Abstract
The superfamily of soluble N-ethylmaleimide-sensitive factor attachment protein receptor (SNARE) proteins mediates membrane fusion during vesicular transport between endosomes and the plasma membrane in eukaryotic cells, playing a vital role in plant development and responses to biotic and abiotic stresses. Peanut (Arachis hypogaea L.) is a major oilseed crop worldwide that produces pods below ground, which is rare in flowering plants. To date, however, there has been no systematic study of SNARE family proteins in peanut. In this study, we identified 129 putative SNARE genes from cultivated peanut (A. hypogaea) and 127 from wild peanut (63 from Arachis duranensis, 64 from Arachis ipaensis). We sorted the encoded proteins into five subgroups (Qa-, Qb-, Qc-, Qb+c- and R-SNARE) based on their phylogenetic relationships with Arabidopsis SNAREs. The genes were unevenly distributed on all 20 chromosomes, exhibiting a high rate of homolog retention from their two ancestors. We identified cis-acting elements associated with development, biotic and abiotic stresses in the promoters of peanut SNARE genes. Transcriptomic data showed that expression of SNARE genes is tissue-specific and stress inducible. We hypothesize that AhVTI13b plays an important role in the storage of lipid proteins, while AhSYP122a, AhSNAP33a and AhVAMP721a might play an important role in development and stress responses. Furthermore, we showed that three AhSNARE genes (AhSYP122a, AhSNAP33a and AhVAMP721) enhance cold and NaCl tolerance in yeast (Saccharomyces cerevisiae), especially AhSNAP33a. This systematic study provides valuable information about the functional characteristics of AhSNARE genes in the development and regulation of abiotic stress responses in peanut.
1. Introduction
Vesicular trafficking is a fundamental system maintaining cellular functions in all eukaryotic cells and is essential for development and adaptation. Communication routes among distinct organelles require vesicular trafficking, through which proteins and soluble cargo are sorted to their correct compartments [1]. The immobile and multicellular nature of plants requires the constant monitoring of changes in the environment and rapid reprogramming of metabolic and gene expression profiles adapted to adverse conditions, such as soil salinity, drought, extreme temperatures, nutrient imbalances and the presence of toxic metals. Cellular responses to various environmental stresses rely on the regulation of vesicular trafficking to ensure proper localization of proteins specialized in sensing stress stimuli and effecting the appropriate response. Membrane fusion, the last step of vesicular trafficking, is promoted by complex formation of soluble N-ethylmaleimide-sensitive factor attachment receptor proteins (SNAREs) [2]. SNAREs are located in different subcellular organelles and take part in a series of fundamental processes, for instance, cytokinesis, cytoskeleton organization, symbiosis, growth, and biotic and abiotic stress responses [3,4,5,6]. For instance, the SYP4 (SYNTAXIN OF PLANTS 4) and SYP6 groups are located in TGN and positively affect tolerance to salinity and osmotic stresses [7,8]. The identified immune protein of the powdery mildew RPW8.2 is transported by AtVAMP721/722 (vesicle-associated membrane protein 721/722) vesicles [9].
SNAREs are well conserved within eukaryotes, with genomic studies demonstrating that plants have considerably more SNARE genes than other eukaryotes. The SNARE family consists of 21–25 and 35–36 family members in yeast (Saccharomyces cerevisiae) and humans (Homo sapiens), respectively, while there are 36 SNAREs in the liverwort Marchantia polymorpha, 64 in Arabidopsis (Arabidopsis thaliana) and 60 in rice (Oryza sativa) [10,11,12]. SNARE proteins contain 100–400 amino acids identified by a coiled-coil SNARE domain and can be divided into R-SNARE and Q-SNARE proteins according to the conserved amino acid residues (Arg or Glu) in their central structure. Q-SNAREs are often located on the cytoplasmic membrane and are also known as t-SNAREs; R-SNAREs are located on vesicle membranes and are also called v-SNAREs [3,13,14]. Q-SNAREs can be further divided into four subgroups: Qa-, Qb-, Qc- and Qb+c-SNAREs [15]. Most SNAREs connect with lipid bilayers via a C-terminal transmembrane domain, but some exceptions attach to membranes via a lipid anchor [16,17]. When vesicles fuse, four different SNAREs (Qa-Qb-Qc-R or Qa-Qb+c-R) interact to form a stable complex called the SNARE complex, facilitating fusion of the phospholipid bilayer [18].
The plant vesicular trafficking system is considerably complex. It is characterized by expanded protein families that increase the specificity of vesicular trafficking as well as by plant-specific features that have evolved to perform specific functions in the plant lifestyle [5,11,19]. Novel Plant-specific SNARE (NPSN) and SYP7 Qc-SNARE proteins are specific to plants [20]. These additional proteins may allow plants to mount a coordinated response to the environment and confer special properties to some plants. Many crops, including wheat (Triticum aestivum L.), maize (Zea mays L.), rice, beans (Phaseolus vulgaris L.) and peanuts, are major sources of protein for humans and livestock. These crops accumulate storage proteins in subcellular organelles such as the endoplasmic reticulum and vacuoles. Therefore, the accumulation, transport and localization of many storage proteins are also dependent on the vesicular transport system. There is potential to improve yield, nutrition and stress tolerance by improving the vesicular transport system [21].
Cultivated peanut (Arachis hypogaea L.) originates from South America. It is a major oil crop grown in semi-arid and arid hilly areas with high tolerance to drought and infertile conditions [22]; yield per unit is higher than that of other oil crops [23]. Peanut growth is very sensitive to environmental stress, which seriously affects yield and quality. To date, the molecular characterization of SNAREs in peanut has identified few genes [24], and a systematic study of the peanut SNARE family has not been performed. Cultivated peanut is an allotetraploid (AABB), and molecular evidence indicates that Arachis duranensis Krapov. & W.C Greg. and Arachis ipaensis Krapov. & W.C Greg. are the donors of the A and B genomes, respectively [25]. Whole-genome sequences and annotation of A. hypogaea ‘Tifrunner’ [22,26] now make it possible to identify all the putative SNARE genes in cultivated peanut and to analyze their potential functions in different tissues and under abiotic stress conditions.
To promote future research on the SNARE family in this species, we identified all SNARE genes in the genomes of peanut (A. hypogaea, A. duranensis and A. ipaensis). We also analyzed the structure, chromosomal location and conserved domains of SNARE genes using bioinformatics methods. In addition, we analyzed the transcriptional profiles of SNARE genes at different developmental stages and performed a comparative analysis of expression levels after different treatments to characterize the mechanisms governing vesicular transport in response to different abiotic stresses and identify candidate genes for peanut improvement. Importantly, we verified the function of the putative SNARE genes’ response to salt and chilling stress in Saccharomyces cerevisiae.
2. Results
2.1. Identification and Classification of SNAREs in Peanut
We identified 129 SNARE proteins from A. hypogaea L. after screening the peanut protein database with the conserved domain PF05739 (Table S2). We also identified SNAREs in the two diploid ancestors of cultivated peanut: 63 in A. duranensis and 64 in A. ipaensis (Table S3). All candidate SNAREs contained the conserved domain PF05739 in peanut, similar to Arabidopsis, but also contained other conserved domains such as syntaxin, sec20, use1, longin and synaptobrevin (Tables S2 and S3).
Among the SNAREs identified in A. hypogaea, 34 were syntaxins—the prototype family of SNARE proteins—including Syntaxin1, Syntaxin2, Syntaxin5, Syntaxin18 and SynN. Syntaxin18 and Syntaxin5 are found in the SNARE complex of the endoplasmic reticulum (ER) and play an important role in transport between the intermediate compartment of the ER and the cis-Golgi vesicle [27]. The N-terminal region of Syntaxin18 is especially important for the formation of ER aggregates [28]; Syntaxin5 (Syn5) participates in the assembly of transitional ER and the Golgi, lipid droplet fusion and cytokinesis [29]. Another conserved family with 32 members identified in cultivated peanut was Synaptobrevin or R-SNAREs, which localize at vesicle membranes [7]. Some of these R-SNAREs contained a longin domain, which is a motif of proteins transporting vesicles from the ER to the plasma membrane via the Golgi apparatus. SNARE-conserved domain GS27 was identified in AhMEMB1 (a Qb-SNARE membrin), a member of the Qb subfamily of SNARE proteins involved in regulating the early secretory pathway of eukaryotic cells [30].
As shown in Table S2, the length of SNARE genes ranged from 516 bp (AhVAMP713b) to 13,843 bp (AhTYN12d), encoding proteins with 124 (AhBET11a) to 1883 (AhTYN11b) amino acids in length; these proteins were mostly between 124 (AhMEMB11b) and 426 (AhSYP111b) amino acids, except for those of the AhTYN subfamily. The predicted molecular weights (MW) of the proteins were mostly between 12.8 kDa (AhSFT12c) and 48.3 kDa (AhNPSN12b); however, those belonging to the AhTYN subfamily were far beyond this range at 105.8~125.4 kDa. Predicted PIs (isoelectric points) varied from 4.75 (AhSYP23c) to 9.99 (AhSYP81b), with an average of 7.46, among which 69 AhSNAREs had pI > 7 and 60 AhSNAREs had pI < 7. Subcellular localization predictions suggested that peanut SNAREs are predominantly located at the plasma membrane, ER/Golgi and trans-Golgi network (TGN)/vacuole/endosome (Table S2).
