The Blinin Accumulation Promoted by CbMYB32 Involved in Conyza blinii Resistance to Nocturnal Low Temperature
Abstract
1. Introduction
2. Results
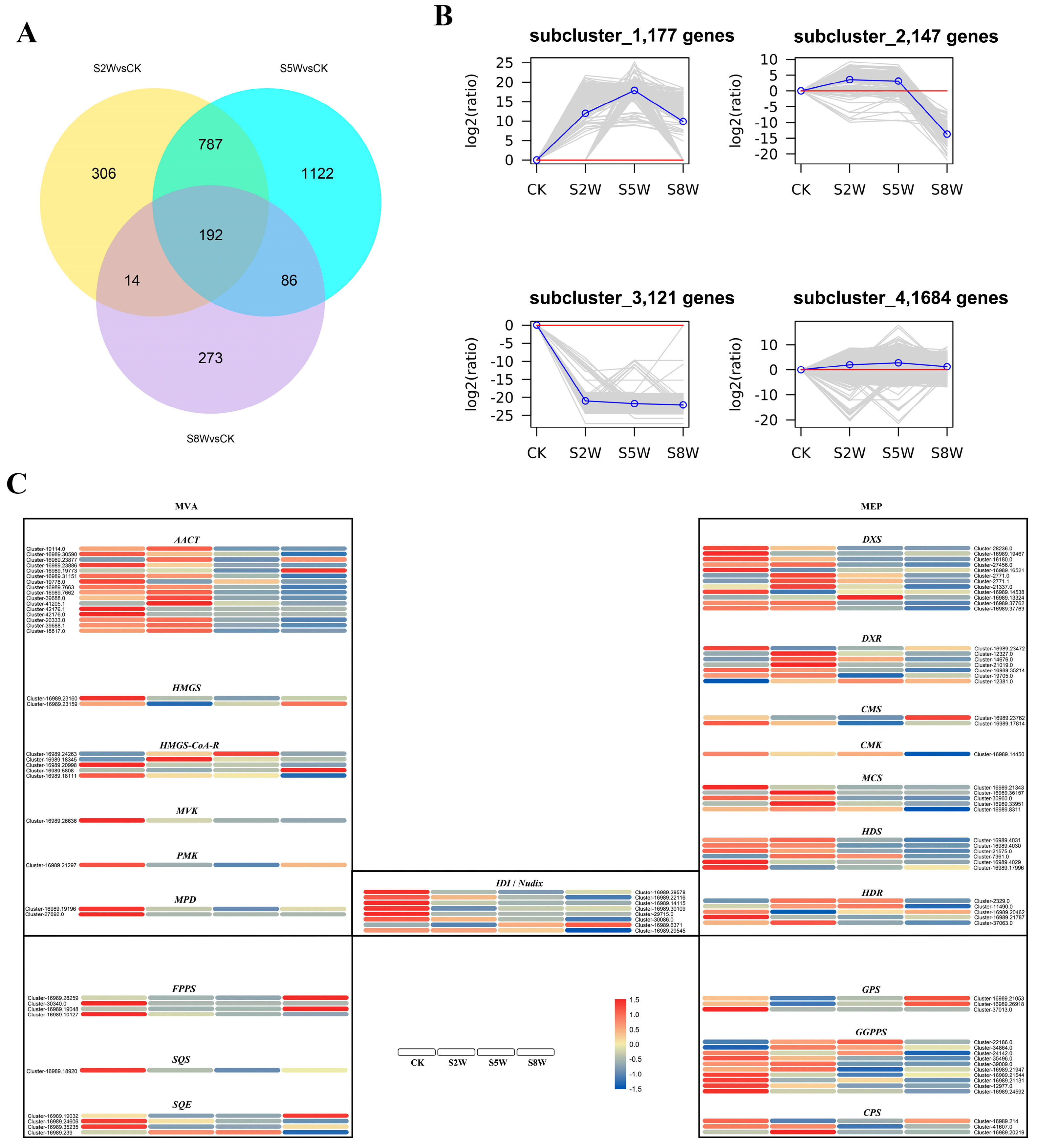
2.1. RNA Sequencing Reveals the Key Process Response to NLT
2.2. Gene-Terpenoid Association Analysis
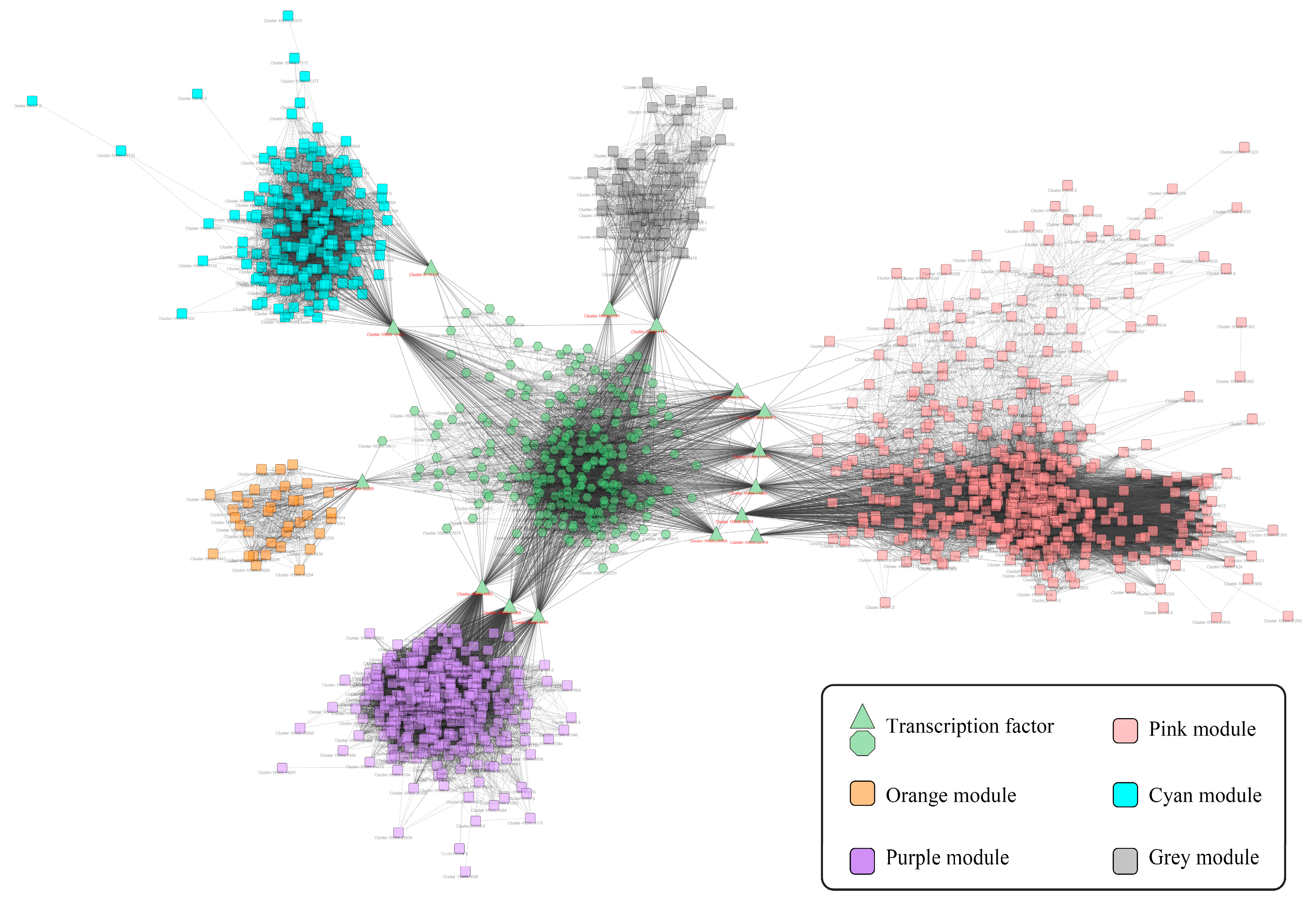
2.3. The Co-Expression Network of TFs and Gene-Terpenoid
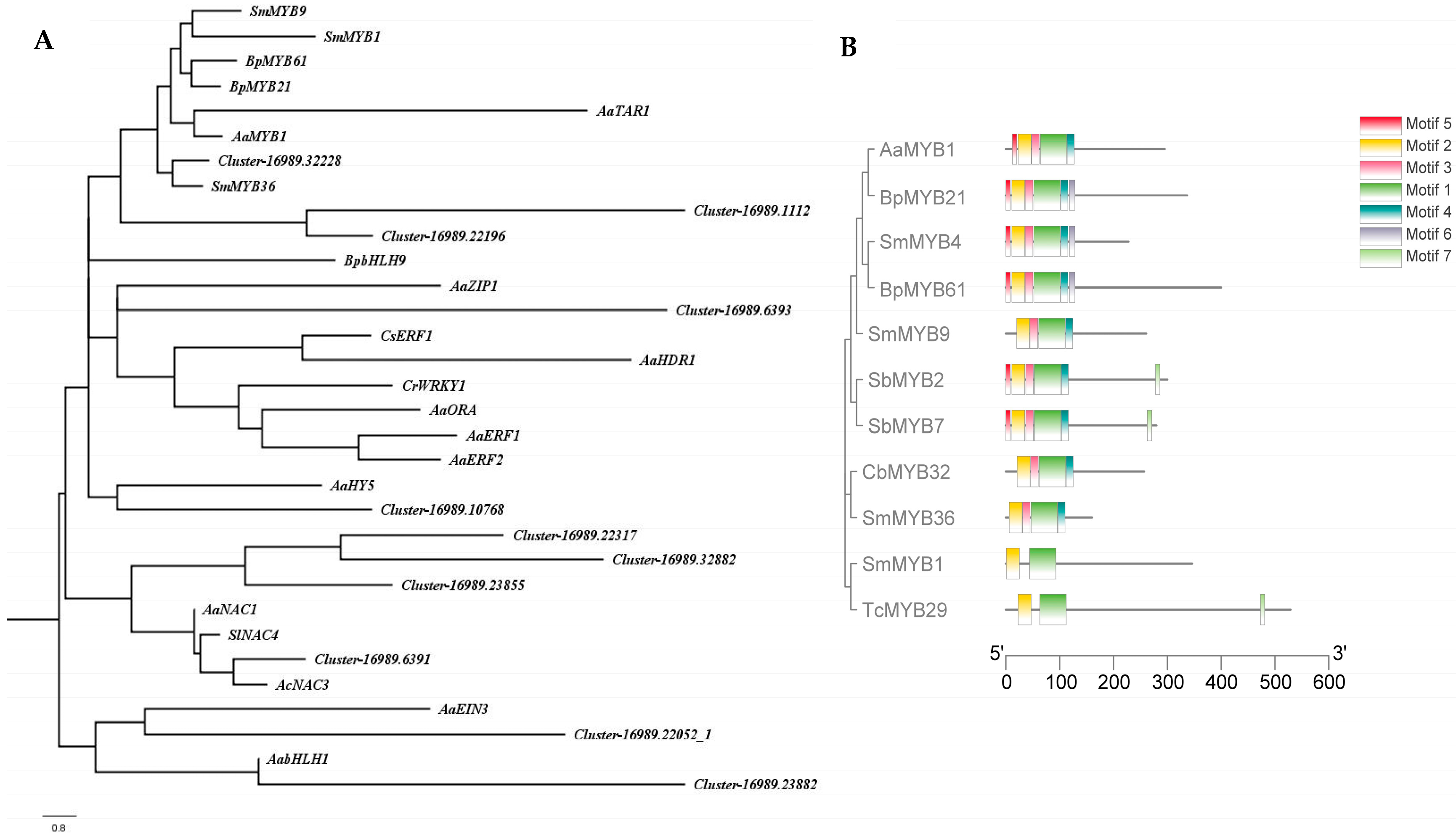
2.4. Screening of Transcription Factors Involved in Terpenoid Metabolism under NLT
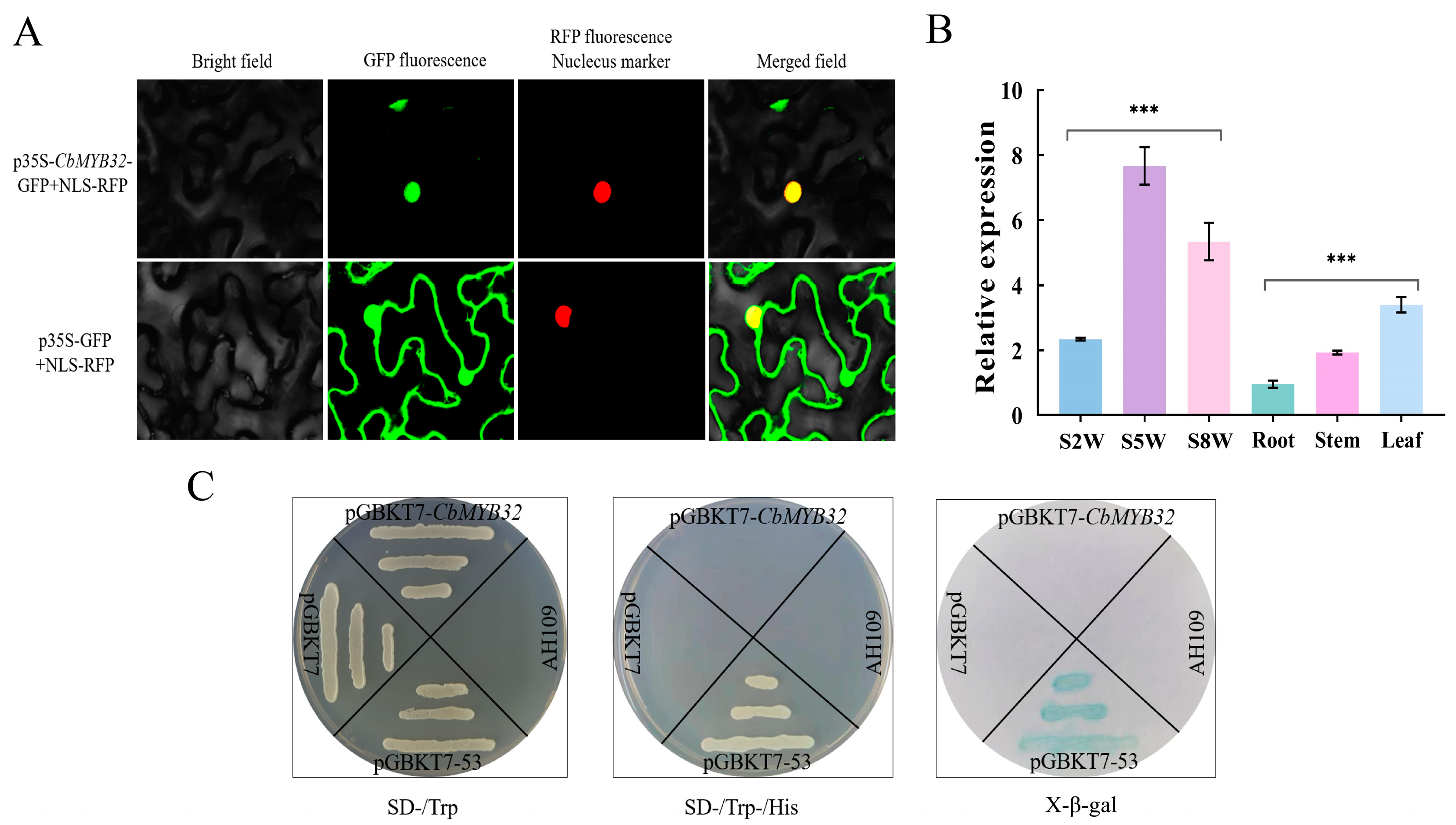
2.5. The Expression Pattern Analysis of CbMYB32
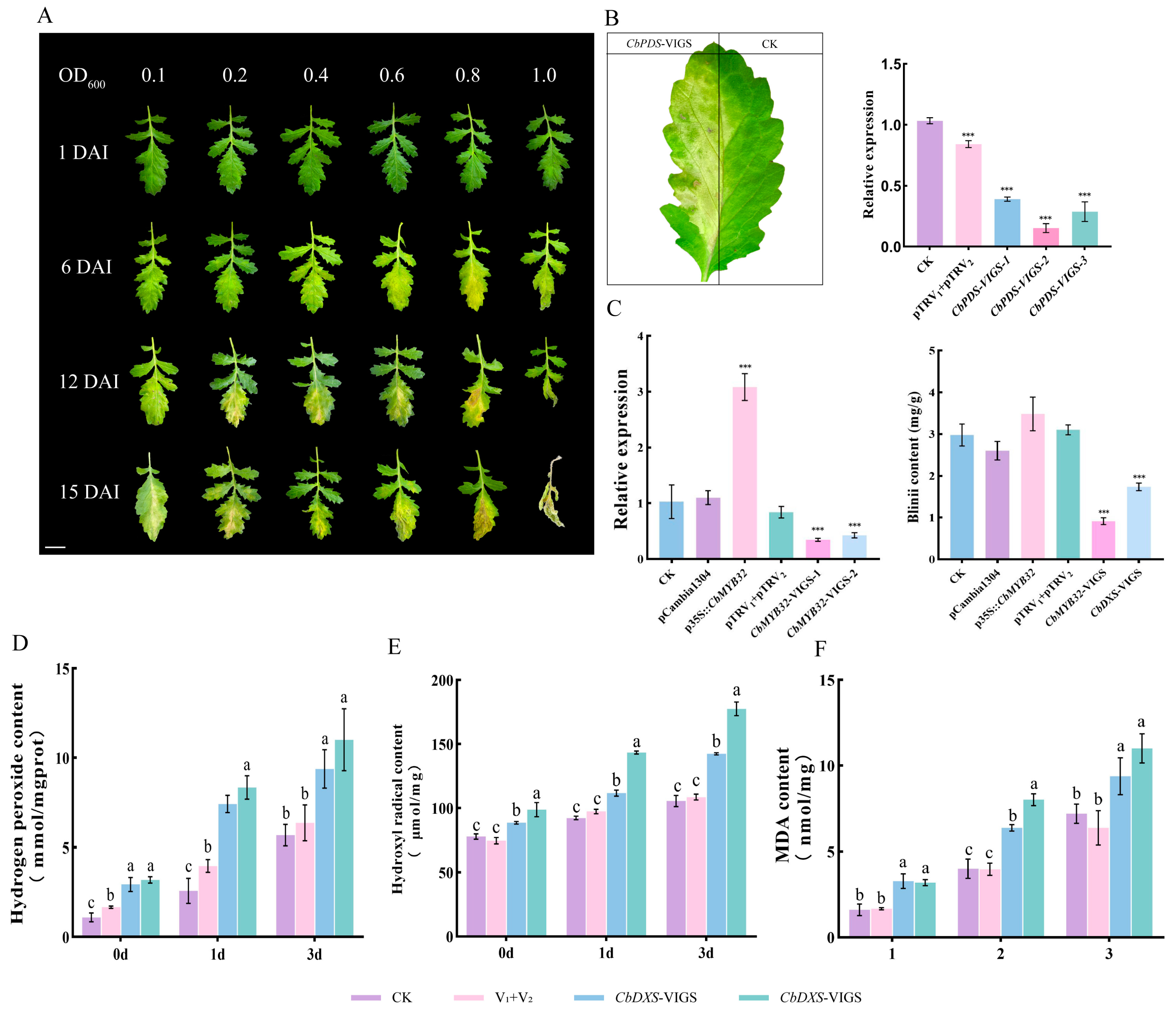
2.6. CbMYB32 Regulates Blinin Metabolism under NLT
2.7. Blinin Maintains Stable Reactive Oxygen Metabolism under NLT
2.8. Effect of SA on the Genes of Terpenoids Metabolic Pathway Enzymes
3. Discussion
4. Materials and Methods
4.1. Plant Cultivation and NLT Treatment
4.2. RNA-Seq and Different Expression Gene Analysis
4.3. Differential Transcription Factor Screening and Co-Expression Network Analysis
4.4. Phylogenetic Tree
4.5. Gene Cloning and Vector Construction
4.6. Subcellular Localization of the CbMYB32
4.7. Transcriptional Activation of the CbMYB32 from C. blinii
4.8. Agrobacterium Tumefaciens-Mediated Transient Transformation of C. blinii Leaves
4.9. Physiological Indices Measurements
4.10. Plant Hormone Treatment and RT-qPCR
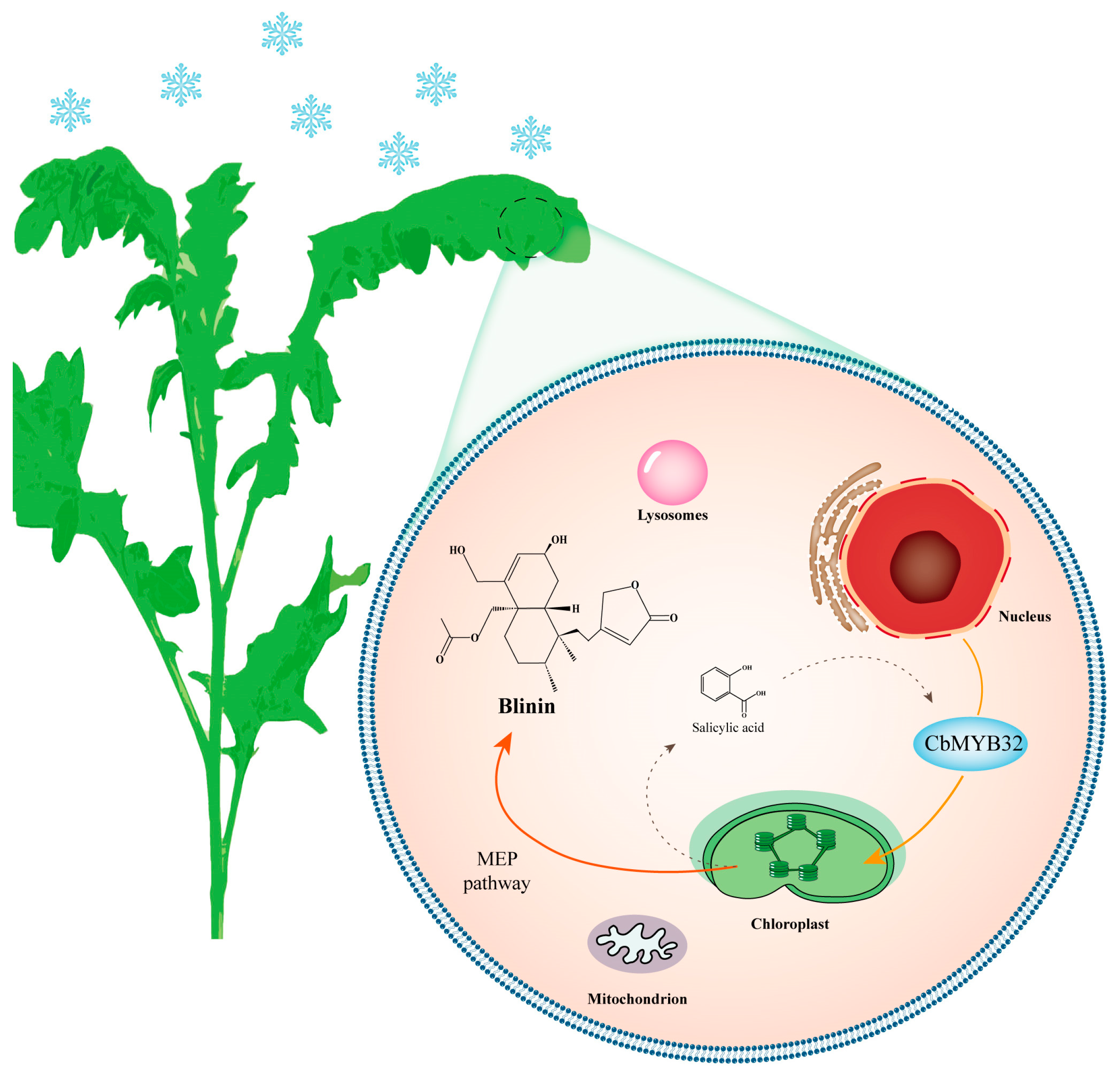
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Acknowledgments
Conflicts of Interest
References
- Quinn, P.J. Effects of temperature on cell membranes. Symp. Soc. Exp. Biol. 1988, 42, 237–258. [Google Scholar] [PubMed]
