Identifying Natural Bioactive Peptides from the Common Octopus (Octopus vulgaris Cuvier, 1797) Skin Mucus By-Products Using Proteogenomic Analysis
Abstract
1. Introduction
2. Results
2.1. Protein Concentration by BCA and SDS-PAGE
2.2. Mass Spectrometry (MS)
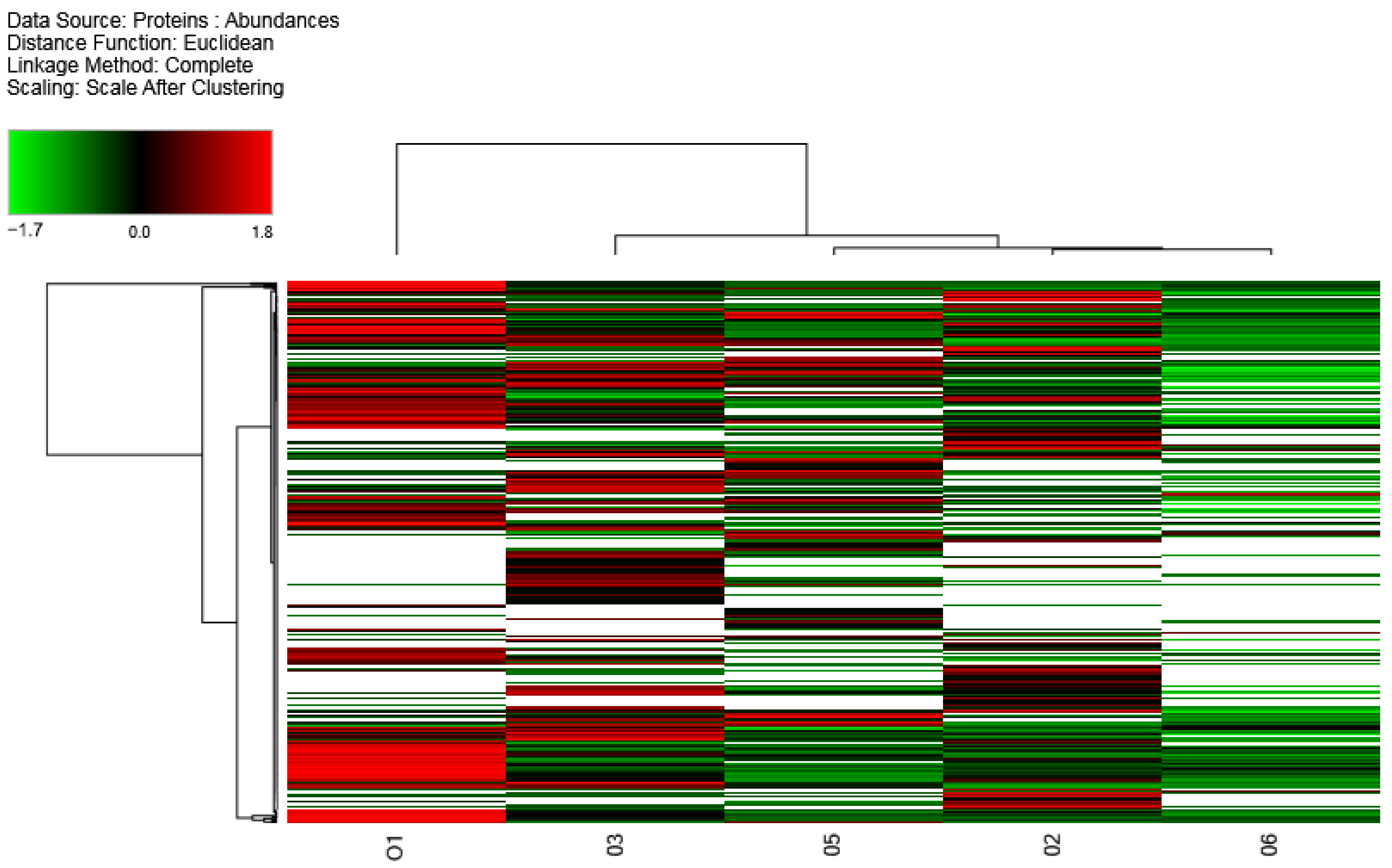
2.3. Label-Free Quantification (LFQ) of O. vulgaris Mucus Samples
2.4. Functional Analysis: Gene Ontologies and Pathways Analysis
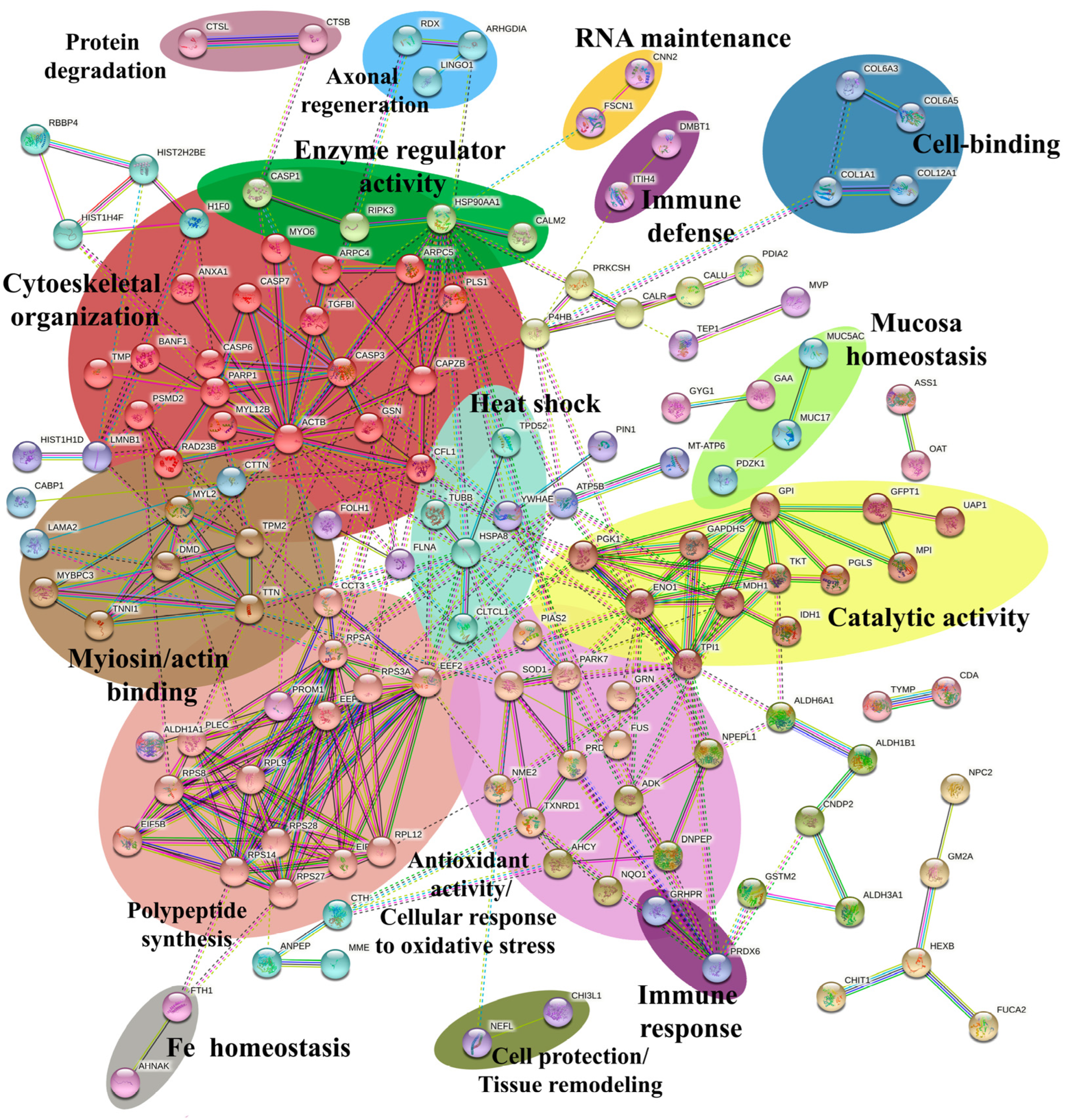
2.5. Network Analysis
2.6. Potential Bioactive Peptides Identification
3. Discussion
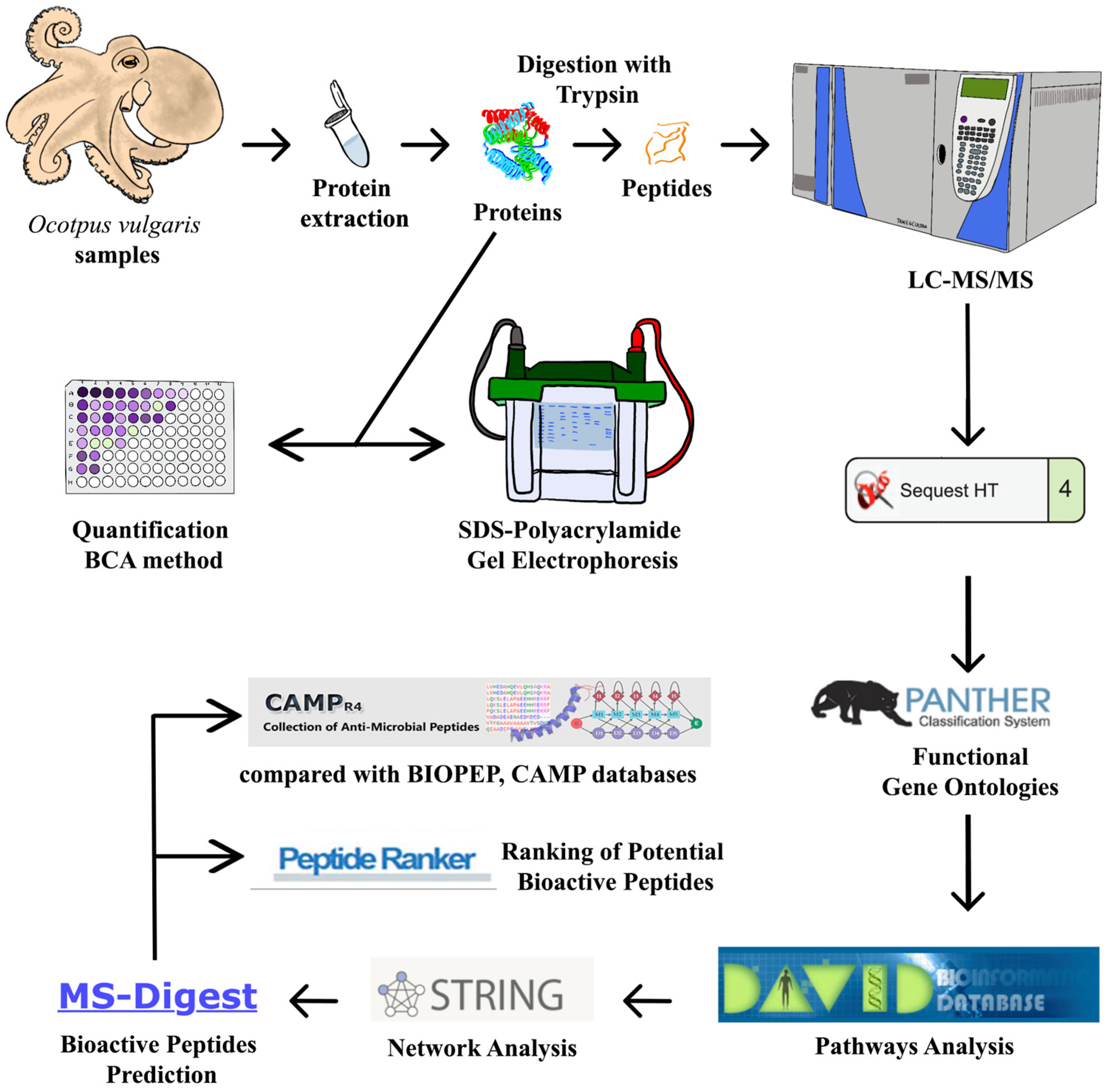
4. Materials and Methods
4.1. Animal Capture and Maintenance
4.2. Sample Collection
4.3. Skin Mucus Protein Extraction
4.4. SDS-Polyacrylamide Gel Electrophoresis (SDS-PAGE)
4.5. Protein Digestion with Trypsin
4.6. LC-MS/MS
4.7. Processing of the Mass Spectrometry Data
4.8. Euclidean Hierarchical Clustering
4.9. Functional Gene Ontologies and Pathways Analysis
4.10. Network Analysis
4.11. Bioactive Peptides Prediction
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Jereb, P.; Roper, C.F.E. No. 2. Chambered nautiluses and sepioids (Nautilidae, Sepiidae, Sepiolidae, Sepiadariidae, Idiosepiidae and Spirulidae). In Cephalopods of the World: An Annotated and Illustrated Catalogue of Cephalopod Species Known to Date; Food & Agriculture Organization of the United Nations: Rome, Italy, 2005; Volume 1, pp. 204–212. [Google Scholar]
- Allam, B.; Espinosa, E.P. Mucosal immunity in mollusks. In Mucosal Health in Aquaculture; Academic Press: Cambridge, MA, USA, 2015; pp. 325–370. [Google Scholar]
- Parra, D.; Reyes-Lopez, F.E.; Tort, L. Mucosal immunity and B cells in teleosts: Effect of vaccination and stress. Front. Immunol. 2015, 6, 354. [Google Scholar] [CrossRef]