2.2. Peanut SNARE Genes Exhibit a High Rate of Homoeolog Retention
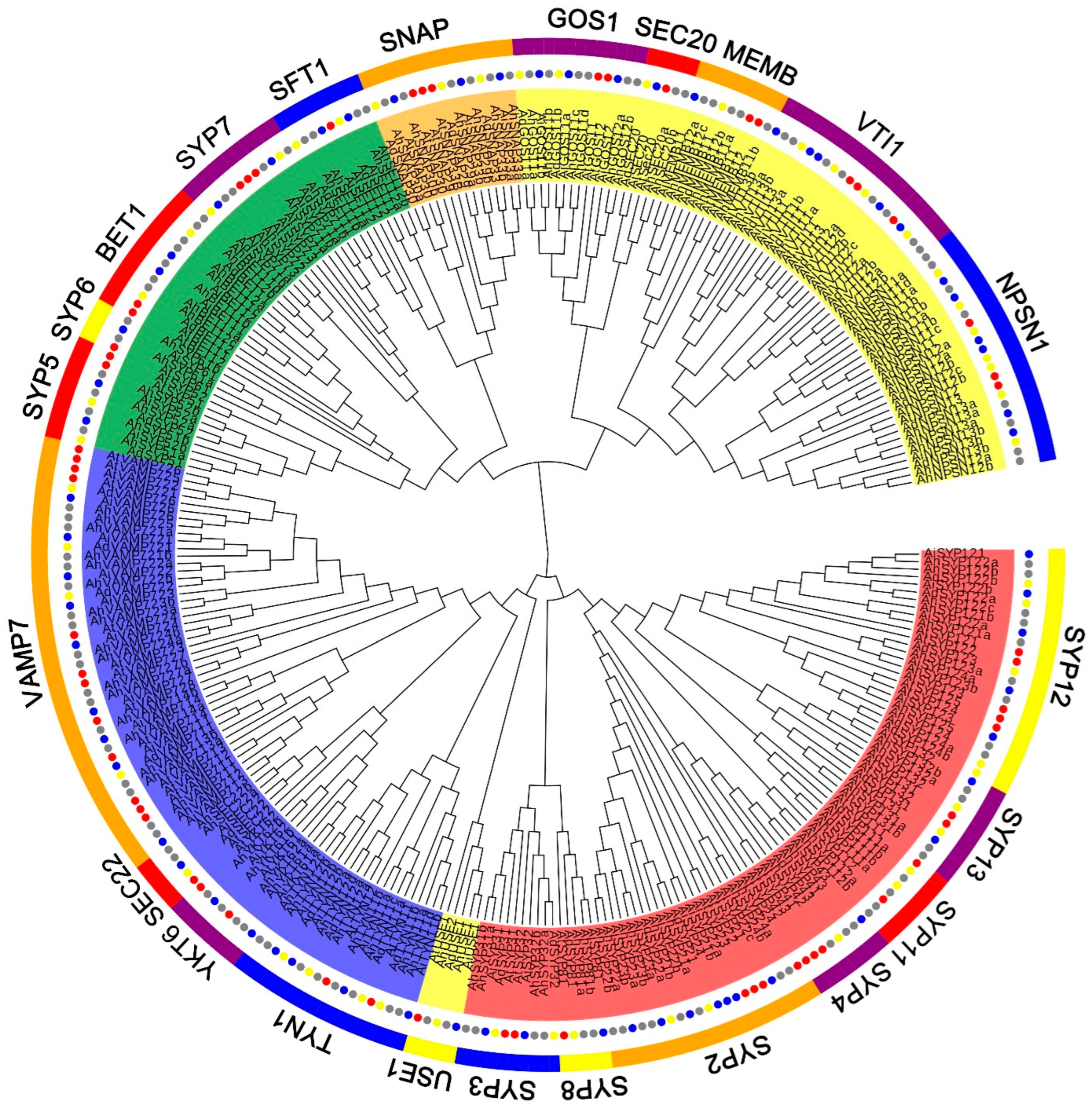
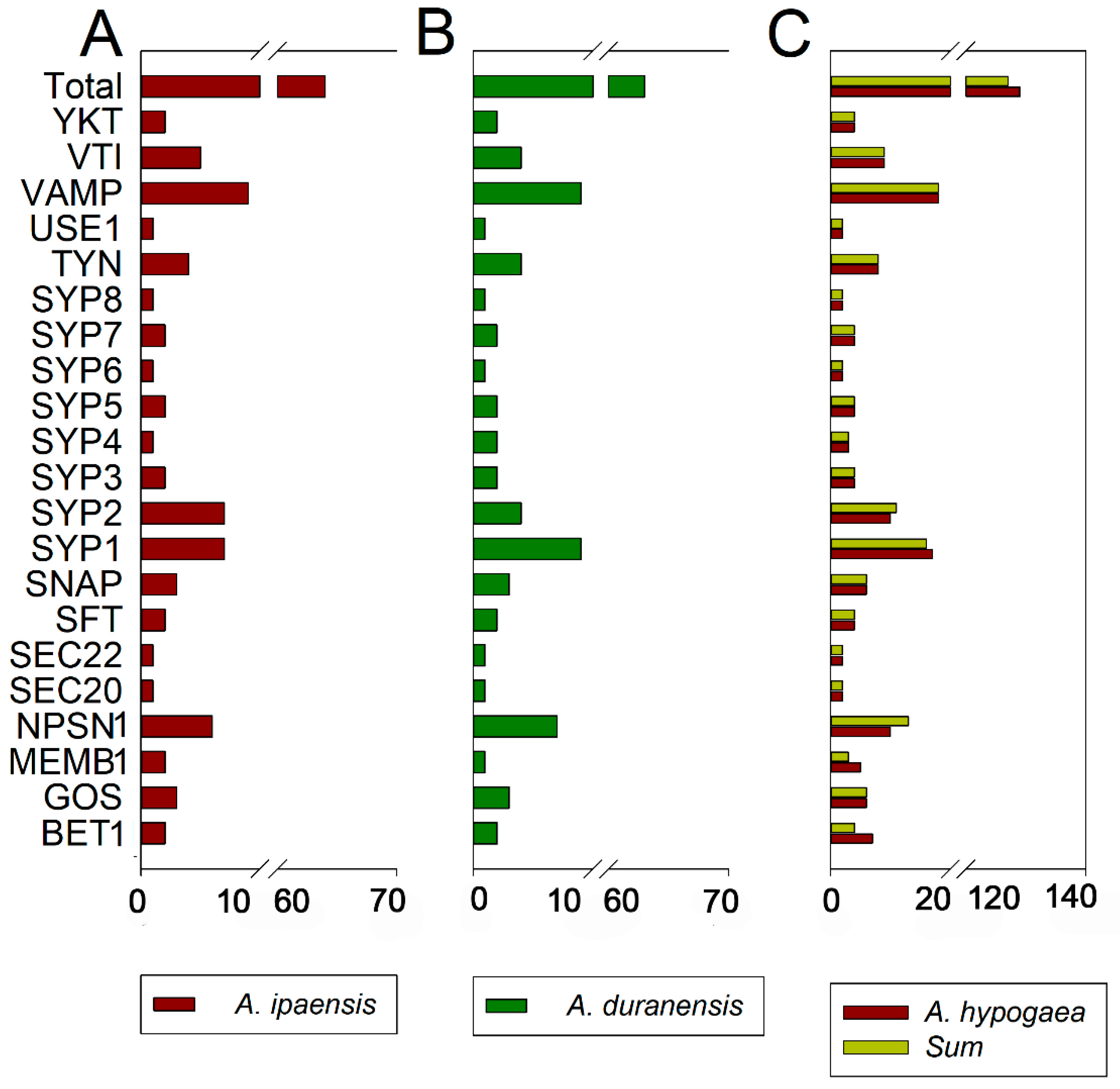
To gain insight into the relationship and classification of AhSNARE genes, we reconstructed a maximum-likelihood phylogenetic tree with 129 AhSNARE proteins from cultivated peanut, 63 AdSNAREs from A. duranensis, 64 AiSNAREs from A. ipaensis and 64 AtSNAREs from Arabidopsis. The cultivated peanut genome retained all major SNARE gene subfamilies: BET1, GOS1, MEMB1, NPSN1, SEC20, SEC22, SFT, SNAP, SYP1(SYP11, SYP12, SYP13), SYP4, SYP5, SYP6, SYP7, SYP8, TYN1, USE1, VAMP, VTI and YKT6 (Figure 1 and Table S2). The above phylogeny roughly followed species phylogeny; A. hypogaea genes displayed a sister–group relationship with Arabidopsis genes in many subclades (Figure 1). Almost all A. hypogaea genes were closely related to their two homologous ancestral sequences; sequence similarity was more than 85%. Many subclades showed the expected 2:1 ratio of cultivated peanut-to-Arabidopsis genes, the sum of two ancestral subfamily members (SEC 20, SEC 22, SFT, SNAP); however, some ratios were lower than expected (e.g., NPSN) (Figure 2). These results suggested that expansion of SNARE homologs in A. hypogaea mainly resulted from heterologous hybridization. The AhBET Qc-SNARE subfamily expanded and experienced duplications in cultivated peanut after hybridization. The increased number of BET subfamily members in peanut suggests the possibility of novel functions for one or more family members [21].
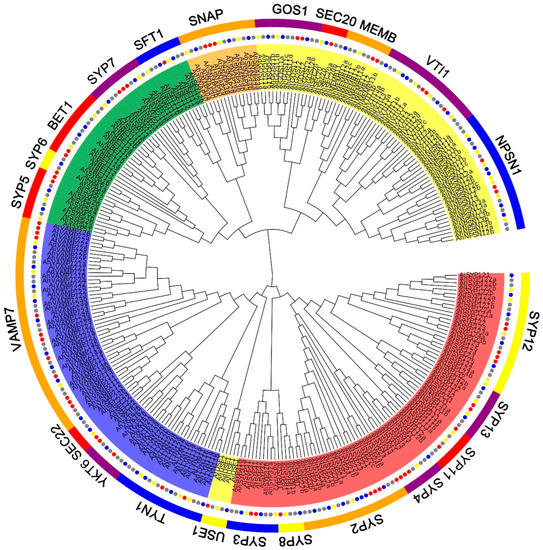
Figure 1.
Phylogenetic tree of the SNARE family in A. hypogaea, A. duranensis, A. ipaensis and Arabidopsis. Maximum-likelihood tree based on full-length SNARE proteins sequences reconstructed using the nearest-neighbor-interchange model. SNARE protein sequences from A. hypogaea, A. duranensis, A. ipaensis and Arabidopsis are marked with gray, blue, green and red circles, respectively. Qa-, Qb-, Qc-, Qb+c and R-SNARE are marked with red, yellow, green, orange and blue backgrounds, respectively. The ribbon in the outer ring indicates different subfamilies.
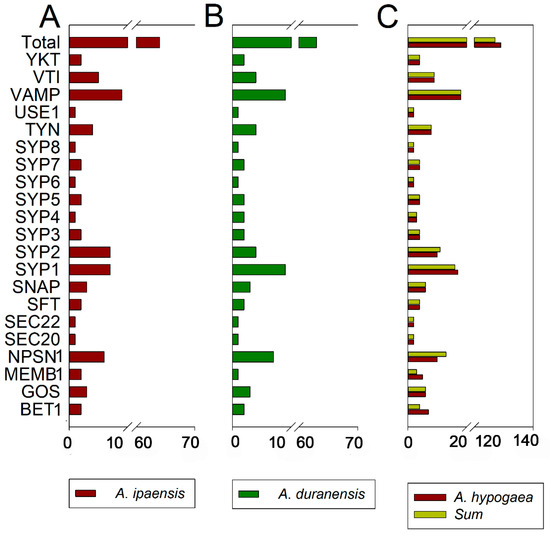
Figure 2.
Number of SNARE genes identified per SNARE-type subfamily. (A) A. ipaensis; (B) A. duranensis; (C) A. hypogaea. The sum of genes in A. ipaensis and A. duranensis (green–yellow) is also shown (C).
2.3. Conserved Domain and Gene Structure Analysis
Analysis of conserved motifs and gene structures of each AhSNARE using TBtools revealed that members of the same subfamily share similar motifs (Figure 3B). Motifs 1 and 8 corresponded to the longin and synaptobrevin domains, respectively, which were present in R-SNAREs; Motif 20 was the syntaxin domain, which was present in Qa-SNAREs; Motifs 23–24 were V-SNARE-C, which were present in Qb-SNAREs; Motif 10 was Qc-SNARE. SYP2 possessed Motifs 7, 9 and 20; SNAP possessed Motifs 10 and 22.

Figure 3.
Phylogenetic relationships, gene structures and conserved motifs of AhSNARE genes in cultivated peanut. (A) Phylogenetic relationships; (B) conversed motifs; (C) exon–intron structures. Yellow boxes represent exons; gray lines represent introns.
A bioinformatics analysis of gene structure can be useful for recognizing patterns of intron acquisition and loss during evolution of a family whose structure and function are well known. A summary of the exon–intron structure of each AhSNARE gene is shown in Figure 3C. The numbers and lengths of introns showed patterns reflecting the organization of the AhSNARE family in the phylogenetic tree (Figure 3A). However, genes from different groups varied widely in size and intron arrangement. In the SYP1 clade, genes of the AhSYP11 and AhSYP12 subfamily had only one or no intron, while there were 12 introns in genes of the AhSYP13 subfamily. The SYP1 clade is reported to split into two groups, SYP12 and SYP13, among embryophytes. A common feature of the SYP13 is that all genes have multiple introns, almost all of which occur at similar positions within the coding region [31]. Furthermore, there were 23 introns in genes of the AhTYN1 subclade, and the structures of these genes showed many variations.
