Bromodomain and Extraterminal Domain (BET) Protein Inhibition Hinders Glioblastoma Progression by Inducing Autophagy-Dependent Differentiation
Abstract
1. Introduction
2. Results
2.1. BRD2 and BRD4 Are Expressed in GBM Cells
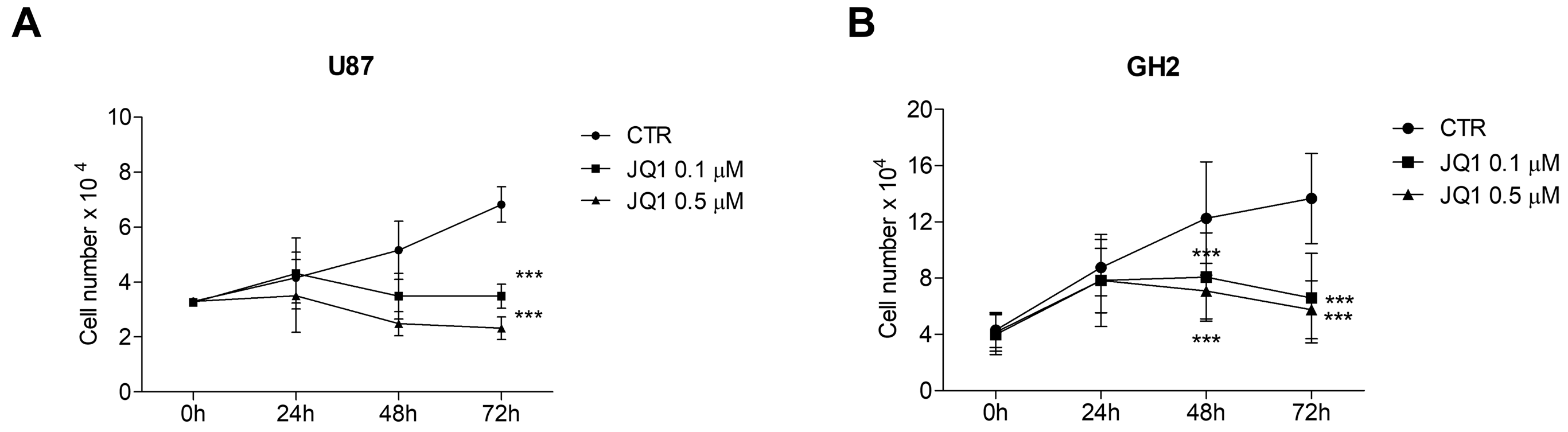
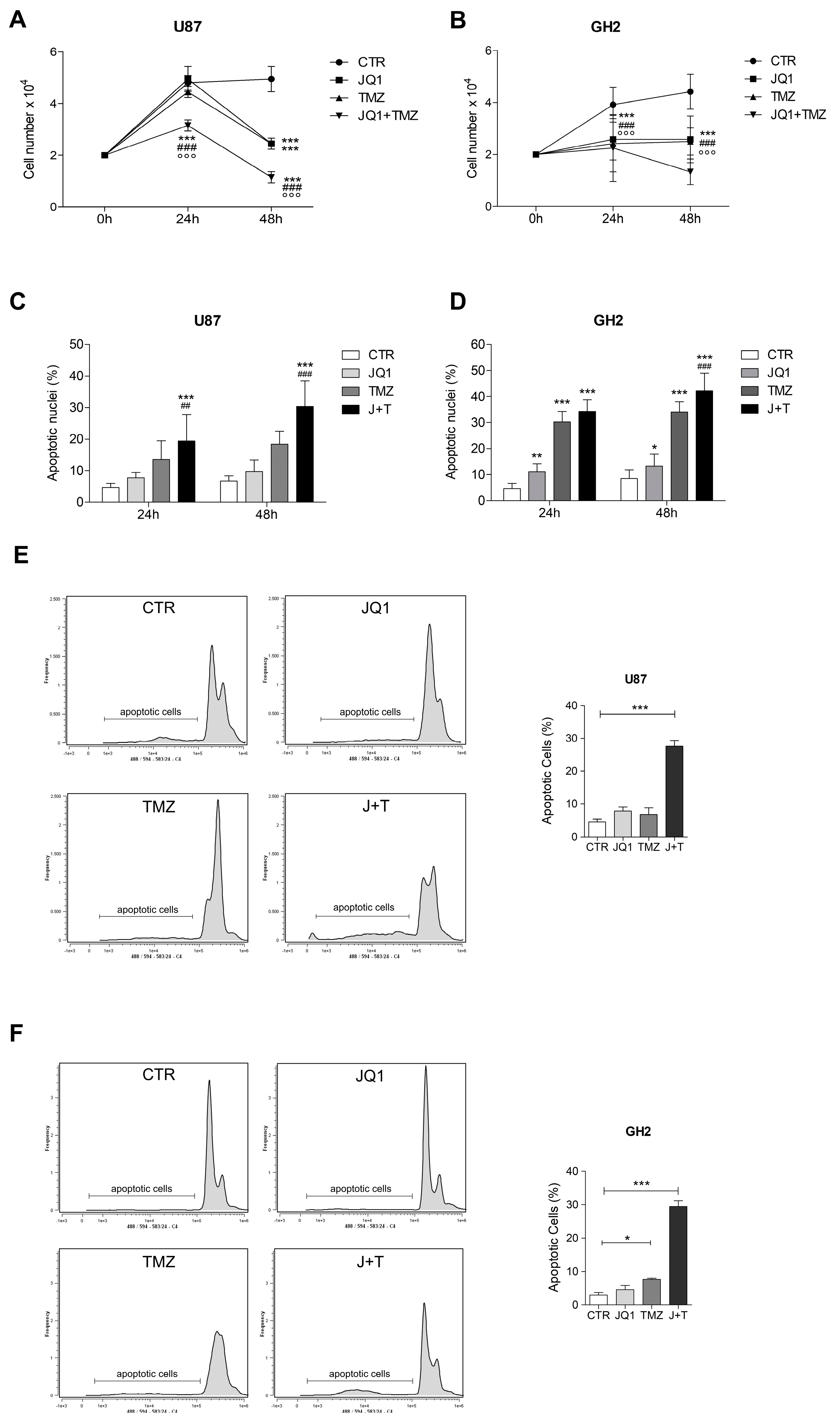
2.2. The BETi JQ1 Hampers Proliferation and Induces Apoptosis in Combination with Temozolomide in GBM Cells
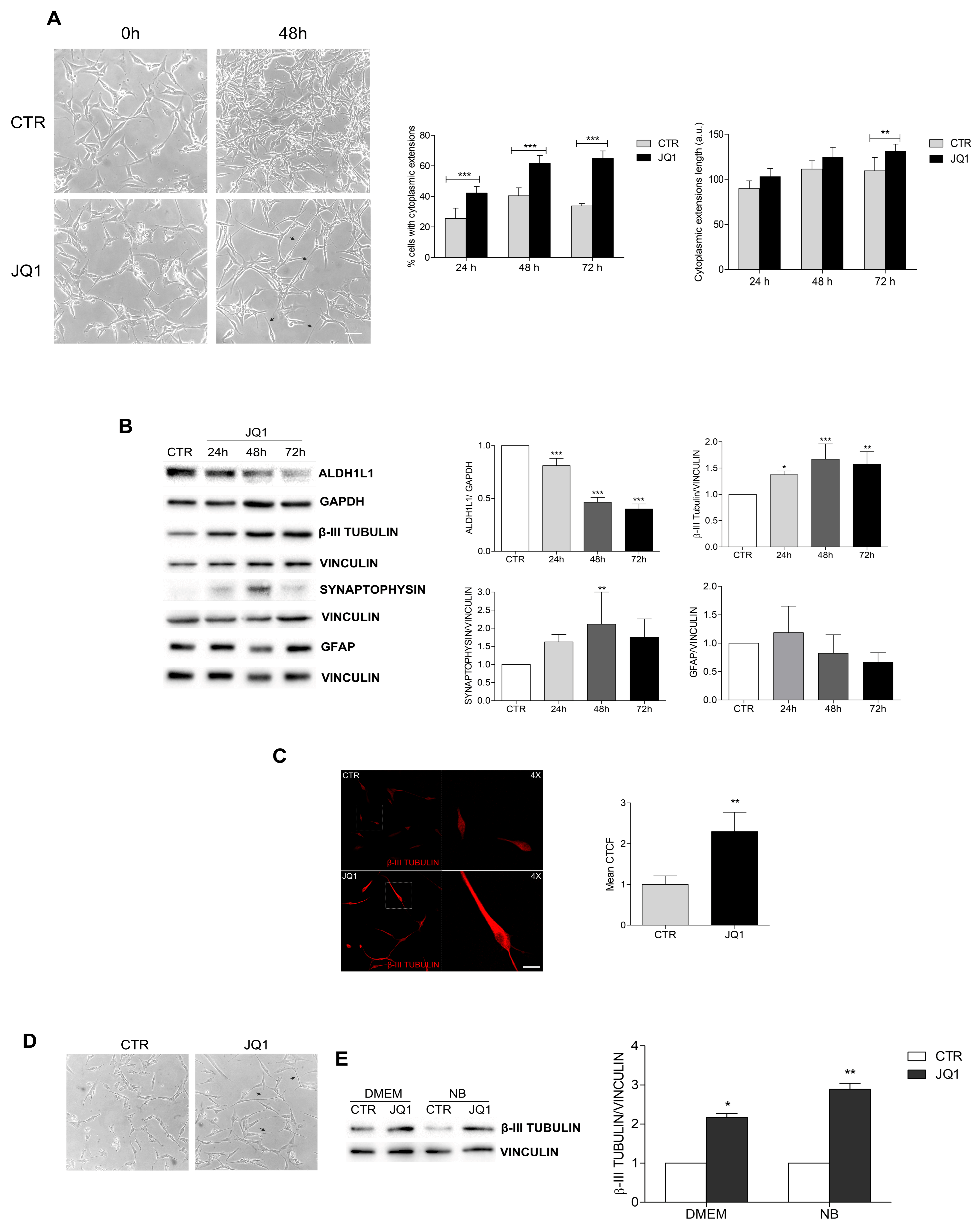
2.3. BET Inhibition Induces the Differentiation of GBM Cells
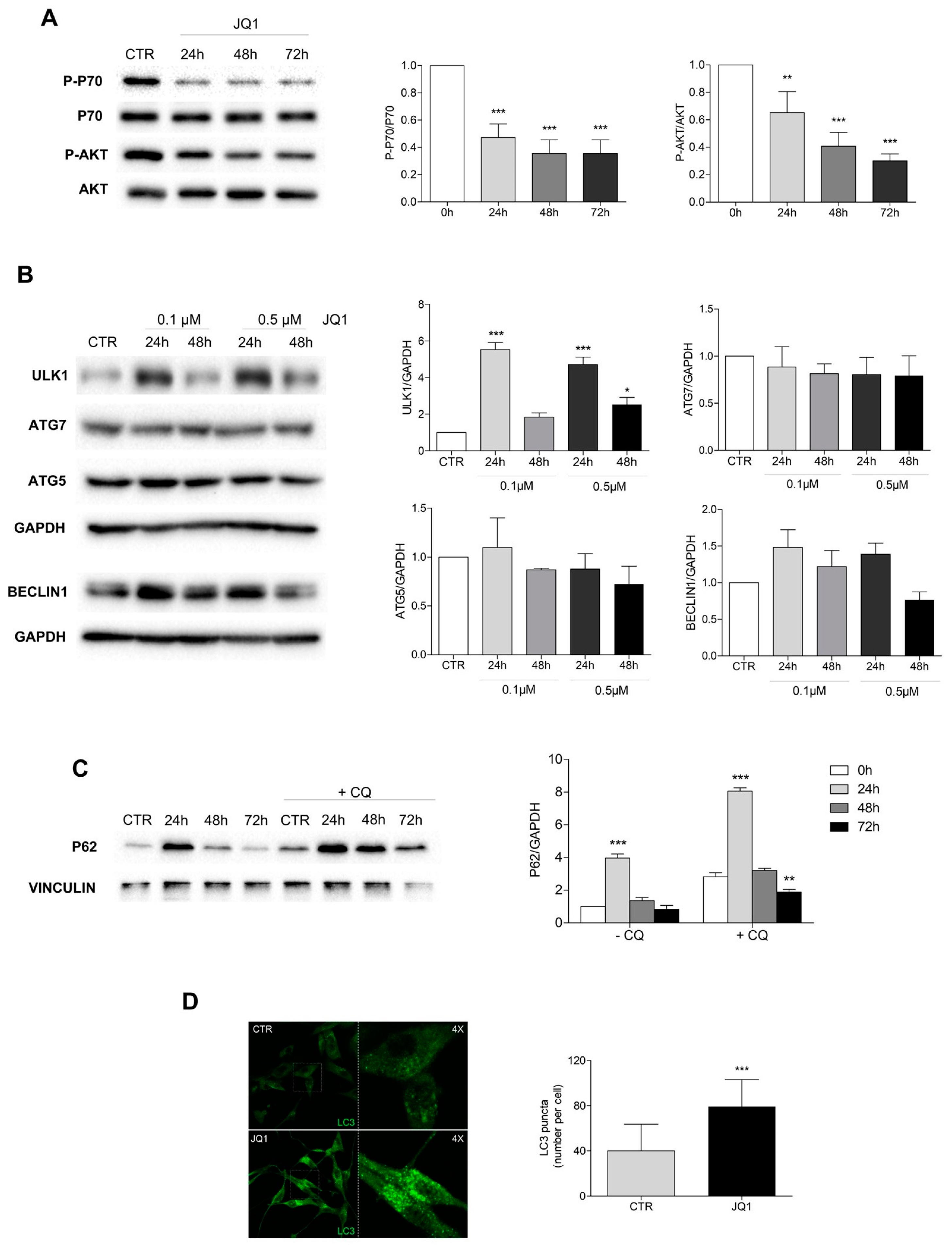
2.4. PI3K/Akt1/mTOR Pathway Is Inhibited and Autophagy Is Modulated by JQ1
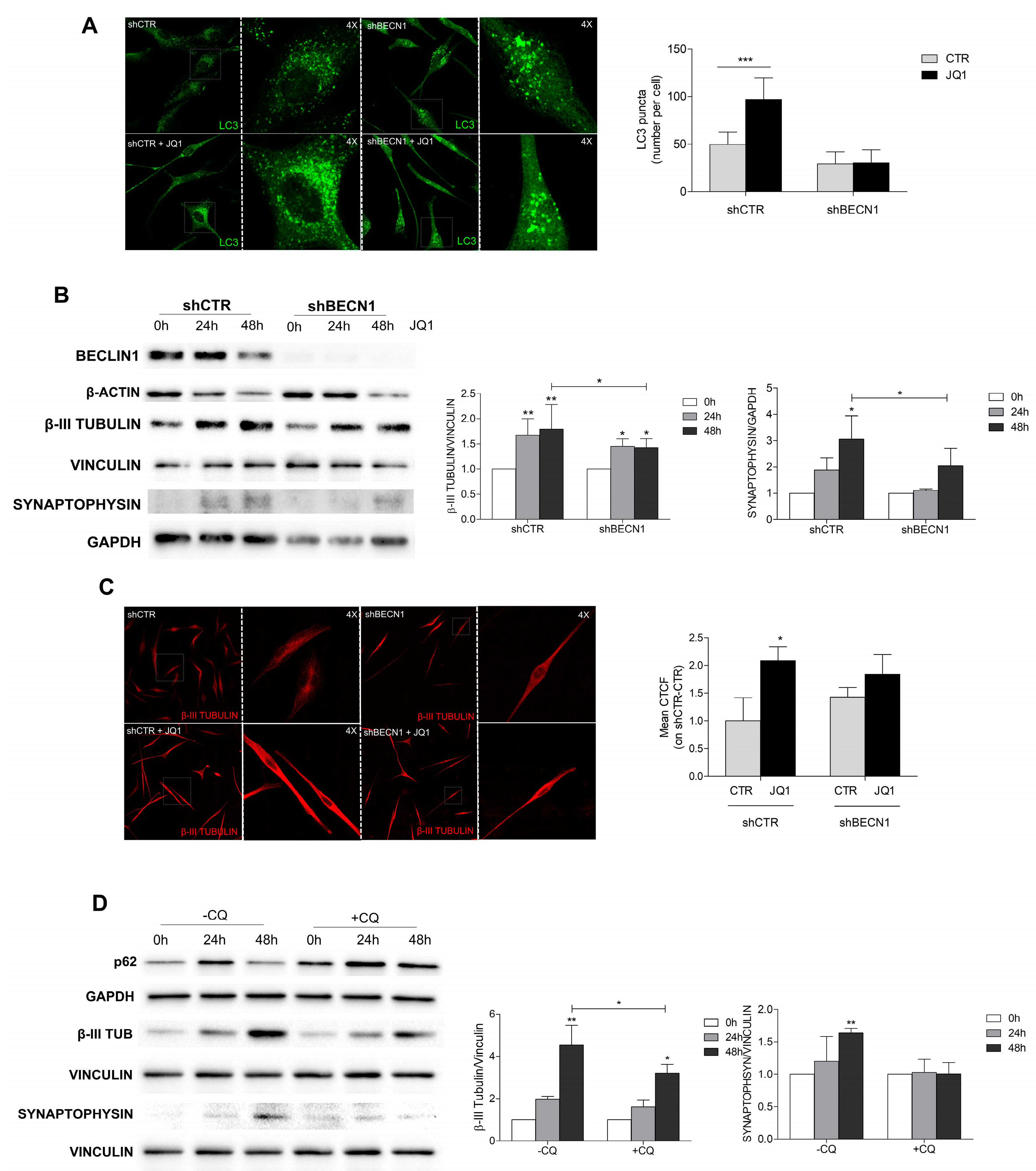
2.5. JQ1-Induced Cell Differentiation Is Dependent on Autophagy
3. Discussion
4. Materials and Methods
4.1. Cell Culture and Treatments
4.2. Cell Lysis and Western Blotting
4.3. Immunocytochemistry and Confocal Analysis
4.4. Morphological Analysis
4.5. Proliferation and Apoptosis Assays
4.6. Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Louis, D.N.; Perry, A.; Reifenberger, G.; von Deimling, A.; Figarella-Branger, D.; Cavenee, W.K.; Ohgaki, H.; Wiestler, O.D.; Kleihues, P.; Ellison, D.W. The 2016 World Health Organization Classification of Tumors of the Central Nervous System: A summary. Acta Neuropathol. 2016, 131, 803–820. [Google Scholar] [CrossRef] [PubMed]
- Wen, P.Y.; Weller, M.; Lee, E.Q.; Alexander, B.M.; Barnholtz-Sloan, J.S.; Barthel, F.P.; Batchelor, T.T.; Bindra, R.S.; Chang, S.M.; Antonio Chiocca, E.; et al. Glioblastoma in adults: A Society for Neuro-Oncology (SNO) and European Society of Neuro-Oncology (EANO) consensus review on current management and future directions. Neuro. Oncol. 2020, 22, 1073–1113. [Google Scholar] [CrossRef] [PubMed]