- Schmidtberg, H. Ultrastructural studies of the suckers of newly hatched Eledone moschata and Octopus vulgaris (Mollusca; Cephalopoda). In Advancing Research on Living and Fossil Cephalopods; Springer: Boston, MA, USA, 1999; pp. 203–221. [Google Scholar]
- Anadón, R. Functional histology: The tissues of common coleoid cephalopods. In Handbook of Pathogens and Diseases in Cephalopods; Springer Nature: Cham, Switzerland, 2019; pp. 39–85. [Google Scholar]
- Schachter, H.; Williams, D. Biosynthesis of mucus glycoproteins. In Mucus in Health and Disease—II; Chantler, E., Elder, J., Elstein, M., Eds.; Plenum Press: New York, NY, USA, 1982; pp. 3–28. [Google Scholar]
- Guardiola, F.A.; Cuartero, M.; del Mar Collado-González, M.; Arizcún, M.; Diaz Banos, F.G.; Meseguer, J.; Cuesta, A.; Esteban, M.A. Description and comparative study of physico-chemical parameters of the teleost fish skin mucus. Biorheology 2015, 52, 247–256. [Google Scholar] [CrossRef] [PubMed]
- Fernández-Alacid, L.; Sanahuja, I.; Ordóñez-Grande, B.; Sánchez-Nuño, S.; Viscor, G.; Gisbert, E.; Herrera, M.; Ibarz, A. Skin mucus metabolites in response to physiological challenges: A valuable non-invasive method to study teleost marine species. Sci. Total Environ. 2018, 644, 1323–1335. [Google Scholar] [CrossRef]
- Guardiola, F.A.; Cuesta, A.; Abellán, E.; Meseguer, J.; Esteban, M.A. Comparative analysis of the humoral immunity of skin mucus from several marine teleost fish. Fish Shellfish Immunol. 2014, 40, 24–31. [Google Scholar] [CrossRef]
- Benoist, L.; Houyvet, B.; Henry, J.; Corre, E.; Zanuttini, B.; Zatylny-Gaudin, C. In-depth in silico search for cuttlefish (Sepia officinalis) antimicrobial peptides following bacterial challenge of haemocytes. Mar. Drugs 2020, 18, 439. [Google Scholar] [CrossRef]
- Suárez, L.; Pereira, A.; Hidalgo, W.; Uribe, N. Antibacterial, Antibiofilm and Anti-Virulence Activity of Biactive Fractions from Mucus Secretion of Giant African Snail Achatina fulica against Staphylococcus aureus Strains. Antibiotics 2021, 10, 1548. [Google Scholar] [CrossRef] [PubMed]