2.4. Chromosomal Mapping and Duplication Analysis of Peanut SNARE Genes
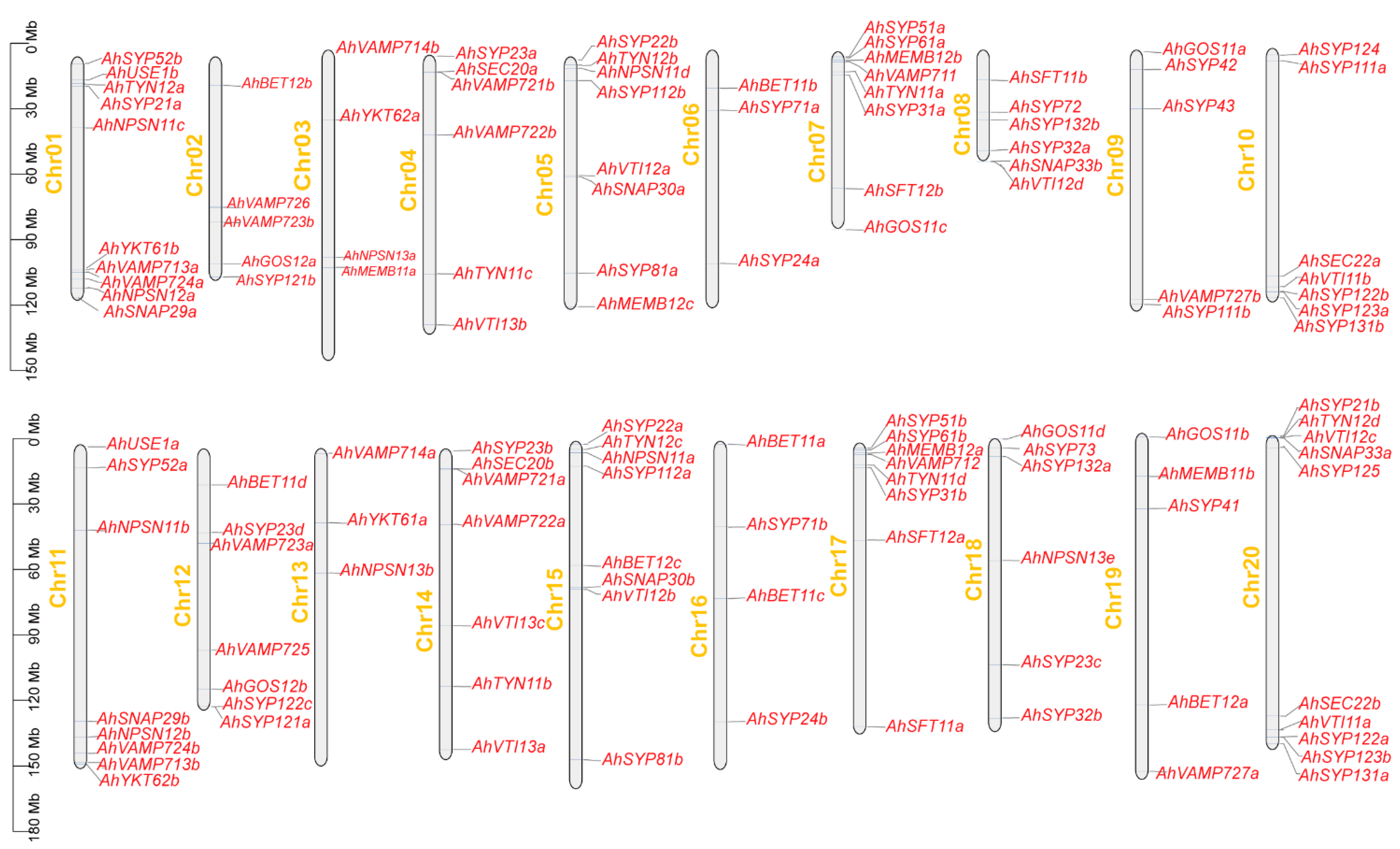
To determine the distribution of AhSNARE genes on chromosomes, we drew a physical map using TBtools onto which we mapped all 129 AhSNARE genes. These genes were distributed unequally on the 20 chromosomes (Figure 4), with 80% of AhSNAREs located in the distal regions of chromosomes. The fewest SNARE genes were on chromosomes 6 and 13, which only had three genes each, while chromosomes 1 and 20 harbored the highest number of SNARE genes (10 genes each). Homoeologous chromosomes from the A and B genomes, such as chromosomes 4 and 14 (6 genes), 7 and 17 (8 genes), and 9 and 19 (5 genes), typically had similar numbers of SNARE genes.
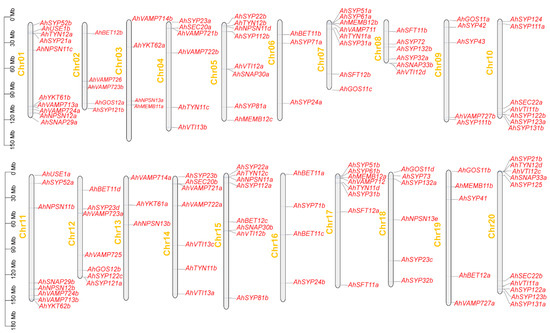
Figure 4.
Chromosomal location and distribution of 129 AhSNARE genes. Chromosome size is indicated by relative length. Scale bar represents megabases (Mb). Physical locations of AhSNARE genes are indicated on each chromosome.
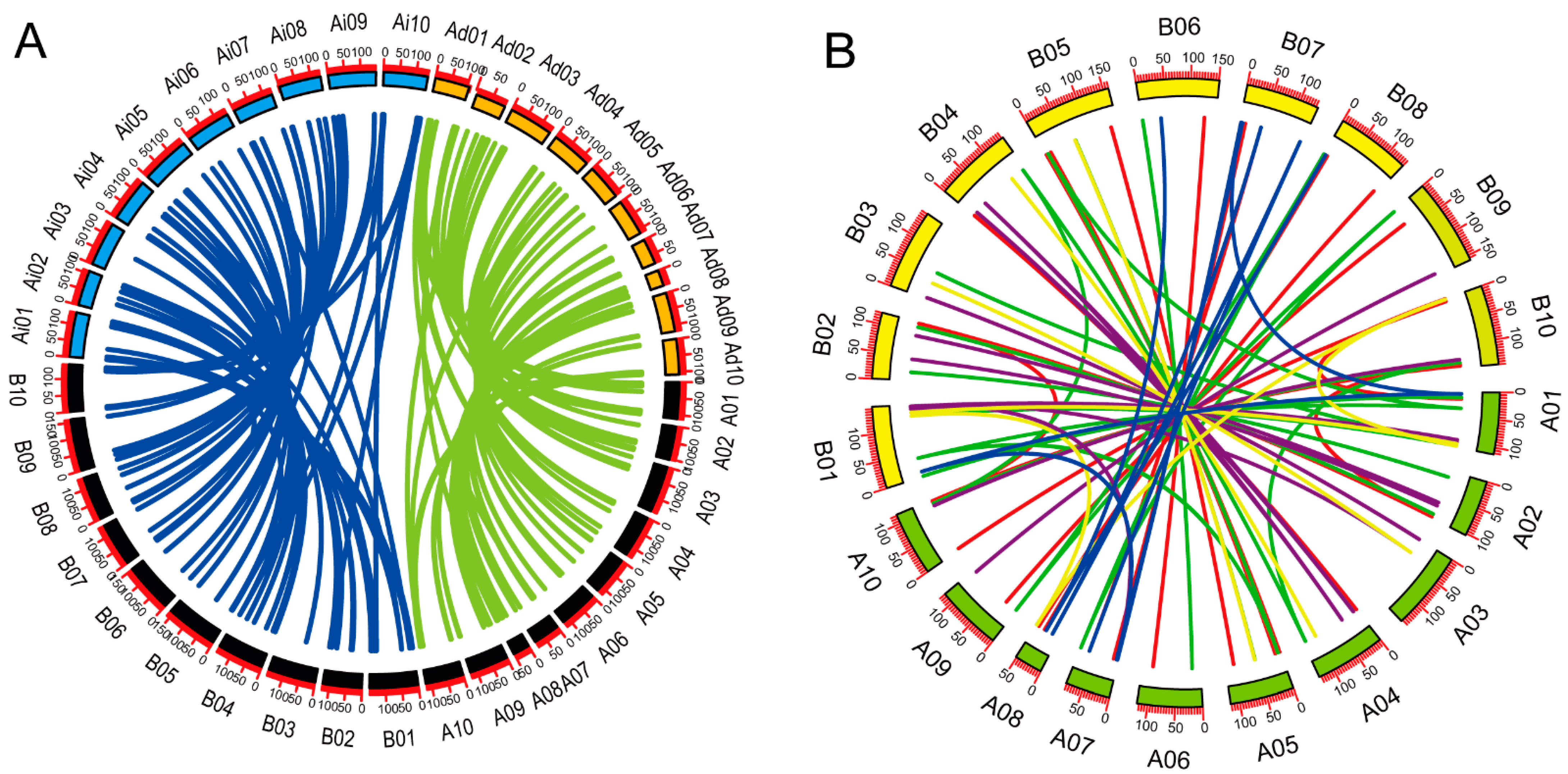
To confirm duplication events between genes, were performed collinearity analysis between A. hypogaea and A. duranensis, A. hypogaea and A. ipaensis (Figure 5A), and between the A and B genomes of A. hypogaea (Figure 5B). We observed high rates of gene retention: 91.5% of A. hypogaea genes were homologous to those of the proposed ancestors. The Ka/Ks ratios for each pair of homologous genes were generally less than 1 (Table S4), except those for AdBET11-AhBET11b (Ka/Ks = 2.042), AdVAMP721-AhVAMP721b (Ka/Ks = 1.161), AiVAMP711-AhVAMP711b (Ka/Ks = 2.042) and AiSFT12-AhSFT12a (Ka/Ks = 1.664). We identified 10 dispersed duplicated genes in A. hypogaea (AhBET11d, AhSYP121b, AhVAMP714b, AhBET12c, AhBET11c, AhBET12b, AhNPSN13b, AhMEMB11b, AhSYP112b and AhMEMB12c). According to the phylogenetic tree, three pairs of dispersed duplicated genes belonged to the Qb-SNARE subfamily, four belonged to the Qc-SNARE subfamily, one belonged to the R-SNARE subfamily and two belonged to the Qa-SNARE subfamily.
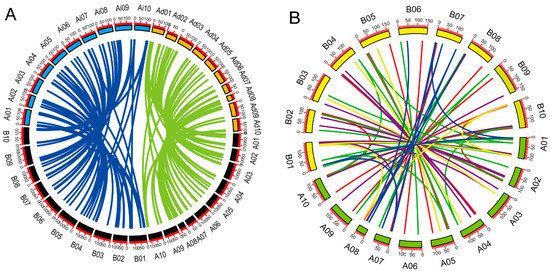
Figure 5.
Collinearity analysis of SNARE genes. (A) Synteny analysis of SNARE genes in A. hypogaea–A. duranensis and A. hypogaea–A. ipaensis. Green lines indicate syntenic genes between A. duranensis and A. hypogaea. Blue lines indicate syntenic genes between A. duranensis and A. hypogaea. Black boxes represent A. hypogaea chromosomes. Colored boxes represent chromosomes of A. duranensis and A. ipaensis. Ad, A. duranensis; Ai, A. ipaensis; Ah, A. hypogaea. (B) Synteny analysis of SNARE genes within A. hypogaea. Colored lines represent syntenic relationships between different type of SNAREs. Red, Qa-SNARE; green, Qb-SNARE; blue, Qb+c-SNARE; purple, R-SNARE. Scale bar represents megabases (Mb). Chromosome numbers are indicated on the top of each bar. Green boxes represent A genome; yellow boxes represent B genome.
2.5. Analysis of Cis-Acting Elements in Promoters of AhSNARE Genes
To gain insight into the cis-acting elements of AhSNAREs, we retrieved sequences 2000 bp upstream of the translation start position of all AhSNAREs from the peanut genome database. We identified many response-related elements within these regions (Table S5), such as MYB recognition, TGAACG-motif, ARE (Antioxidant response element), ABRE (ABA-response element), STRE (Hot responsive element), TC-rich repeats and LTR (Low temperature response element). These cis-acting regulatory elements are associated with plant biotic and abiotic stress responses. The TGAACG-motif and ABRE are cis-acting regulatory elements involved in responsiveness to methyl jasmonate and abscisic acid (ABA), respectively. We also identified some development-related elements, such as CAT-box and the GCN4_motif. These results suggest that AhSNAREs are involved in responses to environmental stress, as well as growth and development of peanut.
2.6. Differential Expression Profiles of SNARE Genes in Different Tissues and at Different Developmental Stages of Peanut Pod
Peanut plants bear fruit underground; groundnut seeds contain 22% to 30% protein and 35% to 60% oil. To understand the functions of AhSNARE genes in peanut development and growth, we analyzed their expression profiles at different developmental stages of peanut pods and in different tissues. We obtained transcriptome data for 118 out of 129 SNARE genes, with 90% of these genes being expressed in at least one tissue or at one developmental stage, with a wide expression range from 1 to 185 FPKM. The remaining 10% of SNARE genes displayed very low expression levels below 1 FPKM (Figure 6) and were considered not expressed. We also analyzed the expression levels of homoeologous SNARE gene pairs from the A and B subgenomes. Unlike results reported from the peanut genome [26], more homoeologous gene pairs exhibited expression bias towards the A subgenome than towards the B subgenome.