- Janjua, T.I.; Rewatkar, P.; Ahmed-Cox, A.; Saeed, I.; Mansfeld, F.M.; Kulshreshtha, R.; Kumeria, T.; Ziegler, D.S.; Kavallaris, M.; Mazzieri, R.; et al. Frontiers in the treatment of glioblastoma: Past, present and emerging. Adv. Drug Deliv. Rev. 2021, 171, 108–138. [Google Scholar] [CrossRef] [PubMed]
- Khaddour, K.; Johanns, T.M.; Ansstas, G. The landscape of novel therapeutics and challenges in glioblastoma multiforme: Contemporary state and future directions. Pharmaceuticals 2020, 13, 389. [Google Scholar] [CrossRef]
- Weller, M.; Cloughesy, T.; Perry, J.R.; Wick, W. Standards of care for treatment of recurrent glioblastoma-are we there yet? Neuro. Oncol. 2013, 15, 4–27. [Google Scholar] [CrossRef] [PubMed]
- Witthayanuwat, S.; Pesee, M.; Supaadirek, C.; Supakalin, N.; Thamronganantasakul, K.; Krusun, S. Survival analysis of Glioblastoma Multiforme. Asian Pacific J. Cancer Prev. 2018, 19, 2613–2617. [Google Scholar]
- Gimple, R.C.; Bhargava, S.; Dixit, D.; Rich, J.N. Glioblastoma stem cells: Lessons from the tumor hierarchy in a lethal cancer. Genes Dev. 2019, 33, 591–609. [Google Scholar] [CrossRef]
- Mattei, V.; Santilli, F.; Martellucci, S.; Monache, S.D.; Fabrizi, J.; Colapietro, A.; Angelucci, A.; Festuccia, C. The importance of tumor stem cells in glioblastoma resistance to therapy. Int. J. Mol. Sci. 2021, 22, 3863. [Google Scholar] [CrossRef]
- Osuka, S.; Van Meir, E.G. Overcoming therapeutic resistance in glioblastoma: The way forward. J. Clin. Investig. 2017, 127, 415–426. [Google Scholar] [CrossRef]
- Galli, R.; Binda, E.; Orfanelli, U.; Cipelletti, B.; Gritti, A.; De Vitis, S.; Fiocco, R.; Foroni, C.; Dimeco, F.; Vescovi, A. Isolation and characterization of tumorigenic, stem-like neural precursors from human glioblastoma (Cancer Research (October 2004) 64 (7011–7021). Cancer Res. 2004, 64, 7011–7021. [Google Scholar] [CrossRef]