- Pollock, C.J.; Eagles, C.F. Low temperature and the growth of plants. Symp. Soc. Exp. Biol. 1988, 42, 157–180. [Google Scholar] [PubMed]
- Yamori, W.; Noguchi, K.; Hikosaka, K.; Terashima, I. Cold-Tolerant Crop Species Have Greater Temperature Homeostasis of Leaf Respiration and Photosynthesis Than Cold-Sensitive Species. Plant Cell Physiol. 2008, 50, 203–215. [Google Scholar] [CrossRef]
- Lin, D.; Kong, R.; Chen, L.; Wang, Y.; Wu, L.; Xu, J.; Piao, Z.; Lee, G.; Dong, Y. Chloroplast development at low temperature requires the pseudouridine synthase gene TCD3 in rice. Sci. Rep. 2020, 10, 8518. [Google Scholar] [CrossRef] [PubMed]
- Guo, Z.; Cai, L.; Liu, C.; Chen, Z.; Guan, S.; Ma, W.; Pan, G. Low-temperature stress affects reactive oxygen species, osmotic adjustment substances, and antioxidants in rice (Oryza sativa L.) at the reproductive stage. Sci. Rep. 2022, 12, 6224. [Google Scholar] [CrossRef] [PubMed]
- Suzuki, N.; Mittler, R. Reactive oxygen species and temperature stresses: A delicate balance between signaling and destruction. Physiol. Plant. 2006, 126, 45–51. [Google Scholar] [CrossRef]
- Zhang, K.-M.; Yu, H.-J.; Shi, K.; Zhou, Y.-H.; Yu, J.-Q.; Xia, X.-J. Photoprotective roles of anthocyanins in Begonia semperflorens. Plant Sci. 2010, 179, 202–208. [Google Scholar] [CrossRef]
- Flutsch, S.; Wang, Y.; Takemiya, A.; Vialet-Chabrand, S.R.M.; Klejchova, M.; Nigro, A.; Hills, A.; Lawson, T.; Blatt, M.R.; Santelia, D. Guard Cell Starch Degradation Yields Glucose for Rapid Stomatal Opening in Arabidopsis. Plant Cell 2020, 32, 2325–2344. [Google Scholar] [CrossRef] [PubMed]