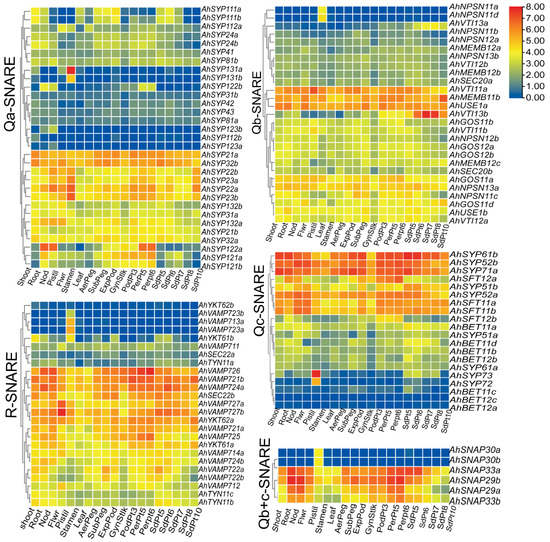
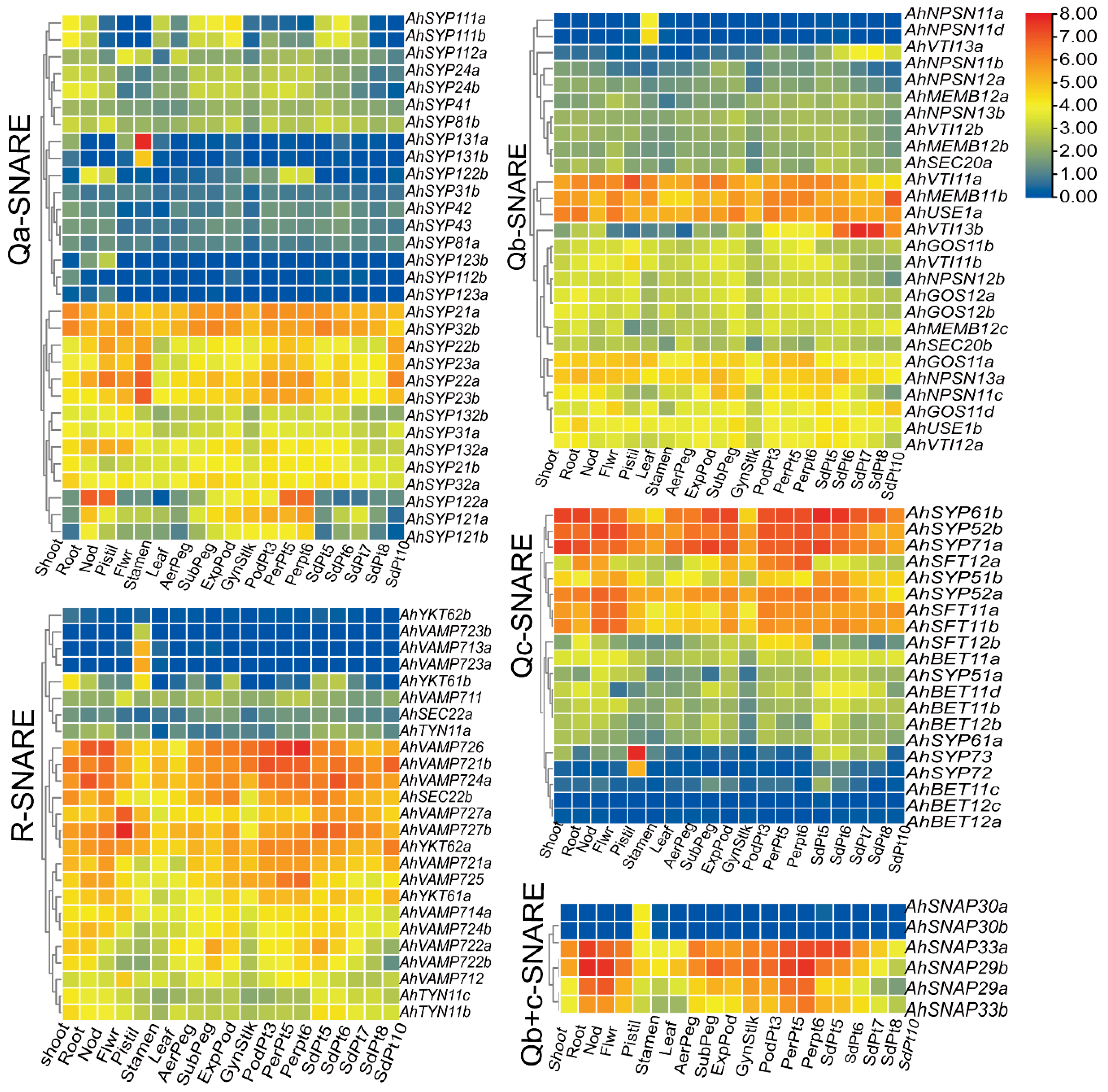
Figure 6.
Expression profiles of SNARE genes in seven peanut tissues and at 12 different developmental stages of peanut. The heatmap was generated using TBtools software, and fragments per kilobase of transcript per million fragments (FPMK) values for SNARE genes were Log2-transformed. The color box represents lower values (blue) to higher values (red).
We used phylogenetic classification to draw five heatmaps of the 118 expressed AhSNARE genes to evaluate their transcript abundance in nine tissues and at 12 pod developmental stages (Figure 6). The expression profile of each SNARE gene was different, although certain genes in different groups had similar expression patterns, which may provide clues as to their interactions. Several genes in each group were highly expressed in all tissues and at all developmental stages analyzed (e.g., AhSYP32 and AhSYP21 in Qa-SNARE; AhUSE1a, AhMEMB11b and AhVTI11a in Qb-SNARE; AhSYP61b, AhSYP52b and AhSYP71a/b in Qc-SNARE; AhVAMP726, AhVAMP 724a and AhVAMP721 in R-SNARE), suggesting that these genes are likely to play a regulatory role during the entire growth period of peanut. Several genes showed higher expression in roots, nodules and pericarp (AhSYP122 in Qa-SNARE; AhSFT12 in Qc-SNARE; AhSNAP29b and AhSNAP33a in Qa+c-SNARE). Qa-SNARE AhSYP131, Qc-SNARE AhSYP73, Qb-SNARE AhNPSN11b/d, Qb+c-SNARE SNAP30a/b and V-SNARE AhVAMP713a/b were specifically expressed in pistils. AhSYP22a/b, AhSYP23a/b, AhMEMB11b and AhYKT62a reached their highest expression levels during the latest stage of peanut pod development. As the seed matured, the expression level of AhVTI13a/b, expressed in the TGN/endosome/vacuole, rose markedly. However, lower expression of AhSFT12a/b and AhSYP121a/b was observed during the later stage of peanut seed development, with higher expression during the early stage of peanut seed development. The gene encoding Qa-SNARE AhSYP122a, which is located at the plasma membrane, showed high expression levels in Pattee 5 pericarp and Pattee 6 pericarp stages but showed very low transcript levels in peanut kernel.
2.7. Expression Patterns of AhSNARE Genes under Salt and Low-Temperature Stresses
SNARE genes play a vital role in abiotic and biotic stresses [3,32,33,34,35]. To understand the roles of SNARE genes in peanut, we summarized their expression patterns by analyzing transcriptomic data of peanut seedlings exposed to diverse stresses (salt, drought, high/low temperature, ABA, sodium nitroprusside, H2O2 and methyl jasmonate) for 24 h (Figure S1). SNARE genes in roots and leaves were regulated differentially by different stresses; most homologous gene pairs had similar expression patterns, and the expression difference was obvious at low temperature. Expression levels of AhSYP122a/b in the Qa-SNARE subfamily, AhSFT12a/b in Qc-SNARE, SNAP29a/b in Qb+c-SNARE and AhVAMP725/726 in R-SNARE were significantly higher after low-temperature treatment for 24 h. Few SNARE genes showed a significant difference in their expression levels under salt stress compared to control conditions (Figure S1). However, salt and chilling stress limit the productivity of peanut, damage photosystem complexes and change the fatty acid composition of peanut leaves [36,37].
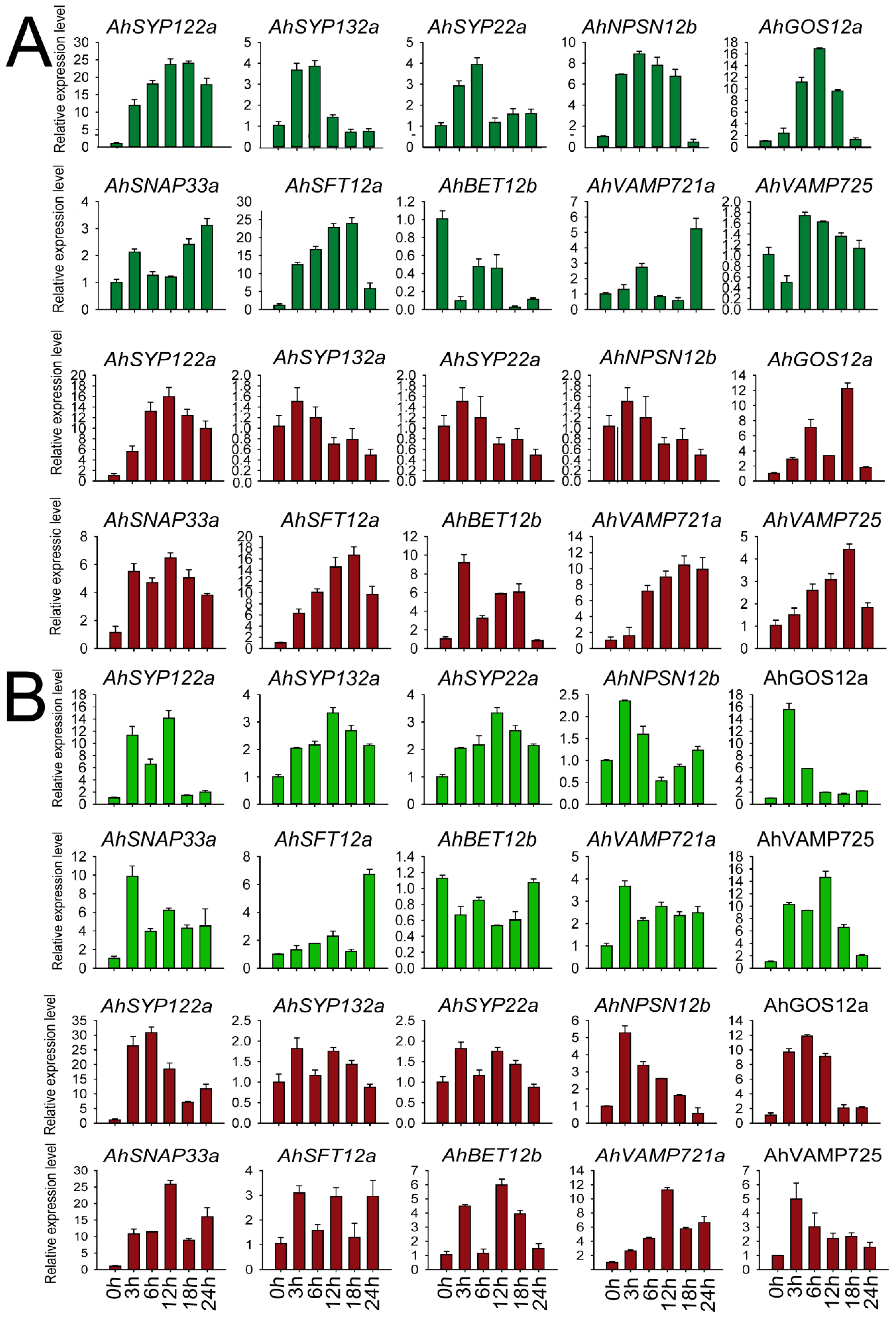
We selected members of each subfamily to validate peanut SNARE expression patterns after salt and chilling stress. As shown in Figure 7, we examined 10 genes from different subfamilies—AhSYP122a (Qa), AhSYP132a (Qa), AhSYP22a (Qa), AhNPSN12b (Qb), GOS12a (Qb), AhSNAP33a (Qb+c), AhSFT12a (Qc), BET12b (Qc), VAMP725 (R) and VAMP721a (R)—at five time points (0, 3, 12, 18 and 24 h) in leaves and roots of FH1. These AhSNARE genes showed different expression profiles in response to low-temperature and salt treatments. Under low-temperature treatment, AhSFT12a and AhSYP122a were highly expressed in both roots and leaves, with expression values reaching 15–20 times those of untreated samples at 18 h. Expression levels of AhSYP132a and AhSYP22a displayed an early (3 h and 6 h) upregulation followed by downregulation in leaves under low-temperature treatment; however, there was little difference in their expression in roots. The expression levels of AhGOS12a, AhSYP122a, AhVAMP725 and AhVAMP721a reached a maximum value at 18 h in roots under low-temperature treatment before dropping. AhBET12b expression was suppressed in leaves under low-temperature treatment but was elevated in roots. After salt treatment, the expression level of AhSYP122a, AhNPSN12b, AhGOS12a, AhSNAP33a, AhVAMP721a and AhVAMP725 was more than five times that of the control group in roots. AhSYP122a, AhNPSN12b and AhGOS12a transcript levels responded quickly to salt treatment, with expression levels reaching a maximum at 3 h before declining. The expression of AhSYP132a and AhSYP22a was not significantly different in roots throughout salt treatment and reached a maximum at 12 h after salt treatment. As shown in Figure 7, AhSYP122a, AhSNAP33a and AhVAMP721a expression was more than ten times that before treatment under both salt and low-temperature stress, and we speculated that these genes play a key role in abiotic stress responses.