- Piccirillo, S.G.M.; Binda, E.; Fiocco, R.; Vescovi, A.L.; Shah, K. Brain cancer stem cells. J. Mol. Med. 2009, 87, 1087–1095. [Google Scholar] [CrossRef] [PubMed]
- Jin, J.; Grigore, F.; Chen, C.C.; Li, M. Self-renewal signaling pathways and differentiation therapies of glioblastoma stem cells (Review). Int. J. Oncol. 2021, 59, 45. [Google Scholar] [CrossRef]
- Nicholson, J.G.; Fine, H.A. Diffuse glioma heterogeneity and its therapeutic implications. Cancer Discov. 2021, 11, 575–590. [Google Scholar] [CrossRef]
- Pastori, C.; Daniel, M.; Penas, C.; Volmar, C.H.; Johnstone, A.L.; Brothers, S.P.; Graham, R.M.; Allen, B.; Sarkaria, J.N.; Komotar, R.J.; et al. BET bromodomain proteins are required for glioblastoma cell proliferation. Epigenetics 2014, 9, 611–620. [Google Scholar] [CrossRef] [PubMed]
- Du, Z.; Song, X.; Yan, F.; Wang, J.; Zhao, Y.; Liu, S. Genome-wide transcriptional analysis of BRD4-regulated genes and pathways in human glioma U251 cells. Int. J. Oncol. 2018, 52, 1415–1426. [Google Scholar] [CrossRef] [PubMed]
- Tao, Z.; Li, X.; Wang, H.; Chen, G.; Feng, Z.; Wu, Y.; Yin, H.; Zhao, G.; Deng, Z.; Zhao, C.; et al. BRD4 regulates self-renewal ability and tumorigenicity of glioma-initiating cells by enrichment in the Notch1 promoter region. Clin. Transl. Med. 2020, 10, e181. [Google Scholar] [CrossRef]
- Delmore, J.E.; Issa, G.C.; Lemieux, M.E.; Rahl, P.B.; Shi, J.; Jacobs, H.M.; Kastritis, E.; Gilpatrick, T.; Paranal, R.M.; Qi, J.; et al. BET bromodomain inhibition as a therapeutic strategy to target c-Myc. Cell 2011, 146, 904–917. [Google Scholar] [CrossRef]