- Nakabayashi, R.; Yonekura-Sakakibara, K.; Urano, K.; Suzuki, M.; Yamada, Y.; Nishizawa, T.; Matsuda, F.; Kojima, M.; Sakakibara, H.; Shinozaki, K.; et al. Enhancement of oxidative and drought tolerance in Arabidopsis by overaccumulation of antioxidant flavonoids. Plant J. 2014, 77, 367–379. [Google Scholar] [CrossRef] [PubMed]
- Lee, W.J.; Jeong, C.Y.; Kwon, J.; Van Kien, V.; Lee, D.; Hong, S.W.; Lee, H. Drastic anthocyanin increase in response to PAP1 overexpression in fls1 knockout mutant confers enhanced osmotic stress tolerance in Arabidopsis thaliana. Plant Cell Rep. 2016, 35, 2369–2379. [Google Scholar] [CrossRef] [PubMed]
- Atanasov, A.G.; Waltenberger, B.; Pferschy-Wenzig, E.M.; Linder, T.; Wawrosch, C.; Uhrin, P.; Temml, V.; Wang, L.; Schwaiger, S.; Heiss, E.H.; et al. Discovery and resupply of pharmacologically active plant-derived natural products: A review. Biotechnol. Adv. 2015, 33, 1582–1614. [Google Scholar] [CrossRef] [PubMed]
- Palaniyandi, K.; Wu, J. Low temperature enhanced the podophyllotoxin accumulation vis-a-vis its biosynthetic pathway gene(s) expression in Dysosma versipellis (Hance) M. Cheng—A pharmaceutically important medicinal plant. Process Biochem. 2020, 95, 197–203. [Google Scholar] [CrossRef]
- Peng, X.; Wu, H.; Chen, H.; Zhang, Y.; Qiu, D.; Zhang, Z. Transcriptome profiling reveals candidate flavonol-related genes of Tetrastigma hemsleyanum under cold stress. BMC Genom. 2019, 20, 687. [Google Scholar] [CrossRef]
- Wang, S.; Liang, W.; Yao, L.; Wang, J.; Gao, W. Effect of temperature on morphology, ginsenosides biosynthesis, functional genes, and transcriptional factors expression in Panax ginseng adventitious roots. J. Food Biochem. 2019, 43, e12794. [Google Scholar] [CrossRef]