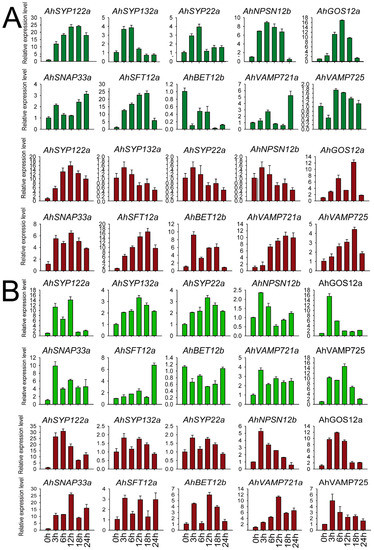
Figure 7.
Expression of AhSNARE genes in response to salt and chilling stress. Relative expression levels of 10 AhSNARE genes in roots (red) and leaves (green) after 0 h, 3 h, 6 h, 12 h and 24 h of salt (A) or low-temperature treatment (B) as determined by RT-qPCR. Data were calculated from three biological replicates. The error bars show the standard deviation of the three biological replicates.
2.8. AhSYP122a, AhSNAP33a and AhVAMP721a Enhance Saccharomyces Cerevisiae Growth under Cold and NaCl Stress
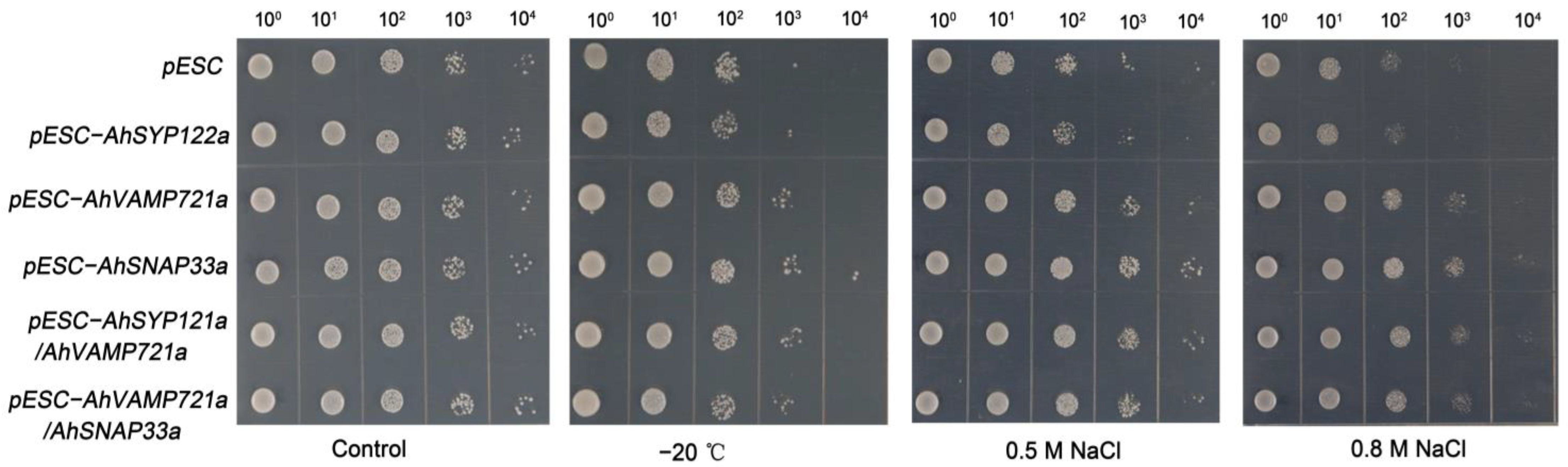
To study the potential function of AhSNAREs under stress conditions, we cloned the full-length coding sequences of three genes: AhSYP122a, AhSNAP33a and AhVAMP721a. We investigated the function of these three AhSNAREs under cold and NaCl stress in budding yeast (Saccharomyces cerevisiae). Accordingly, we individually transformed yeast strain BY4741 with the plasmids pESC-AhSYP122a, pESC-AhVAMP721a, pESC-AhSNAP33a, pESC-AhSYP122a/AhSNAP33a or pESC-AhVAMP721a/AhSNAP33a allowing the expression of each selected gene. We then exposed the resulting positive yeast colonies to cold stress at −20 °C for 1 h, or to growth on medium containing 0.5 M NaCl or 0.8 M NaCl for 5 h. As illustrated in Figure 8, there was no substantial difference between BY4741 expressing AhSNARE genes and BY4741 harboring the empty vector pESC under normal conditions. However, BY4741 containing pESC-AhSYP122a, pESC-AhVAMP721a or pESC-AhSNAP33a, particularly the latter, had a higher viability than yeast containing empty vector pESC after NaCl or freezing (−20 °C) stress. In addition, BY4741 heterologously expressing two SNARE genes (pESC-AhSYP122a/AhSNAP33a or pESC-AhVAMP722a/AhSNAP33a) had slightly greater viability under NaCl or freezing (−20 °C) than yeast cells expressing only one SNARE gene. This result suggests that heterologous expression of AhSYP122a, AhSNAP33a or AhVAMP721a enhanced tolerance to abiotic stress in yeast.
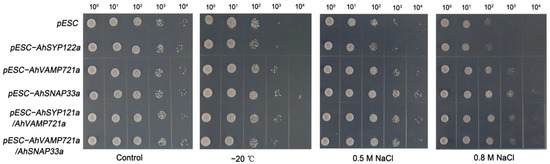
Figure 8.
Growth of transformed yeast harboring pESC-AhSYP122a, pESC-AhVAMP721a, pESC-AhSNAP33a, pESC-AhSYP122a/AhSNAP33a or pESC-AhVAMP721a/AhSNAP33a vectors under −20 °C for 1 h, and 0.5 M or 0.8 M NaCl for 5 h. The experiment was repeated three times.
3. Discussion
SNAREs are known to perform crucially important roles in vesicular trafficking and participate in multiple growth and stress responses. Therefore, they are expected to be a target for crop breeding and improvement. The SNARE multigene family has been studied and analyzed in several plant species, for instance, Arabidopsis, rice [31], tomato (Solanum lycopersicum) [13], olive rape (Brassica napus) [38] and wheat (Triticum aestivum) [39]. Peanut is a special leguminous crop with subterranean pods and high oil and protein contents. Therefore, it is important to identify all putative SNARE genes in cultivated peanut and analyze their potential functions.
Here, we identified 129 SNARE genes from A. hypogaea, 63 from A. duranensis and 64 from A. ipaensis, which were assigned to 21 conserved subfamilies. Studies on the SNARE family in other species have indicated that the number of SNAREs may be relevant to plant polyploidy. For instance, there are 63 SNAREs in tomato (AB) [28] and 173 in wheat (AABBDD) [39]. In cultivated peanut (A. hypogaea), 90.0% of peanut SNAREs can be assigned to 1:1 homoeologous genes from A. duranensis and A. ipaensis [25]. This is a considerably high homoeologous retention rate. The high numbers of AhSNARE genes might reflect the allotetraploid genome of peanut. Five out of twenty-one subfamilies did not appear as pairage in our study (Table S2 and Figure 2). For instance, the number of AhNPSN1 subfamily members was lower in A. hypogaea than in the probable progenitors combined, suggesting loss of these genes during long-term evolution or insufficient sequencing depth. By contrast, the number of AhBET subfamily members in A. hypogaea was three more than the sum from the two ancestral genomes. AtBET11 (also named Bs14b) and AtBET12 (also named Bs14a) localize to the Golgi membrane and affect protein ER–Golgi anterograde transport. Previous studies have shown that AtBET11 and AtBET12 are necessary for the growth and development of pollen tubes, with reduced seed fertilization, defective embryo development, and reduced pollen tube length and secondary tube formation observed in the Arabidopsis bet11 bet12 double mutant [40]. SNARE proteins act in multi-protein complexes; R-SNARE, Qa-, Qb- and Qc-SNARE self-assemble into a complex of four coiled-coil helices to complete the fusion of vesicles and target membranes [41]. Changes in gene dosage may lead to changes in the possible combinations of proteins in these complexes, which may result in phenotypic or functional effects [42]. A large number of AhBET genes may be related to the special flower development of peanut. Through gene duplication and collinearity analysis, we determined that 91.5% of SNARE genes are homologous to their ancestral genes. Once more, this result demonstrates a high rate of homoeolog retention after the hybridization of the A and B genomes (Figure 5) and the biological importance of the SNARE gene family in general.
We identified a large number of cis-acting elements related to various stresses, phytohormones and developmental processes in the AhSNARE promoters of peanut (Table S5). Most of the SNARE proteins studied participate in responses to abiotic and biotic stress [32,43,44] and the regulation of development [5,29,45]. The peanut has one of the most interesting growth habits: it produces flowers aerially and bears fruit underground. The ovary is not surrounded by petals but is located instead at the base of the hypanthium. After fertilization, the embryo undergoes a few rounds of cell division and then becomes dormant. The meristem of the ovary then begins to elongate and forms a “peg” structure, with the ovary just behind the lignified top. Our expression data suggested that AhSYP131b, AhSYP72, AhNPSN11a/b, AhVAMP713 and AhVAMP723 were specifically expressed in pistils. It is possible that SNARE genes play a key role in the special reproductive mode of peanuts. Interestingly, AhSYP131b, AhNPSN11a/b and AhVAMP723 are also related to pollen tube development [46,47]. Once the tip is a few centimeters below the soil surface, the pod begins to develop. Most of the cell division occurs in the distal region 1–3 mm from the tip [48]. In our study, AhSYP11 showed the highest expression during the development of the peg (Figure 6). AtSYP111 (also named KNOLLE), the first SNARE family member discovered, and its interacting protein KEULE (Sec1/Munc18), play key roles in cell-plate formation and their encoding genes are specifically expressed during cell division [49]. In the knolle and keule mutants, many unfused vesicles aggregate at the cell plate [49,50,51]. Several studies showed that indole-3-acetic acid (IAA) plays the most important role in the development of gynophore and peanut fruits [48,52]. Immunogold electron microscopy (EM) study of Arabidopsis root apices displayed that there was a large percentage of IAA clustered within vesicles and membranous compartments [53]. Furthermore, the expression of AhSYP122a and AhVAMP725 was higher in pericarp. Fibrous materials were deposited in the pericarp throughout maturation. Cellulose, synthesized by cellulose synthase complexes (CSCs), were transported to the plasma membrane by SYP61 labeled vesicles; the SYP121 complex was also contained in these vesicles.