- Bandopadhayay, P.; Piccioni, F.; O’Rourke, R.; Ho, P.; Gonzalez, E.M.; Buchan, G.; Qian, K.; Gionet, G.; Girard, E.; Coxon, M.; et al. Neuronal differentiation and cell-cycle programs mediate response to BET-bromodomain inhibition in MYC-driven medulloblastoma. Nat. Commun. 2019, 10, 2400. [Google Scholar] [CrossRef]
- Aird, F.; Kandela, I.; Mantis, C.; Iorns, E.; Denis, A.; Williams, S.R.; Perfito, N.; Errington, T.M. Replication study: BET bromodomain inhibition as a therapeutic strategy to target c-Myc. Elife 2017, 6, e21253. [Google Scholar] [CrossRef]
- Jermakowicz, A.M.; Rybin, M.J.; Suter, R.K.; Sarkaria, J.N.; Zeier, Z.; Feng, Y.; Ayad, N.G. The novel BET inhibitor UM-002 reduces glioblastoma cell proliferation and invasion. Sci. Rep. 2021, 11, 23370. [Google Scholar] [CrossRef]
- Lam, F.C.; Morton, S.W.; Wyckoff, J.; Vu Han, T.L.; Hwang, M.K.; Maffa, A.; Balkanska-Sinclair, E.; Yaffe, M.B.; Floyd, S.R.; Hammond, P.T. Enhanced efficacy of combined temozolomide and bromodomain inhibitor therapy for gliomas using targeted nanoparticles. Nat. Commun. 2018, 9, 1991. [Google Scholar] [CrossRef]
- Berenguer-Daizé, C.; Astorgues-Xerri, L.; Odore, E.; Cayol, M.; Cvitkovic, E.; Noel, K.; Bekradda, M.; MacKenzie, S.; Rezai, K.; Lokiec, F.; et al. OTX015 (MK-8628), a novel BET inhibitor, displays in vitro and in vivo antitumor effects alone and in combination with conventional therapies in glioblastoma models. Int. J. Cancer 2016, 139, 2047–2055. [Google Scholar] [CrossRef] [PubMed]
- Tancredi, A.; Gusyatiner, O.; Bady, P.; Buri, M.C.; Lomazzi, R.; Chiesi, D.; Messerer, M.; Hegi, M.E. BET protein inhibition sensitizes glioblastoma cells to temozolomide treatment by attenuating MGMT expression. Cell Death Dis. 2022, 13, 1037. [Google Scholar] [CrossRef] [PubMed]