- Tur, R.; Domingues, P.; Almansa, E.; Lago, M.; García-Fernández, P.; Pérez-Rial, E. Patente Nacional por “Procedimiento Para el Cultivo de Paralarvas del Pulpo Común Octopus vulgaris”. España ES2714930, 2020. [Google Scholar]
- Gestal, C.; Pascual, S.; Guerra, Á.; Fiorito, G.; Vieites, J.M. (Eds.) Handbook of Pathogens and Diseases in Cephalopods; Springer Nature: Cham, Switzerland, 2019. [Google Scholar]
- Keil, B. Specificity of Proteolysis, 1st ed.; Springer: Berlin/Heidelberg, Germany, 1992. [Google Scholar]
- Mooney, C.; Haslam, N.J.; Pollastri, G.; Shields, D.C. Towards the improved discovery and design of functional peptides: Common features of diverse classes permit generalized prediction of bioactivity. PLoS ONE 2012, 7, e45012. [Google Scholar] [CrossRef] [PubMed]
- Albertin, C.B.; Simakov, O.; Mitros, T.; Wang, Z.Y.; Pungor, J.R.; Edsinger-Gonzales, E.; Brenner, S.; Ragsdale, C.W.; Rokhsar, D.S. The octopus genome and the evolution of cephalopod neural and morphological novelties. Nature 2015, 524, 220–224. [Google Scholar] [CrossRef]
- Kim, B.M.; Kang, S.; Ahn, D.H.; Jung, S.H.; Rhee, H.; Yoo, J.S.; Lee, J.E.; Lee, S.; Han, Y.H.; Ryu, K.B.; et al. The genome of common long-arm octopus Octopus minor. Gigascience 2018, 7, giy119. [Google Scholar] [CrossRef]
- Belcaid, M.; Casaburi, G.; McAnulty, S.J.; Schmidbaur, H.; Suria, A.M.; Moriano-Gutierrez, S.; Pankey, M.S.; Oakley, T.H.; Kremer, N.; Koch, E.J.; et al. Symbiotic organs shaped by distinct modes of genome evolution in cephalopods. Proc. Natl. Acad. Sci. USA 2019, 116, 3030–3035. [Google Scholar] [CrossRef] [PubMed]
- Zarrella, I.; Herten, K.; Maes, G.E.; Tai, S.; Yang, M.; Seuntjens, E.; Ritschard, E.A.; Zach, M.; Styfhals, R.; Sanges, R.; et al. The survey and reference assisted assembly of the Octopus vulgaris genome. Sci. Data 2019, 6, 13. [Google Scholar] [CrossRef]