Peanut is rich in protein and lipid, and its lipid content reaches its maximum in the later stages of development. The expression data in this study also showed that AhVTI13b in the Qb-SNARE subfamily was expressed at high levels during the late stage of seed development and might play a vital role in peanut lipoprotein storage. Previous studies demonstrated that AtVTI12, a homolog of AhVTI13, mediates trafficking to storage vacuoles [21]. SNARE proteins of different classes interact with each other to promote vesicle fusion [54]. Co-expression of genes from different groups provides a basis for analyzing the potential interaction of their encoding proteins, for example, AhSYP122, AhSYP132, AhSNAP33 and AhVAMP725 show higher expression in roots, nodules and pericarp than in other tissues. A recent study showed that the infection thread (IT) is not elongated in roots of birdsfoot trefoil (Lotus japonicus) knocked down for LjSYP132 by RNA interference (LjSYP132a-RNAi, LjSYP132b-RNAi) [55]. Nitrogen fixation activity and the number of bacteroides severely decreased in nodules formed on LjVAMP72a/72b-RNAi knockdown roots, and arbuscular mycorrhization (AM) was also curtailed. Kim S. et al. found that the SYP132–VAMP721/722 interaction is involved in immunity to Pseudomonas syringae in tomato [56]. In Arabidopsis, most vesicular trafficking to the plasma membrane is driven by SYP121/122 through their assembly with the R-SNAREs VAMP721/722 and the Qb+c-SNARE SNAP33 [57]. Wang and Zhang found that SYP121 and SYP122 are hub genes that play a key role in plant growth-to-defense transition [58].
Plant responses to abiotic stress rely on the regulation of vesicular trafficking to ensure that proteins specialized for sensing stimuli and responding to stress are correctly localized. Drakakaki et al. (2012) isolated the SYP61 labeled compartment and carried out proteomic analysis of its contents; the SYP121-complex, cellulose synthases, and PIP 2;1 were identified [59]. Previous studies have shown that plant cells produce a large amount of reactive oxygen species under high salt environment, and the inhibition of VAMP7 gene expression leads to the failure of vesicles containing H2O2 fused with tonoplast and the formation of giant vesicles containing H2O2 in the cytoplasm. Under salt stress, the vesicles are very active, and Na+ accumulates not only in the main vacuole but also in the small vesicles around the vacuole [60]. Lu et al. (2020) also found that vesicular transport is involved in the secretion of salt in the leaves of Limonium bicolor [32]. Peanut is a thermophilic, drought-tolerant plant that is relatively sensitive to salt and low temperature. We studied the transcript levels of AhSNARE genes under salt and low-temperature conditions, which provided clues for exploring their function. Improper low-temperature stress can increase cell membrane permeability and membrane lipid peroxidation. Low temperature has a serious effect on peanut seedlings, with many differentially expressed genes identified at low temperature; however, there was no distinct difference in expression under salt treatment (Figure S1). RT-qPCR analysis over a 24 h time course revealed that many AhSNARE genes were highly expressed at 3–6 h in response to stress and their expression levels decreased at 24 h into salt treatment (Figure 7); expression levels of AhSYP122a, AhSNAP33a, AhVAMP725 and AhVAMP722a were different after salt and low-temperature treatments. Most SNARE proteins reported to date participate in responses to abiotic and biotic stress [3]. VAMP71 proteins play a vital role in the localization of reactive oxygen species, and suppression of AtVAMP7C expression increases tolerance to salt stress [34,61]. SYP121/122 are redundant in their function, facilitating the majority of secretory trafficking to the plasma membrane but mediating different secretory cargoes [62]. SYP121/122 are involved in responses to pathogen invasion and abiotic stress, and the AtSYP121-AtSNAP33-AtVAMP721 SNARE complex participates in transport of K+ channels to the plasma membrane in Arabidopsis [57]. Furthermore, AtSYP121 together with AtSYP61 regulates the water potential of cells by coordinating the transport of aquaporin AtPIP2;7 (plasma membrane intrinsic protein 2;7) [59,63]. Heterologous expression of AhSYP122a, AhVAMP721a and AhSNAP33a increased cold and salt tolerance in yeast (Figure 8). However, heterologously expressing two SNARE genes (pESC-AhSYP122a/AhSNAP33a or pESC-AhVAMP722a/AhSNAP33a) had slightly greater viability than that in one SNARE gene. This is due to the snares’ needs to form a complex to function [7]. Salinas-Cornejo et al. (2021) reported that tomato overexpressing SlSNAP33.2 displays more tolerance to salt stress than control tomato plants [64]. In addition, when wild soybean (Glycine soja) SNAP33 was heterologously expressed in Arabidopsis, the transgenic plants showed improved salt and drought tolerance [65]. In Arabidopsis, AtSYP121 and AtVAMP721/722 are necessary for both biotic and abiotic stress responses [66]. AhSNAREs are engaged in multiple stress responses. Therefore, we believe that AhSNARE is a multifunctional gene family that plays vital roles in peanut growth and development as well as in defense responses to various environmental stresses.
4. Materials and Methods
4.1. Identification of the SNARE Family in Peanut
Genome sequences, functional annotations and coding sequence (CDS) predictions for Arachis hypogaea, Arachis duranensis and Arachis ipaensis were downloaded from Peanutbase (https://peanutbase.org/peanut_genome, accessed on 24 May 2022). SNARE family proteins were identified in the peanut protein database using HMMER 3.0 software with the HMM (Hidden Markov Model chain) file of the domain (PF05739) downloaded from the Pfam database. Incomplete and redundant amino acid sequences were removed. Splice variants were deleted, and only the longest variant was retained for further analysis. SNARE protein sequences from Arabidopsis were obtained from the TAIR database (https://www.arabidopsis.org, accessed on 22 May 2022). Additionally, each protein was manually validated using BLASTp (https://blast.ncbi.nlm.nih.gov, accessed on 24 May 2022), the online CD-search tool (http://www.ncbi.nlm.nih.gov/Structure/cdd/wrpsb.cgi, accessed on 28 May 2022) and the SMART tool (embl-heidelberg.de, accessed on 28 May 2022). Candidate SNAREs were named according to their most probable homolog in Arabidopsis, preceded by an abbreviation for the species name (Ah for A. hypogaea, Ad for A. duranensis and Ai for A. ipaensis); genes belonging to one subfamily but found in different genomes were numbered consecutively. The subcellular localization of SNAREs was predicted using Plant-mPLoc (http://www.csbio.sjtu.edu.cn/bioinf/plant-multi/, accessed on 2 June 2022) and ProtComp (http://linux1.softberry.com/berry.phtml, accessed on 2 June 2022).
4.2. Phylogenetic Analysis, Gene Structure and Conserved Motif Analyses of the SNARE Family
Alignment of full-length SNARE protein sequences from A. hypogaea, A. ipaensis, A. duranensis and Arabidopsis was performed using the MUSCLE program in the MEGA 7 software [67]. On the basis of this alignment, an unrooted phylogenetic tree was reconstructed using the maximum-likelihood method and the nearest-neighbor-interchange model. The resulting tree file was visualized using Evoview version 2.0 (https://www.evolgenius.info/evolview-v2, accessed on 24 July 2022). Conserved motifs in SNARE proteins were analyzed using an online tool (http://meme-suite.org/, accessed on 28 May 2022), allowing any number of repetitions with an optimum motif width of 6–50 residues and up to 30 motifs. Gene structure information was downloaded from the peanut database and assessed using an online tool (http://gsds.cbi.pku.edu.cn, accessed on 22 May 2022). Motifs were then identified using TBtools, and schematic diagrams of conserved motifs in the SNARE proteins were drawn accordingly [68].
4.3. Chromosome Location and Gene Duplications in the SNARE Family
The lengths of each chromosome and the position of every SNARE gene were obtained from gff3 annotation files in the peanut database, and the chromosomal distribution and density of genes were determined using TBtools. The following two criteria were considered when analyzing SNARE gene duplications: (1) the length of the shorter sequence covers at least 70% of the longer sequence; (2) the identity of the two sequences is at least 70%. SNARE gene duplications were screened using the MCScanX algorithm with default settings (e ≤ 1 × 10−10) [69]. Diagrams were drawn using advanced Circos in TBtools [68].
4.4. Promoter Analysis of SNARE Genes in Peanut
Sequences encompassing 2000 bp upstream of the start codon of each AhSNARE gene were obtained from the peanut genome using TBtools, and analysis of cis-acting elements was performed using the online server PlantCARE [70]. The distribution of motifs in each promoter was drawn using TBtools [68].
4.5. RNA-Seq Data and Differential Expression of SNARE Genes
The differential expression of SNARE genes in different tissues and at different stages of peanut pod development were analyzed. Specifically, 22 different transcriptome samples were obtained from Genbank BioProject PRJNA291488 [71]. Transcriptome assembly and expression value estimation (Fragments per kilobase million, FPKM) were performed using StringTie (v1.3.4) with default parameters, as reported by Liu et al. [72].
RNA-seq data of roots and leaves of FH1 peanut (‘Fenghua No. 1′) were derived from previous work (Genbank PRJNA553073). Raw sequences were compared to the genome of A. hypogaea ‘Tifrunner’ (https://www.peanutbase.org/, accessed on 24 May 2022) using HISAT2 software [73]. Gene expression levels and FPKM values of all genes were obtained using the Featurecounts tool in the Subread software [74]. The expression profiles of SNARE genes in different tissues and under diverse stress were extracted and averaged for plotting heatmaps using TBtools.
4.6. Plant Materials and Growth Conditions
Peanut seeds (FH1) of uniform size were selected and surface sterilized with 0.1% mercuric chloride; four seeds per bottle were planted in Murashige and Skoog (MS) medium and placed under 16-h light (26 °C)/8-h dark (20 °C) conditions in a culture room for 2 weeks. Seedlings showing the same growth vigor were selected for 8 °C treatment and 200 mM NaCl treatment; each treatment was repeated three times.
4.7. RNA Isolation and Expression of SNARE Genes after Stress
After treatment, leaves and roots were separately harvested, frozen immediately in liquid nitrogen and stored at −80 °C. Total RNA was extracted using an FastPure® Plant Total RNA Isolation Kit (Vazyme, Nanjing, China). Ten genes showing significant differential expression after salt and low-temperature treatments were examined using RT-qPCR. Specific primers were designed using Beacon Designer 7.0 software (Table S1). The housekeeping gene TUA5 was used as an internal reference with primers, as described by Liu et al. [72]. RT-qPCR was performed using a HiScript III RT SuperMix kit (Vazyme, Nanjing, China), as described by Lu et al. [32]. The 2−∆∆Ct method was used for calculating the relative expression of each SNARE gene [36,75].
4.8. Functional Identification Using a Yeast Expression System
Full-length cDNA sequences of AhSYP122a, AhSNAP33a and AhVAMP721a were obtained from the genome database of A. hypogaea. PCR fragments were amplified from cDNA using specific primers (Table S1) designed using CE Design software (https://crm.vazyme.com/cetool/simple.html, accessed on 22 July 2022); then, they were purified and cloned into the EcoR I sites of pESC-URA (Sangon Biotech, Shanghai, China) using a ClonExpress II One Step Cloning Kit (Vazyme Biotech, Nanjing, China) to generate pESC-AhSYP122a, pESC-AhSNAP33a and pESC-AhVAMP721a plasmids. The AhSNAP33a and AhVAMP721a coding sequences were inserted at another site of the pESC-AhSYP122a vector to generate pESC-AhSYP122a/AhSNAP33a and pESC-AhSYP122a/AhVAMP721a plasmids, respectively.
The resulting plasmids were introduced into Saccharomyces cerevisiae strain BY4741 using the lithium acetate/polyethylene glycol (LiAc/PEG) method. Single colonies of transformant yeasts were picked and inoculated into liquid synthetic defined SD–Ura medium for 2 days at 28 °C (OD600 = 0.6). Cultures were then diluted with fresh pre-warmed SD–Ura medium (dilution 1:10) and serially diluted in 10-fold steps; 3-µL aliquots of each dilution were dropped onto SD–Ura medium (2% [w/v] galactose) plates with or without different concentrations of NaCl for 5 h and at −20 °C for 1 h, respectively. Growth of yeast cells was observed and recorded after 48 h at 29 °C.