- Sakamaki, J.I.; Wilkinson, S.; Hahn, M.; Tasdemir, N.; O’Prey, J.; Clark, W.; Hedley, A.; Nixon, C.; Long, J.S.; New, M.; et al. Bromodomain Protein BRD4 Is a Transcriptional Repressor of Autophagy and Lysosomal Function. Mol. Cell 2017, 66, 517.e9–532.e9. [Google Scholar] [CrossRef]
- Simpson, J.E.; Gammoh, N. The impact of autophagy during the development and survival of glioblastoma: Role of autophagy in glioblastoma. Open Biol. 2020, 10, 200184. [Google Scholar] [CrossRef] [PubMed]
- Batara, D.C.R.; Choi, M.C.; Shin, H.U.; Kim, H.; Kim, S.H. Friend or foe: Paradoxical roles of autophagy in gliomagenesis. Cells 2021, 10, 1411. [Google Scholar] [CrossRef]
- Colella, B.; Faienza, F.; Di Bartolomeo, S. EMT regulation by autophagy: A new perspective in glioblastoma biology. Cancers 2019, 11, 312. [Google Scholar] [CrossRef]
- Colella, B.; Colardo, M.; Iannone, G.; Contadini, C.; Saiz-Ladera, C.; Fuoco, C.; Barilà, D.; Velasco, G.; Segatto, M.; Di Bartolomeo, S. Mtor inhibition leads to src-mediated egfr internalisation and degradation in glioma cells. Cancers 2020, 12, 2266. [Google Scholar] [CrossRef]
- Colardo, M.; Segatto, M.; Di Bartolomeo, S. Targeting rtk-pi3k-mtor axis in gliomas: An update. Int. J. Mol. Sci. 2021, 22, 4899. [Google Scholar] [CrossRef]
- Ryskalin, L.; Gaglione, A.; Limanaqi, F.; Biagioni, F.; Familiari, P.; Frati, A.; Esposito, V.; Fornai, F. The autophagy status of cancer stem cells in gliobastoma multiforme: From cancer promotion to therapeutic strategies. Int. J. Mol. Sci. 2019, 20, 3824. [Google Scholar] [CrossRef]
- Ferrucci, M.; Biagioni, F.; Lenzi, P.; Gambardella, S.; Ferese, R.; Calierno, M.T.; Falleni, A.; Grimaldi, A.; Frati, A.; Esposito, V.; et al. Rapamycin promotes differentiation increasing βIII-tubulin, NeuN, and NeuroD while suppressing nestin expression in glioblastoma cells. Oncotarget 2017, 8, 29574–29599. [Google Scholar] [CrossRef] [PubMed]