- Prado-Álvarez, M.; Dios, S.; García-Fernández, P.; Tur, R.; Hachero-Cruzado, I.; Domingues, P.; Almansa, E.; Varó, I.; Gestal, C. De novo transcriptome reconstruction in aquacultured early life stages of the cephalopod Octopus vulgaris. Sci. Data 2022, 9, 609. [Google Scholar] [CrossRef]
- Huang, L.; Huang, Q.Y.; Chen, H.B.; Huang, F.S.; Huang, H.Q. Differential proteins of the optic ganglion in Octopus vulgaris under methanol stress revealed using proteomics. Appl. Biochem. Biotechnol. 2011, 165, 978–988. [Google Scholar] [CrossRef] [PubMed]
- Baldascino, E.; Di Cristina, G.; Tedesco, P.; Hobbs, C.; Shaw, T.J.; Ponte, G.; Andrews, P.L. The gastric ganglion of Octopus vulgaris: Preliminary characterization of gene-and putative neurochemical-complexity, and the effect of Aggregata octopiana digestive tract infection on gene expression. Front. Physiol. 2017, 8, 1001. [Google Scholar] [CrossRef] [PubMed]
- Fingerhut, L.C.; Strugnell, J.M.; Faou, P.; Labiaga, Á.R.; Zhang, J.; Cooke, I.R. Shotgun proteomics analysis of saliva and salivary gland tissue from the common octopus Octopus vulgaris. J. Proteome Res. 2018, 17, 3866–3876. [Google Scholar] [CrossRef] [PubMed]
- Varó, I.; Cardenete, G.; Hontoria, F.; Monroig, Ó.; Iglesias, J.; Otero, J.J.; Almansa, E.; Navarro, J.C. Dietary effect on the proteome of the common octopus (Octopus vulgaris) paralarvae. Front. Physiol. 2017, 8, 309. [Google Scholar] [CrossRef]
- Varó, I.; Prado-Álvarez, M.; Ortea, I.; Morales, A.E.; García-Fernández, P.; Domingues, P.; Tur, R.; Dios, S.; Gestal, C. Proteogenomic Study of the Effect of an Improved Mixed Diet of Live Preys on the Aquaculture of Octopus vulgaris Paralarvae. Front. Mar. Sci. 2022, 8, 817701. [Google Scholar] [CrossRef]
