André, B. K63-linked ubiquitin chains as a specific signal for protein sorting into the multivesicular body pathway. J. Cell Biol. 2009, 185, 493–502. [Google Scholar] [CrossRef] [PubMed]
- Bianchi, F.; Klooster, J.S.V.; Ruiz, S.J.; Poolman, B. Regulation of Amino Acid Transport in Saccharomyces cerevisiae. Microbiol. Mol. Biol. Rev. 2019, 83, e00024-19. [Google Scholar] [CrossRef]
- Donaton, M.C.V.; Holsbeeks, I.; Lagatie, O.; Van Zeebroeck, G.; Crauwels, M.; Winderickx, J.; Thevelein, J.M. The Gap1 general amino acid permease acts as an amino acid sensor for activation of protein kinase A targets in the yeast Saccharomyces cerevisiae. Mol. Microbiol. 2003, 50, 911–929. [Google Scholar] [CrossRef] [PubMed]
- Zattas, D.; Hochstrasser, M. Ubiquitin-dependent protein degradation at the yeast endoplasmic reticulum and nuclear envelope. Crit. Rev. Biochem. Mol. Biol. 2015, 50, 1–17. [Google Scholar] [CrossRef]
- Braun, R.J. Ubiquitin-dependent proteolysis in yeast cells expressing neurotoxic proteins. Front. Mol. Neurosci. 2015, 8, 8. [Google Scholar] [CrossRef] [PubMed]
- Sherman, F. Guide to yeast genetics and molecular biology. Methods Enzymol. 1991, 194, 1–863. [Google Scholar]
- Gietz, D.; Jean, A.S.; Woods, R.A.; Schiestl, R.H. Improved method for high efficiency transformation of intact yeast cells. Nucleic Acids Res. 1992, 20, 1425. [Google Scholar] [CrossRef] [PubMed]
- Sherman, F.; Fink, G.; Hicks, J. Methods in Yeast Genetics: A Laboratory Course Manual; Cold Spring Harbor Laboratory Press: New York, NY, USA, 1987. [Google Scholar]
- Kim, S.; Chen, J.; Cheng, T.; Gindulyte, A.; He, J.; He, S.; Li, Q.; Shoemaker, B.A.; Thiessen, P.A.; Yu, B.; et al. PubChem in 2021: New data content and improved web interfaces. Nucleic Acids Res. 2021, 49, D1388–D1395. [Google Scholar] [CrossRef]
- Morris, G.M.; Huey, R.; Lindstrom, W.; Sanner, M.F.; Belew, R.K.; Goodsell, D.S.; Olson, A.J. AutoDock4 and AutoDockTools4: Automated docking with selective receptor flexibility. J. Comput. Chem. 2009, 30, 2785–2791. [Google Scholar] [CrossRef]
- Labbé, C.M.; Rey, J.; Lagorce, D.; Vavruša, M.; Becot, J.; Sperandio, O.; Villoutreix, B.; Tuffery, P.; Miteva, M. MTiOpenScreen: A web server for structure-based virtual screening. Nucleic Acids Res. 2015, 43, W448–W454. [Google Scholar] [CrossRef]
- Pettersen, E.F.; Goddard, T.D.; Huang, C.C.; Couch, G.S.; Greenblatt, D.M.; Meng, E.C.; Ferrin, T.E. UCSF Chimera? A visualization system for exploratory research and analysis. J. Comput. Chem. 2004, 25, 1605–1612. [Google Scholar] [CrossRef]
- Leduc, A.; He, C.H.; Ramotar, D. Disruption of the Saccharomyces cerevisiae cell-wall pathway gene SLG1 causes hypersensitivity to the antitumor drug bleomycin. Mol. Genet. Genom. 2003, 269, 78–89. [Google Scholar] [CrossRef]
- Vongsamphanh, R.; Fortier, P.-K.; Ramotar, D. Pir1p Mediates Translocation of the Yeast Apn1p Endonuclease into the Mitochondria To Maintain Genomic Stability. Mol. Cell. Biol. 2001, 21, 1647–1655. [Google Scholar] [CrossRef] [PubMed]



Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mohanty, A.; Alhaj Sulaiman, A.; Moovarkumudalvan, B.; Ali, R.; Aouida, M.; Ramotar, D. The Yeast Permease Agp2 Senses Cycloheximide and Undergoes Degradation That Requires the Small Protein Brp1-Cellular Fate of Agp2 in Response to Cycloheximide. Int. J. Mol. Sci. 2023, 24, 6975. https://doi.org/10.3390/ijms24086975
Mohanty A, Alhaj Sulaiman A, Moovarkumudalvan B, Ali R, Aouida M, Ramotar D. The Yeast Permease Agp2 Senses Cycloheximide and Undergoes Degradation That Requires the Small Protein Brp1-Cellular Fate of Agp2 in Response to Cycloheximide. International Journal of Molecular Sciences. 2023; 24(8):6975. https://doi.org/10.3390/ijms24086975
Chicago/Turabian StyleMohanty, Ashima, Abdallah Alhaj Sulaiman, Balasubramanian Moovarkumudalvan, Reem Ali, Mustapha Aouida, and Dindial Ramotar. 2023. "The Yeast Permease Agp2 Senses Cycloheximide and Undergoes Degradation That Requires the Small Protein Brp1-Cellular Fate of Agp2 in Response to Cycloheximide" International Journal of Molecular Sciences 24, no. 8: 6975. https://doi.org/10.3390/ijms24086975
APA StyleMohanty, A., Alhaj Sulaiman, A., Moovarkumudalvan, B., Ali, R., Aouida, M., & Ramotar, D. (2023). The Yeast Permease Agp2 Senses Cycloheximide and Undergoes Degradation That Requires the Small Protein Brp1-Cellular Fate of Agp2 in Response to Cycloheximide. International Journal of Molecular Sciences, 24(8), 6975. https://doi.org/10.3390/ijms24086975