- Brunel, A.; Hombourger, S.; Barthout, E.; Battu, S.; Kögel, D.; Antonietti, P.; Deluche, E.; Saada, S.; Durand, S.; Lalloué, F.; et al. Autophagy inhibition reinforces stemness together with exit from dormancy of polydisperse glioblastoma stem cells. Aging 2021, 13, 18106–18130. [Google Scholar] [CrossRef]
- Catalano, M.; D’Alessandro, G.; Lepore, F.; Corazzari, M.; Caldarola, S.; Valacca, C.; Faienza, F.; Esposito, V.; Limatola, C.; Cecconi, F.; et al. Autophagy induction impairs migration and invasion by reversing EMT in glioblastoma cells. Mol. Oncol. 2015, 9, 1612–1625. [Google Scholar] [CrossRef] [PubMed]
- Filippakopoulos, P.; Qi, J.; Picaud, S.; Shen, Y.; Smith, W.B.; Fedorov, O.; Morse, E.M.; Keates, T.; Hickman, T.T.; Felletar, I.; et al. Selective inhibition of BET bromodomains. Nature 2010, 468, 1067–1073. [Google Scholar] [CrossRef] [PubMed]
- Taniguchi, Y. The bromodomain and extra-terminal domain (BET) family: Functional anatomy of BET paralogous proteins. Int. J. Mol. Sci. 2016, 17, 1849. [Google Scholar] [CrossRef]
- Filippakopoulos, P.; Picaud, S.; Mangos, M.; Keates, T.; Lambert, J.P.; Barsyte-Lovejoy, D.; Felletar, I.; Volkmer, R.; Müller, S.; Pawson, T.; et al. Histone recognition and large-scale structural analysis of the human bromodomain family. Cell 2012, 149, 214–231. [Google Scholar] [CrossRef]
- Cheung, K.L.; Kim, C.; Zhou, M.M. The Functions of BET Proteins in Gene Transcription of Biology and Diseases. Front. Mol. Biosci. 2021, 8, 728777. [Google Scholar] [CrossRef]
- Li, J.; Yang, B.; Zhou, Q.; Wu, Y.; Shang, D.; Guo, Y.; Song, Z. Autophagy promotes hepatocellular carcinoma cell invasion through activation of epithelial—Mesenchymal transition. Carcinogenesis 2018, 34, 1343–1351. [Google Scholar] [CrossRef]