- García-Fernández, P.; Prado-Álvarez, M.; Nande, M.; Perales-Raya, C.; Almansa, E.; Varó, I.; Gestal, C. Global impact of diet and temperature over aquaculture of Octopus vulgaris paralarvae from a transcriptomic approach. Sci. Rep. 2019, 9, 10312. [Google Scholar] [CrossRef]
- Fernández-Boo, S.; Gervais, O.; Prado-Alvarez, M.; Chollet, B.; Claverol, S.; Lecadet, C.; Dubreuil, C.; Arzul, I. Is pallial mucus involved in Ostrea edulis defenses against the parasite Bonamia ostreae? J. Invertebr. Pathol. 2020, 169, 107259. [Google Scholar] [CrossRef]
- Wells, J. Cutaneous respiration in Octopus vulgaris. J. Exp. Biol. 1996, 199, 2477–2483. [Google Scholar]
- Bertók, L.; Chow, D. (Eds.) Natural Immunity; Elsevier: Amsterdam, The Netherlands, 2005; ISBN 9780080460468. [Google Scholar]
- Richter, K.; Haslbeck, M.; Buchner, J. The heat shock response: Life on the verge of death. Mol. Cell. 2010, 40, 253–266. [Google Scholar] [CrossRef]
- Hayes, J.J.; Clark, D.J.; Wolffe, A.P. Histone contributions to the structure of DNA in the nucleosome. Proc. Natl. Acad. Sci. USA 1991, 88, 6829–6833. [Google Scholar] [CrossRef] [PubMed]
- Masso-Silva, J.A.; Diamond, G. Antimicrobial peptides from fish. Pharmaceuticals 2014, 7, 265–310. [Google Scholar] [CrossRef] [PubMed]
- Alex, A.; Antunes, A. Pyrosequencing characterization of the microbiota from Atlantic intertidal marine sponges reveals high microbial diversity and the lack of co-occurrence patterns. PLoS ONE 2015, 10, e0127455. [Google Scholar] [CrossRef]
- Almeida, D.; Domínguez-Pérez, D.; Matos, A.; Agüero-Chapin, G.; Osório, H.; Vasconcelos, V.; Campos, A.; Antunes, A. Putative antimicrobial peptides of the posterior salivary glands from the cephalopod Octopus vulgaris revealed by exploring a composite protein database. Antibiotics 2020, 9, 757. [Google Scholar] [CrossRef] [PubMed]