5. Conclusions
We identified 129 SNARE genes in A. hypogaea, 63 in A. duranensis and 64 in A. ipaensis, respectively, which we classified into five groups based on their phylogenetic relationship with their homologs in Arabidopsis. AhSNAREs are unevenly scattered on the chromosomes and exhibit a high rate of homoeolog retention. The exon–intron and motif structures of the SNARE family are highly conserved. Promoters of AhSNAREs contain cis-elements associated with development and stress responses. Comprehensive analysis of AhSNARE gene expression patterns suggested that AhVTI13b plays an important role in the storage of lipid proteins; AhSYP131b, AhSYP72, AhNPSN11a/b, AhVAMP713 and AhVAMP723 may play a role in the specific developmental patterns of peanut; and AhSYP122a-AhSNAP33a-AhVAMP721a may play a crucial role in stress responses. This study provides a reference for peanut breeding and further exploration of the mechanism by which SNAREs respond to abiotic stress and development in peanuts.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/ijms24087103/s1.
Author Contributions
S.W. and G.L.: supervision and data curation. C.L.: writing—original draft. C.L., Z.P. and Y.L.: writing—review and editing. All authors have read and agreed to the published version of the manuscript.
Funding
Shandong Province key research and development plan (2021TZXD005). Special Project of Centra Government for Local Science and Technology Development of Shandong Province (YDZX20203700003989).
Institutional Review Board Statement
No applicable.
Informed Consent Statement
No applicable.
Data Availability Statement
The data and materials that were analyzed in the current study are available from the corresponding author upon reasonable request.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Wang, X.; Xu, M.; Gao, C.; Zeng, Y.; Cui, Y.; Shen, W.; Jiang, L. The roles of endomembrane trafficking in plant abiotic stress responses. J. Integr. Plant Biol. 2020, 62, 55–69. [Google Scholar] [CrossRef] [PubMed]
- Paul, P.; Simm, S.; Mirus, O.; Scharf, K.-D.; Fragkostefanakis, S.; Schleiff, E. The complexity of vesicle transport factors in plants examined by orthology search. PLoS ONE 2014, 9, e97745. [Google Scholar] [CrossRef]
- Kwon, C.; Lee, J.H.; Yun, H.S. SNAREs in plant biotic and abiotic stress responses. Mol. Cells 2020, 43, 501–508. [Google Scholar] [PubMed]
- Lian, Q.; Meng, Y.; Zhao, X.; Xu, Y.; Wang, Y.; Day, B.; Ma, Q. ShNPSN11, a vesicle-transport-related gene, confers disease resistance in tomato to Oidium neolycopersici. Biochem. J. 2020, 477, 3851–3866. [Google Scholar] [CrossRef] [PubMed]
- Luo, C.; Shi, Y.; Xiang, Y. SNAREs regulate vesicle trafficking during root growth and development. Front. Plant Sci. 2022, 13, 853251. [Google Scholar] [CrossRef]
- Lu, C.; Yuan, F.; Guo, J.; Han, G.; Wang, C.; Chen, M.; Wang, B. Current understanding of role of vesicular transport in salt secretion by salt glands in recretohalophytes. Int. J. Mol. Sci. 2021, 22, 2203. [Google Scholar] [CrossRef]
- Uemura, T.; Ueda, T.; Ohniwa, R.L.; Nakano, A.; Takeyasu, K.; Sato, M.H. Systematic analysis of SNARE molecules in Arabidopsis: Dissection of the post-Golgi network in plant cells. Cell Struct. Funct. 2004, 29, 49–65. [Google Scholar] [CrossRef]
- Zhu, J.; Gong, Z.; Zhang, C.; Song, C.-P.; Damsz, B.; Inan, G.; Koiwa, H.; Zhu, J.-K.; Hasegawa, P.M.; Bressan, R.A. OSM1/SYP61: A syntaxin protein in Arabidopsis controls abscisic acid-mediated and non-abscisic acid-mediated responses to abiotic stress. Plant Cell 2002, 14, 3009–3028. [Google Scholar] [CrossRef]
- Kim, H.; O’Connell, R.; Maekawa-Yoshikawa, M.; Uemura, T.; Neumann, U.; Schulze-Lefert, P. The powdery mildew resistance protein RPW8.2 is carried on VAMP721/722 vesicles to the extrahaustorial membrane of haustorial complexes. Plant J. 2014, 79, 835–847. [Google Scholar] [CrossRef]
- Jahn, R.; Scheller, R.H. SNAREs—Engines for membrane fusion. Nat. Rev. Mol. Cell Biol. 2006, 7, 631–643. [Google Scholar] [CrossRef]
- Kanazawa, T.; Era, A.; Minamino, N.; Shikano, Y.; Fujimoto, M.; Uemura, T.; Nishihama, R.; Yamato, K.T.; Ishizaki, K.; Nishiyama, T.; et al. SNARE molecules in marchantia polymorpha: Unique and conserved features of the membrane fusion machinery. Plant Cell Physiol. 2016, 57, 307–324. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.-J.; Brandizzi, F. News and Views into the SNARE Complexity in Arabidopsis. Front. Plant Sci. 2012, 3, 28. [Google Scholar] [CrossRef] [PubMed]
- Salinas-Cornejo, J.; Madrid-Espinoza, J.; Ruiz-Lara, S. Identification and transcriptional analysis of SNARE vesicle fusion regulators in tomato (Solanum lycopersicum) during plant development and a comparative analysis of the response to salt stress with wild relatives. J. Plant Physiol. 2019, 242, 153018. [Google Scholar] [CrossRef] [PubMed]
- Sanderfoot, A.A.; Assaad, F.F.; Raikhel, N.V. The Arabidopsis genome. An abundance of soluble N-ethylmaleimide-sensitive factor adaptor protein receptors. Plant Physiol. 2000, 124, 1558–1569. [Google Scholar] [CrossRef] [PubMed]
- Sutter, J.U.; Campanoni, P.; Blatt, M.R.; Paneque, M. Setting SNAREs in a different wood. Traffic 2006, 7, 627–638. [Google Scholar] [CrossRef] [PubMed]
- Mcnew, J.A.; Sogaard, M.; Lampen, N.M.; Machida, S.; Ye, R.R.; Lacomis, L.; Tempst, P.; Rothman, J.E.; Sllner, T.H. Ykt6p, a prenylated SNARE essential for endoplasmic reticulum-Golgi transport. J. Biol. Chem. 1997, 272, 17776–17783. [Google Scholar] [CrossRef]
- Vogel, K.; Roche, P.A. SNAP-23 and SNAP-25 are palmitoylated in vivo. Biochem. Biophys. Res. Commun. 1999, 258, 407–410. [Google Scholar] [CrossRef]
- Malsam, J.; Kreye, S.; Söllner, T.H. Membrane fusion: SNAREs and regulation. Cell. Mol. Life Sci. 2008, 65, 2814–2832. [Google Scholar] [CrossRef]
- Aniento, F.; Sánchez de Medina Hernández, V.; Dagdas, Y.; Rojas-Pierce, M.; Russinova, E. Molecular mechanisms of endomembrane trafficking in plants. Plant Cell 2022, 34, 146–173. [Google Scholar] [CrossRef]
- Lipka, V.; Chian, K.; Panstruga, R.; Schekman, R.; Goldstein, L.; Rossant, J. SNARE-ware: The role of SNARE-domain proteins in plant biology. Annu. Rev. Cell Dev. Biol. 2007, 23, 147. [Google Scholar] [CrossRef]
- Sanmartín, M.; Ordóñez, A.; Sohn, E.J.; Robert, S.; Sánchez-Serrano, J.J.; Surpin, M.A.; Raikhel, N.V.; Rojo, E. Divergent functions of VTI12 and VTI11 in trafficking to storage and lytic vacuoles in Arabidopsis. Proc. Natl. Acad. Sci. USA 2007, 104, 3645–3650. [Google Scholar] [CrossRef]
- Liu, Y.; Shao, L.; Zhou, J.; Li, R.; Pandey, M.K.; Han, Y.; Cui, F.; Zhang, J.; Guo, F.; Chen, J.; et al. Genomic insights into the genetic signatures of selection and seed trait loci in cultivated peanut. J. Adv. Res. 2022, 42, 237–248. [Google Scholar] [CrossRef] [PubMed]
- Li, P.; Peng, Z.; Xu, P.; Tang, G.; Ma, C.; Zhu, J.; Shan, L.; Wan, S. Genome-wide identification of NAC transcription factors and their functional prediction of abiotic stress response in peanut. Front. Genet. 2021, 12, 630292. [Google Scholar] [CrossRef] [PubMed]
- Yan, H.; Zong, C.; Shan, S. Vesicle Trafficking Patterns in Developing Peanut Fruits Related to Aspergillus flavus Resistancy and Development. Chin. J. Cell Biol. 2014, 36, 12. [Google Scholar]
- Bertioli, D.J.; Cannon, S.B.; Froenicke, L.; Huang, G.; Farmer, A.D.; Cannon, E.K.; Liu, X.; Gao, D.; Clevenger, J.; Dash, S.; et al. The genome sequences of Arachis duranensis and Arachis ipaensis, the diploid ancestors of cultivated peanut. Nat. Genet. 2016, 48, 438–446. [Google Scholar] [CrossRef] [PubMed]
- Bertioli, D.J.; Jenkins, J.; Clevenger, J.; Dudchenko, O.; Gao, D.; Seijo, G.; Leal-Bertioli, S.C.M.; Ren, L.; Farmer, A.D.; Pandey, M.K.; et al. The genome sequence of segmental allotetraploid peanut Arachis hypogaea. Nat. Genet. 2019, 51, 877–884. [Google Scholar] [CrossRef]
- Bubeck, J.; Scheuring, D.; Hummel, E.; Langhans, M.; Viotti, C.; Foresti, O.; Denecke, J.; Banfield, D.K.; Robinson, D.G. The syntaxins SYP31 and SYP81 control ER-Golgi trafficking in the plant secretory pathway. Traffic 2008, 9, 1629–1652. [Google Scholar] [CrossRef]
- Sasvari, Z.; Kovalev, N.; Gonzalez, P.A.; Xu, K.; Nagy, P.D. Assembly-hub function of ER-localized SNARE proteins in biogenesis of tombusvirus replication compartment. PLoS Path. 2018, 14, e1007028. [Google Scholar] [CrossRef]
- Rui, Q.; Tan, X.; Liu, F.; Li, Y.; Liu, X.; Li, B.; Wang, J.; Yang, H.; Qiao, L.; Li, T.; et al. Syntaxin of plants31 (SYP31) and SYP32 is essential for Golgi morphology maintenance and pollen development. Plant Physiol. 2021, 186, 330–343. [Google Scholar] [CrossRef]
- Fusella, A.; Micaroni, M.; Di Giandomenico, D.; Mironov, A.A.; Beznoussenko, G.V. Segregation of the Qb-SNAREs GS27 and GS28 into Golgi vesicles regulates intra-Golgi transport. Traffic 2013, 14, 568–584. [Google Scholar] [CrossRef]
- Sanderfoot, A. Increases in the number of SNARE genes parallels the rise of multicellularity among the green plants. Plant Physiol. 2007, 144, 6–17. [Google Scholar] [CrossRef] [PubMed]
- Lu, C.; Feng, Z.; Yuan, F.; Han, G.; Guo, J.; Chen, M.; Wang, B. The SNARE protein LbSYP61 participates in salt secretion in Limonium bicolor. Environ. Exp. Bot. 2020, 176, 104076. [Google Scholar] [CrossRef]
- Kim, S.J.; Bassham, D.C. TNO1 is involved in salt tolerance and vacuolar trafficking in Arabidopsis. Plant Physiol. 2011, 156, 514–526. [Google Scholar] [CrossRef] [PubMed]
- Leshem, Y.; Melamed-Book, N.; Cagnac, O.; Ronen, G.; Nishri, Y.; Solomon, M.; Cohen, G.; Levine, A. Suppression of Arabidopsis vesicle-SNARE expression inhibited fusion of H2O2-containing vesicles with tonoplast and increased salt tolerance. Proc. Natl. Acad. Sci. USA 2006, 103, 18008–18013. [Google Scholar] [CrossRef]