- Tian, T.; Guo, T.; Zhen, W.; Zou, J.; Li, F. BET degrader inhibits tumor progression and stem-like cell growth via Wnt/β-catenin signaling repression in glioma cells. Cell Death Dis. 2020, 11, 1–13. [Google Scholar] [CrossRef]
- Li, J.; Ma, J.; Meng, G.; Lin, H.; Wu, S.; Wang, J.; Luo, J.; Xu, X.; Tough, D.; Lindon, M.; et al. BET bromodomain inhibition promotes neurogenesis while inhibiting gliogenesis in neural progenitor cells. Stem Cell Res. 2016, 17, 212–221. [Google Scholar] [CrossRef]
- Nakano, I. Stem cell signature in glioblastoma: Therapeutic development for a moving target. J. Neurosurg. 2015, 122, 324–330. [Google Scholar] [CrossRef] [PubMed]
- Li, F.; Yang, C.; Zhang, H.B.; Ma, J.; Jia, J.; Tang, X.; Zeng, J.; Chong, T.; Wang, X.; He, D.; et al. BET inhibitor JQ1 suppresses cell proliferation via inducing autophagy and activating LKB1/AMPK in bladder cancer cells. Cancer Med. 2019, 8, 4792–4805. [Google Scholar] [CrossRef] [PubMed]
- Di Bartolomeo, S.; Nazio, F.; Cecconi, F. The Role of Autophagy During Development in Higher Eukaryotes. Traffic 2010, 11, 1280–1289. [Google Scholar] [CrossRef] [PubMed]
- Rodolfo, C.; Di Bartolomeo, S.; Cecconi, F. Autophagy in stem and progenitor cells. Cell. Mol. Life Sci. 2016, 73, 475–496. [Google Scholar] [CrossRef]
- Adelipour, M.; Saleth, L.R.; Ghavami, S.; Alagarsamy, K.N.; Dhingra, S.; Allameh, A. The role of autophagy in the metabolism and differentiation of stem cells. Biochim. Biophys. Acta Mol. Basis Dis. 2022, 1868, 166412. [Google Scholar] [CrossRef]