- Ferreira, V.P.; Pangburn, M.K.; Cortés, C. Complement control protein factor H: The good, the bad, and the inadequate. Mol. Immunol. 2010, 47, 2187–2197. [Google Scholar] [CrossRef]
- Kobe, B.; Kajava, A.V. The leucine-rich repeat as a protein recognition motif. Curr. Opin. Struct. Biol. 2001, 11, 725–732. [Google Scholar] [CrossRef]
- Kajava, A.V.; Kobe, B. Assessment of the ability to model proteins with leucine–rich repeats in light of the latest structural information. Protein Sci. 2002, 11, 1082–1090. [Google Scholar] [CrossRef]
- Oren, Z.; Shai, Y. A class of highly potent antibacterial peptides derived from pardaxin, a pore–forming peptide isolated from Moses sole fish Pardachirus marmoratus. Eur. J. Biochem. 1996, 237, 303–310. [Google Scholar] [CrossRef]
- Salinas, I. The mucosal immune system of teleost fish. Biology 2015, 4, 525–539. [Google Scholar] [CrossRef]
- Tachapuripunya, V.; Roytrakul, S.; Chumnanpuen, P. Unveiling putative functions of mucus proteins and their tryptic peptides in seven gastropod species using comparative proteomics and machine learning-based bioinformatics predictions. Molecules 2021, 26, 3475. [Google Scholar] [CrossRef] [PubMed]
- Besednova, N.N.; Zaporozhets, T.S.; Kovalev, N.N.; Makarenkova, I.D.; Yakovlev, Y.M. Cephalopods: The potential for their use in medicine. Russ. J. Mar. Biol. 2017, 43, 101–110. [Google Scholar] [CrossRef]
- Maselli, V.; Galdiero, E.; Salzano, A.M.; Scaloni, A.; Maione, A.; Falanga, A.; Naviglio, D.; Guida, M.; Cosmo, A.; Galdiero, S. OctoPartenopin: Identification and preliminary characterization of a novel antimicrobial peptide from the suckers of Octopus vulgaris. Mar. Drugs 2020, 18, 380. [Google Scholar] [CrossRef] [PubMed]
- Liu, S.; Aweya, J.J.; Zheng, L.; Wang, F.; Zheng, Z.; Zhong, M.; Zhang, Y. A Litopenaeus vannamei hemocyanin-derived antimicrobial peptide (peptide B11) attenuates cancer cells’ proliferation. Molecules 2018, 23, 3202. [Google Scholar] [CrossRef] [PubMed]