- Jiang, M.; Liu, J.; Quan, X.; Quan, L.; Wu, S. Different chilling stresses stimulated the accumulation of different types of ginsenosides in Panax ginseng cells. Acta Physiol. Plant. 2016, 38, 210. [Google Scholar] [CrossRef]
- Shen, Q.; Lu, X.; Yan, T.; Fu, X.; Zongyou, l.; Zhang, F.; Pan, Q.; Wang, G.; Sun, X.; Tang, K. The jasmonate-responsive AaMYC2 transcription factor positively regulates artemisinin biosynthesis in Artemisia annua. New Phytol. 2016, 210. [Google Scholar] [CrossRef] [PubMed]
- Song, J.J.; Fang, X.; Li, C.Y.; Jiang, Y.; Li, J.X.; Wu, S.; Guo, J.; Liu, Y.; Fan, H.; Huang, Y.B.; et al. A 2-oxoglutarate-dependent dioxygenase converts dihydrofuran to furan in Salvia diterpenoids. Plant Physiol. 2022, 188, 1496–1506. [Google Scholar] [CrossRef]
- Yang, C.-R.; He, Z.-T.; Li, X.-C.; Zheng, Q.-T.; He, C.-H.; Yang, J.; Morita, T. Blinin, a neoclerodane diterpene from Conyza blinii. Phytochemistry 1989, 28, 3131–3134. [Google Scholar]
- Yang, M.; Zheng, T.; Zhan, J.; Wang, M.; Sun, W.; Zhou, M.; Tang, Z.; Bu, T.; Li, Q.; Chen, H. Conyza blinii responds to the changes of exogenous iron through auxin-terpenoids metabolism pathway. J. Plant Interact. 2022, 17, 485–495. [Google Scholar] [CrossRef]
- Zhan, J.; Yang, Q.; Lin, Z.; Zheng, T.; Wang, M.; Sun, W.; Bu, T.; Tang, Z.; Li, C.; Han, X.; et al. Enhanced antioxidant capacity and upregulated transporter genes contribute to the UV-B-induced increase in blinin in Conyza blinii. Environ. Sci. Pollut. Res. 2021, 28, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Yang, M.; Wang, M.; Zhou, M.; Zhang, Y.; Yu, K.; Wang, T.; Bu, T.; Tang, Z.; Zheng, T.; Chen, H. ABA and SA Participate in the Regulation of Terpenoid Metabolic Flux Induced by Low-Temperature within Conyza blinii. Life 2023, 13, 371. [Google Scholar] [CrossRef] [PubMed]
- Klein, M.; Martinoia, E.; Hoffmann-Thoma, G.; Weissenbock, G. A membrane-potential dependent ABC-like transporter mediates the vacuolar uptake of rye flavone glucuronides: Regulation of glucuronide uptake by glutathione and its conjugates. Plant J. 2000, 21, 289–304. [Google Scholar] [CrossRef] [PubMed]
- van den Brule, S.; Muller, A.; Fleming, A.J.; Smart, C.C. The ABC transporter SpTUR2 confers resistance to the antifungal diterpene sclareol. Plant J. 2002, 30, 649–662. [Google Scholar] [CrossRef] [PubMed]
- Jasinski, M. A Plant Plasma Membrane ATP Binding Cassette-Type Transporter Is Involved in Antifungal Terpenoid Secretion. Plant Cell Online 2001, 13, 1095–1107. [Google Scholar] [CrossRef]
- Loizzo, M.; Tundis, R.; Menichini, F.; Saab, A.; Statti, G.; Menichini, F. Cytotoxic activity of essential oils from Labiatae and Lauraceae families against in vitro human tumor models. Anticancer. Res. 2007, 27, 3293–3299. [Google Scholar] [PubMed]
- Ding, K.; Pei, T.; Bai, Z.; Jia, Y.; Ma, P.; Liang, Z. SmMYB36, a Novel R2R3-MYB Transcription Factor, Enhances Tanshinone Accumulation and Decreases Phenolic Acid Content in Salvia miltiorrhiza Hairy Roots. Sci. Rep. 2017, 7, 5104. [Google Scholar] [CrossRef] [PubMed]