- Klionsky, D.J.; Abdelmohsen, K.; Abe, A.; Abedin, M.J.; Abeliovich, H.; Arozena, A.A.; Adachi, H.; Adams, C.M.; Adams, P.D.; Adeli, K.; et al. Guidelines for the use and interpretation of assays for monitoring autophagy (3rd edition). Autophagy 2016, 12, 382. [Google Scholar]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Colardo, M.; Gargano, D.; Russo, M.; Petraroia, M.; Pensabene, D.; D’Alessandro, G.; Santoro, A.; Limatola, C.; Segatto, M.; Di Bartolomeo, S. Bromodomain and Extraterminal Domain (BET) Protein Inhibition Hinders Glioblastoma Progression by Inducing Autophagy-Dependent Differentiation. Int. J. Mol. Sci. 2023, 24, 7017. https://doi.org/10.3390/ijms24087017
Colardo M, Gargano D, Russo M, Petraroia M, Pensabene D, D’Alessandro G, Santoro A, Limatola C, Segatto M, Di Bartolomeo S. Bromodomain and Extraterminal Domain (BET) Protein Inhibition Hinders Glioblastoma Progression by Inducing Autophagy-Dependent Differentiation. International Journal of Molecular Sciences. 2023; 24(8):7017. https://doi.org/10.3390/ijms24087017
Chicago/Turabian StyleColardo, Mayra, Deborah Gargano, Miriam Russo, Michele Petraroia, Daniele Pensabene, Giuseppina D’Alessandro, Antonio Santoro, Cristina Limatola, Marco Segatto, and Sabrina Di Bartolomeo. 2023. "Bromodomain and Extraterminal Domain (BET) Protein Inhibition Hinders Glioblastoma Progression by Inducing Autophagy-Dependent Differentiation" International Journal of Molecular Sciences 24, no. 8: 7017. https://doi.org/10.3390/ijms24087017
APA StyleColardo, M., Gargano, D., Russo, M., Petraroia, M., Pensabene, D., D’Alessandro, G., Santoro, A., Limatola, C., Segatto, M., & Di Bartolomeo, S. (2023). Bromodomain and Extraterminal Domain (BET) Protein Inhibition Hinders Glioblastoma Progression by Inducing Autophagy-Dependent Differentiation. International Journal of Molecular Sciences, 24(8), 7017. https://doi.org/10.3390/ijms24087017