- Carrera, M.; Ezquerra-Brauer, J.M.; Aubourg, S.P. Characterization of the jumbo squid (Dosidicus gigas) skin by-product by shotgun proteomics and protein-based bioinformatics. Mar. Drugs 2019, 18, 31. [Google Scholar] [CrossRef]
- Gianazza, E.; Eberini, I.; Palazzolo, L.; Miller, I. Hemolymph proteins: An overview across marine arthropods and molluscs. J. Proteom. 2021, 245, 104294. [Google Scholar] [CrossRef]
- Kim, S.K. (Ed.) Marine Mucin. In Marine Pharmacognosy: Trends and Applications; CRC Press: Boca Raton, FL, USA, 2012. [Google Scholar]
- Kang, V.; Lengerer, B.; Wattiez, R.; Flammang, P. Molecular insights into the powerful mucus-based adhesion of limpets (Patella vulgata L.). Open Biol. 2020, 10, 200019. [Google Scholar] [CrossRef]
- Carrera, M.; Cañas, B.; Gallardo, J.M. The sarcoplasmic fish proteome: Pathways, metabolic networks and potential bioactive peptides for nutritional inferences. J. Proteom. 2013, 78, 211–220. [Google Scholar] [CrossRef]
- Sherman, B.T.; Hao, M.; Qiu, J.; Jiao, X.; Baseler, M.W.; Lane, H.C.; Imamichi, T.; Chang, W. DAVID: A web server for functional enrichment analysis and functional annotation of gene lists (2021 update). Nucleic Acids Res. 2022, 10, W216–W221. [Google Scholar] [CrossRef]
- Szklarczyk, D.; Franceschini, A.; Kuhn, M.; Simonovic, M.; Roth, A.; Minguez, P.; Doerks, T.; Stark, M.; Muller, J.; Bork, P.; et al. The STRING database in 2011: Functional interaction networks of proteins, globally integrated and scored. Nucleic Acids Res. 2010, 39 (Suppl. S1), D561–D568. [Google Scholar] [CrossRef]
- Minkiewicz, P.; Iwaniak, A.; Darewicz, M. BIOPEP-UWM database of bioactive peptides: Current opportunities. Int. J. Mol. Sci. 2019, 20, 5978. [Google Scholar] [CrossRef] [PubMed]