- Li, Q.; Fang, X.; Zhao, Y.; Cao, R.; Dong, J.; Ma, P. The SmMYB36-SmERF6/SmERF115 module regulates the biosynthesis of tanshinones and phenolic acids in salvia miltiorrhiza hairy roots. Hortic Res. 2023, 10, uhac238. [Google Scholar] [CrossRef] [PubMed]
- Zeng, T.; Li, J.W.; Xu, Z.Z.; Zhou, L.; Li, J.J.; Yu, Q.; Luo, J.; Chan, Z.L.; Jongsma, M.A.; Hu, H.; et al. TcMYC2 regulates Pyrethrin biosynthesis in Tanacetum cinerariifolium. Hortic Res. 2022, 9, uhac178. [Google Scholar] [CrossRef] [PubMed]
- Pradhan, S.K.; Pandit, E.; Nayak, D.K.; Behera, L.; Mohapatra, T. Genes, pathways and transcription factors involved in seedling stage chilling stress tolerance in indica rice through RNA-Seq analysis. BMC Plant Biol. 2019, 19, 1–17. [Google Scholar] [CrossRef] [PubMed]
- Ji, Q.; Wang, D.W.; Zhou, J.; Xu, Y.L.; Shen, B.Q.; Zhou, F. Genome-wide characterization and expression analyses of the MYB superfamily genes during developmental stages in Chinese jujube. Peerj 2019, 7, 6353. [Google Scholar] [CrossRef]
- Peng, X.J.; Wu, Q.Q.; Teng, L.H.; Tang, F.; Pi, Z.; Shen, S.H. Transcriptional regulation of the paper mulberry under cold stress as revealed by a comprehensive analysis of transcription factors. BMC Plant Biol. 2015, 15, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Sun, X.M.; Matus, J.T.; Wong, D.C.J.; Wang, Z.M.; Chai, F.M.; Zhang, L.L.; Fang, T.; Zhao, L.; Wang, Y.; Han, Y.P.; et al. The GARP/MYB-related grape transcription factor AQUILO improves cold tolerance and promotes the accumulation of raffinose family oligosaccharides. J. Exp. Bot. 2018, 69, 1749–1764. [Google Scholar] [CrossRef]
- Erb, M.; Kliebenstein, D. Plant Secondary Metabolites as Defenses, Regulators, and Primary Metabolites: The Blurred Functional Trichotomy. Plant Physiol. 2020, 184, 00433–02020. [Google Scholar] [CrossRef] [PubMed]