- Yun, H.S.; Kwon, C. Vesicle trafficking in plant immunity. Curr. Opin. Plant Biol. 2017, 40, 34–42. [Google Scholar] [CrossRef]
- Sui, N.; Wang, Y.; Liu, S.; Yang, Z.; Wang, F.; Wan, S. Transcriptomic and physiological evidence for the relationship between unsaturated fatty acid and salt stress in peanut. Front. Plant Sci. 2018, 9, 7. [Google Scholar] [CrossRef]
- Qin, L.; Li, L.; Bi, C.; Zhang, Y.; Wan, S.; Meng, J.; Meng, Q.; Li, X. Damaging mechanisms of chilling and salt stress to Arachis hypogaea L. leaves. Photosynthetica 2011, 49, 37–42. [Google Scholar] [CrossRef]
- Xu, J.; Zhao, X.; Bao, J.; Shan, Y.; Zhang, M.; Shen, Y.; Abubakar, Y.S.; Lu, G.; Wang, Z.; Wang, A. Genome-wide identification and expression analysis of SNARE genes in Brassica napus. Plants 2022, 11, 711. [Google Scholar] [CrossRef]
- Wang, G.; Long, D.; Yu, F.; Zhang, H.; Chen, C.; Wang, Y.; Ji, W. Genome-wide identification, evolution, and expression of the SNARE gene family in wheat resistance to powdery mildew. PeerJ 2021, 9, e10788. [Google Scholar] [CrossRef]
- Bolaños-Villegas, P.; Guo, C.L.; Jauh, G.Y. Arabidopsis Qc-SNARE genes BET11 and BET12 are required for fertility and pollen tube elongation. Bot. Stud. 2015, 56, 21. [Google Scholar] [CrossRef]
- Gu, X.; Brennan, A.; Wei, W.; Guo, G.; Lindsey, K. Vesicle transport in plants: A revised phylogeny of SNARE proteins. Evol. Bioinform. 2020, 16, 1176934320956575. [Google Scholar] [CrossRef] [PubMed]
- Schilling, S.; Kennedy, A.; Pan, S.; Jermiin, L.S.; Melzer, R. Genome-wide analysis of MIKC-type MADS-box genes in wheat: Pervasive duplications, functional conservation and putative neofunctionalization. New Phytol. 2020, 225, 511–529. [Google Scholar] [CrossRef] [PubMed]
- Uemura, T.; Ueda, T.; Nakano, A. The physiological role of SYP4 in the salinity and osmotic stress tolerances. Plant Signal. Behav. 2012, 7, 1118–1120. [Google Scholar] [CrossRef] [PubMed]
- Zouhar, J.; Rojo, E.; Bassham, D.C. AtVPS45 is a positive regulator of the SYP41/SYP61/VTI12 SNARE complex involved in trafficking of vacuolar cargo. Plant Physiol. 2009, 149, 1668–1678. [Google Scholar] [CrossRef]
- Ichikawa, M.; Hirano, T.; Enami, K.; Fuselier, T.; Kato, N.; Kwon, C.; Voigt, B.; Schulze-Lefert, P.; Baluška, F.; Sato, M.H. Syntaxin of plant proteins SYP123 and SYP132 mediate root hair tip growth in Arabidopsis thaliana. Plant Cell Physiol. 2014, 55, 790–800. [Google Scholar] [CrossRef]
- Ruan, H.; Li, J.; Wang, T.; Ren, H. Secretory vesicles targeted to plasma membrane during pollen germination and tube growth. Front. Cell Dev. Biol. 2021, 8, 615447. [Google Scholar] [CrossRef]
- Slane, D.; Reichardt, I.; El Kasmi, F.; Bayer, M.; Jürgens, G. Evolutionarily diverse SYP1 Qa-SNAREs jointly sustain pollen tube growth in Arabidopsis. Plant J. 2017, 92, 375–385. [Google Scholar] [CrossRef]
- Kumar, R.; Pandey, M.K.; Roychoudhry, S.; Nayyar, H.; Kepinski, S.; Varshney, R.K. Peg Biology: Deciphering the molecular regulations involved during peanut peg development. Front. Plant Sci. 2019, 10, 1289. [Google Scholar] [CrossRef]
- Lauber, M.H.; Waizenegger, I.; Steinmann, T.; Schwarz, H.; Mayer, U.; Hwang, I.; Lukowitz, W.; Jürgens, G. The Arabidopsis KNOLLE protein is a cytokinesis-specific syntaxin. J. Cell Biol. 1997, 139, 1485–1493. [Google Scholar] [CrossRef]
- Cheung, A.Y.; Cosgrove, D.J.; Hara-Nishimura, I.; Jürgens, G.; Lloyd, C.; Robinson, D.G.; Staehelin, L.A.; Weijers, D. A rich and bountiful harvest: Key discoveries in plant cell biology. Plant Cell 2022, 34, 53–71. [Google Scholar] [CrossRef]
- Jürgens, G.; Park, M.; Richter, S.; Touihri, S.; Krause, C.; El Kasmi, F.; Mayer, U. Plant cytokinesis: A tale of membrane traffic and fusion. Biochem. Soc. Trans. 2015, 43, 73–78. [Google Scholar] [CrossRef] [PubMed]
- Moctezuma, E. The peanut gynophore: A developmental and physiological perspective. Can. J. Bot. 2011, 81, 183–190. [Google Scholar] [CrossRef]
- Mettbach, U.; Strnad, M.; Mancuso, S.; Baluška, F. Immunogold-EM analysis reveal brefeldin a-sensitive clusters of auxin in Arabidopsis root apex cells. Commun. Integr. Biol. 2017, 10, e1327105. [Google Scholar] [CrossRef] [PubMed]
- Fujiwara, M.; Uemura, T.; Ebine, K.; Nishimori, Y.; Ueda, T.; Nakano, A.; Sato, M.H.; Fukao, Y. Interactomics of Qa-SNARE in Arabidopsis thaliana. Plant Cell Physiol. 2014, 55, 781–789. [Google Scholar] [CrossRef]
- Sogawa, A.; Takahashi, I.; Kyo, M.; Imaizumi-Anraku, H.; Tajima, S.; Nomura, M. Requirements of Qa-SNARE LjSYP132s for nodulation and seed development in Lotus japonicus. Plant Cell Physiol. 2020, 61, 1750–1759. [Google Scholar] [CrossRef]
- Kim, S.; Kim, H.; Park, K.; Cho, D.J.; Kim, M.K.; Kwon, C.; Yun, H.S. Synaptotagmin 5 Controls SYP132-VAMP721/722 Interaction for Arabidopsis immunity to Pseudomonas syringae pv tomato DC3000. Mol. Cells 2021, 44, 670–679. [Google Scholar] [CrossRef]
- Waghmare, S.; Lefoulon, C.; Zhang, B.; Liliekyte, E.; Donald, N.; Blatt, M.R. K(+) Channel-SEC11 binding exchange regulates SNARE assembly for secretory traffic. Plant Physiol. 2019, 181, 1096–1113. [Google Scholar] [CrossRef]
- Wang, T.; Zhang, X. Genome-wide dynamic network analysis reveals the potential genes for MeJA-induced growth-to-defense transition. BMC Plant Biol. 2021, 21, 450. [Google Scholar] [CrossRef]
- Drakakaki, G.; van de Ven, W.; Pan, S.Q.; Miao, Y.S.; Wang, J.Q.; Keinath, N.F.; Weatherly, B.; Jiang, L.W.; Schumacher, K.; Hicks, G.; et al. Isolation and proteomic analysis of the SYP61 compartment reveal its role in exocytic trafficking in Arabidopsis. Cell Res. 2012, 22, 413–424. [Google Scholar] [CrossRef]
- Hamaji, K.; Nagira, M.; Yoshida, K.; Ohnishi, M.; Oda, Y.; Uemura, T.; Goh, T.; Sato, M.H.; Morita, M.T.; Tasaka, M.; et al. Dynamic aspects of ion accumulation by vesicle traffic under salt stress in Arabidopsis. Plant Cell Physiol. 2009, 50, 2023–2033. [Google Scholar] [CrossRef]
- Leshem, Y.; Golani, Y.; Kaye, Y.; Levine, A. Reduced expression of the v-SNAREs AtVAMP71/AtVAMP7C gene family in Arabidopsis reduces drought tolerance by suppression of abscisic acid-dependent stomatal closure. J. Exp. Bot. 2010, 61, 2615–2622. [Google Scholar] [CrossRef] [PubMed]
- Waghmare, S.; Lileikyte, E.; Karnik, R.; Goodman, J.K.; Blatt, M.R.; Jones, A.M.E. SNAREs SYP121 and SYP122 mediate the secretion of distinct cargo subsets. Plant Physiol. 2018, 178, 1679–1688. [Google Scholar] [CrossRef] [PubMed]
- Hachez, C.; Laloux, T.; Reinhardt, H.; Cavez, D.; Degand, H.; Grefen, C.; De Rycke, R.; Inze, D.; Blatt, M.R.; Russinova, E.; et al. Arabidopsis SNAREs SYP61 and SYP121 coordinate the trafficking of plasma membrane aquaporin PIP2;7 to modulate the cell membrane water permeability. Plant Cell 2014, 26, 3132–3147. [Google Scholar] [CrossRef] [PubMed]
- Salinas-Cornejo, J.; Madrid-Espinoza, J.; Verdugo, I.; Pérez-Díaz, J.; Martín-Davison, A.S.; Norambuena, L.; Ruiz-Lara, S. The exocytosis associated SNAP25-Type protein, SlSNAP33, increases salt stress tolerance by modulating endocytosis in tomato. Plants 2021, 10, 1322. [Google Scholar] [CrossRef]
- Nisa, Z.U.; Mallano, A.I.; Yu, Y.; Chen, C.; Duan, X.; Amanullah, S.; Kousar, A.; Baloch, A.W.; Sun, X.; Tabys, D.; et al. GsSNAP33, a novel Glycine soja SNAP25-type protein gene: Improvement of plant salt and drought tolerances in transgenic Arabidopsis thaliana. Plant Physiol. Biochem. 2017, 119, 9–20. [Google Scholar] [CrossRef]
- Sup Yun, H.; Yi, C.; Kwon, H.; Kwon, C. Model for regulation of VAMP721/722-mediated secretion: Growth vs. stress responses. Plant Signal. Behav. 2013, 8, e27116. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Tamura, K. MEGA7: Molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 2016, 33, 1870–1874. [Google Scholar] [CrossRef]
- Chen, C.; Chen, H.; Zhang, Y.; Thomas, H.R.; Frank, M.H.; He, Y.; Xia, R. TBtools: An Integrative Toolkit Developed for Interactive Analyses of Big Biological Data. Mol. Plant 2020, 13, 1194–1202. [Google Scholar] [CrossRef]
- Wang, Y.; Tang, H.; Debarry, J.D.; Tan, X.; Li, J.; Wang, X.; Lee, T.H.; Jin, H.; Marler, B.; Guo, H.; et al. MCScanX: A toolkit for detection and evolutionary analysis of gene synteny and collinearity. Nucleic Acids Res. 2012, 40, e49. [Google Scholar] [CrossRef]
- Lescot, M.; Déhais, P.; Thijs, G.; Marchal, K.; Moreau, Y.; Van de Peer, Y.; Rouzé, P.; Rombauts, S. PlantCARE, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucleic Acids Res. 2002, 30, 325–327. [Google Scholar] [CrossRef]
- Clevenger, J.; Chu, Y.; Scheffler, B.; Ozias-Akins, P. A Developmental Transcriptome Map for Allotetraploid Arachis hypogaea. Front. Plant Sci. 2016, 7, 1446. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Xiao, L.; Chi, J.; Li, R.; Han, Y.; Cui, F.; Peng, Z.; Wan, S.; Li, G. Genome-wide identification and expression of SAUR gene family in peanut (Arachis hypogaea L.) and functional identification of AhSAUR3 in drought tolerance. BMC Plant Biol. 2022, 22, 178. [Google Scholar] [CrossRef]
- Kim, D.; Paggi, J.M.; Park, C.; Bennett, C.; Salzberg, S.L. Graph-based genome alignment and genotyping with HISAT2 and HISAT-genotype. Nat. Biotechnol. 2019, 37, 907–915. [Google Scholar] [CrossRef] [PubMed]
- Liao, Y.; Smyth, G.K.; Shi, W. The Subread aligner: Fast, accurate and scalable read mapping by seed-and-vote. Nucleic Acids Res. 2013, 41, e108. [Google Scholar] [CrossRef] [PubMed]
- Livak, K.; Schmittgen, T. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCt method. Methods 2000, 25, 402–408. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).