- Wu, Z.; Han, S.; Zhou, H.; Tuang, Z.; Wang, Y.; Jin, Y.; Shi, H.; Yang, W. Cold stress activates disease resistance in Arabidopsis thaliana through a salicylic acid dependent pathway. Plant Cell Environ. 2019, 42, 2645–2663. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.; Liu, H.; Ding, Z.; Yan, J.; Yu, H.; Pan, R.; Hu, J.; Guan, Y.; Hua, J. Low Temperature Enhances Plant Immunity via Salicylic Acid Pathway Genes That Are Repressed by Ethylene. Plant Physiol. 2019, 182, 01130–02019. [Google Scholar] [CrossRef] [PubMed]
- Ye, J.; Mao, D.; Cheng, S.; Zhang, X.; Tan, J.; Zheng, J.; Xu, F. Comparative transcriptome analysis reveals the potential stimulatory mechanism of terpene trilactone biosynthesis by exogenous salicylic acid in Ginkgo biloba. Ind. Crop. Prod. 2020, 145, 112104. [Google Scholar] [CrossRef]
- Xu, M.; Dong, J.; Wang, H.; Huang, L. Complementary action of jasmonic acid on salicylic acid in mediating fungal elicitor-induced flavonol glycoside accumulation of Ginkgo biloba cells. Plant Cell Environ. 2009, 32, 960–967. [Google Scholar] [CrossRef]
- Grabherr, M.G.; Haas, B.J.; Yassour, M.; Levin, J.Z.; Thompson, D.A.; Amit, I.; Adiconis, X.; Fan, L.; Raychowdhury, R.; Zeng, Q.; et al. Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat. Biotechnol. 2011, 29, 644–652. [Google Scholar] [CrossRef] [PubMed]
- Zheng, T.; Wang, M.; Zhan, J.; Sun, W.; Yang, Q.; Lin, Z.; Bu, T.; Tang, Z.; Li, C.; Yan, J.; et al. Ferrous iron-induced increases in capitate glandular trichome density and upregulation of CbHO-1 contributes to increases in blinin content in Conyza blinii. Planta 2020, 252, 81. [Google Scholar] [CrossRef] [PubMed]
- Sun, W.-J.; Zhan, J.-Y.; Zheng, T.-R.; Sun, R.; Wang, T.; Tang, Z.-Z.; Bu, T.-L.; Li, C.-L.; Wu, Q.; Chen, H. The jasmonate-responsive transcription factor CbWRKY24 regulates terpenoid biosynthetic genes to promote saponin biosynthesis in Conyza blinii H. Lév. J. Genet. 2018, 97, 1379–1388. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yang, M.; Zhou, M.; Shu, M.; Han, Z.; Ma, R.; Chen, Y.; Zheng, T.; Chen, H. The Blinin Accumulation Promoted by CbMYB32 Involved in Conyza blinii Resistance to Nocturnal Low Temperature. Int. J. Mol. Sci. 2023, 24, 7143. https://doi.org/10.3390/ijms24087143
Yang M, Zhou M, Shu M, Han Z, Ma R, Chen Y, Zheng T, Chen H. The Blinin Accumulation Promoted by CbMYB32 Involved in Conyza blinii Resistance to Nocturnal Low Temperature. International Journal of Molecular Sciences. 2023; 24(8):7143. https://doi.org/10.3390/ijms24087143
Chicago/Turabian StyleYang, Ming, Min Zhou, Mengdan Shu, Zhengqi Han, Ruiqi Ma, Yuting Chen, Tianrun Zheng, and Hui Chen. 2023. "The Blinin Accumulation Promoted by CbMYB32 Involved in Conyza blinii Resistance to Nocturnal Low Temperature" International Journal of Molecular Sciences 24, no. 8: 7143. https://doi.org/10.3390/ijms24087143
APA StyleYang, M., Zhou, M., Shu, M., Han, Z., Ma, R., Chen, Y., Zheng, T., & Chen, H. (2023). The Blinin Accumulation Promoted by CbMYB32 Involved in Conyza blinii Resistance to Nocturnal Low Temperature. International Journal of Molecular Sciences, 24(8), 7143. https://doi.org/10.3390/ijms24087143