- Thomas, S.; Karnik, S.; Barai, R.S.; Jayaraman, V.K.; Idicula-Thomas, S. CAMP: A useful resource for research on antimicrobial peptides. Nucleic Acids Res. 2010, 38, D774–D780. [Google Scholar] [CrossRef] [PubMed]
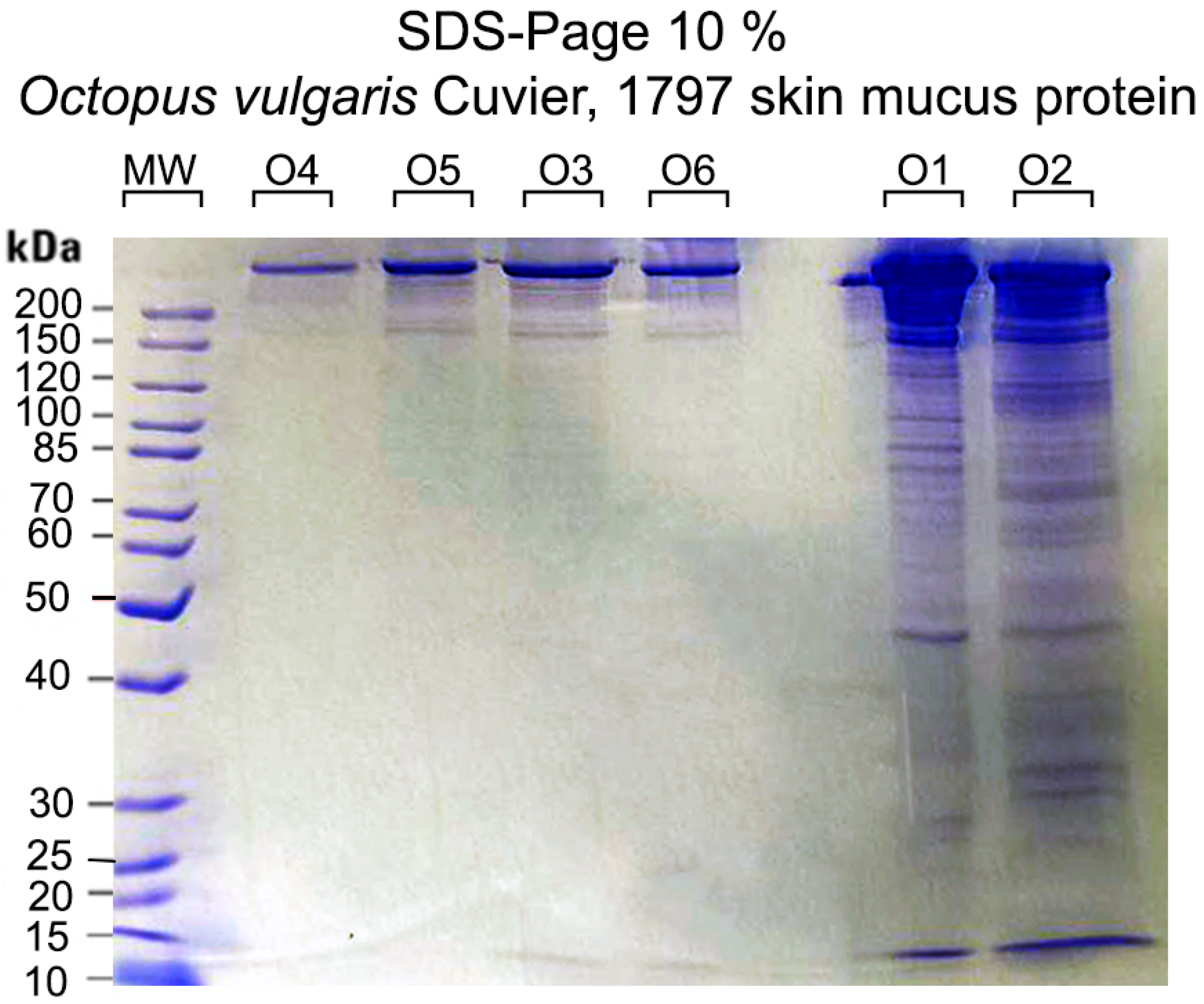


| Sample | Absorbance | Concentration µg/µL |
|---|---|---|
| O1 | 0.67 | 2.43 |
| O2 | 0.43 | 1.44 |
| O3 | 0.18 | 0.42 |
| O4 | 0.10 | 0.07 |
| O5 | 0.14 | 0.23 |
| O6 | 0.18 | 0.42 |
| KEGG Pathway | p-Value |
|---|---|
| Ribosome | 3.90 × 10−2 |
| Phagosome | 8.30 × 10−2 |
| InterPro Motifs | Count | % | p-Value |
|---|---|---|---|
| von Willebrand factor, type D domain | 11 | 2.66 | 5.46 × 10−9 |
| Lamin Tail Domain | 7 | 1.69 | 1.83 × 10−7 |
| Lipid transport protein, N-terminal | 6 | 1.45 | 4.46 × 10−7 |
| Lipid transport protein, beta-sheet shell | 6 | 1.45 | 9.54 × 10−7 |
| Glycoside hydrolase, superfamily | 11 | 2.66 | 1.34 × 10−6 |
| Metallopeptidase, catalytic domain | 9 | 2.18 | 4.34 × 10−5 |
| Uncharacterized domain, cysteine-rich | 6 | 1.45 | 7.74 × 10−5 |
| von Willebrand factor, type A | 11 | 2.66 | 1.08 × 10−4 |
| EF-hand domain | 13 | 3.15 | 2.77 × 10−4 |
| Vitellinogen, beta-sheet N-terminal | 4 | 0.97 | 3.50 × 10−4 |
| Vitellinogen, superhelical | 4 | 0.97 | 4.51 × 10−4 |
| FAS1 domain | 4 | 0.97 | 4.51 × 10−4 |
| Speract/scavenger receptor-related | 5 | 1.21 | 5.00 × 10−4 |
| EF-hand-like domain | 14 | 3.39 | 6.06 × 10−4 |
| Speract/scavenger receptor | 5 | 1.21 | 6.38 × 10−4 |
| Intermediate filament protein, conserved site | 4 | 0.97 | 8.59 × 10−4 |
| Chitin binding domain | 9 | 2.18 | 1.09 × 10−3 |
| Thrombospondin, type 1 repeat | 6 | 1.45 | 1.28 × 10−3 |
| Trypsin Inhibitor-like, cysteine rich domain | 4 | 0.97 | 1.45 × 10−3 |
| Peptidase M12A, astacin | 4 | 0.97 | 1.69 × 10−3 |
| EF-Hand 1, calcium-binding site | 11 | 2.66 | 1.74 × 10−3 |
| Concanavalin A-like lectin/glucanase, subgroup | 9 | 2.18 | 1.80 × 10−3 |
| MAM domain | 4 | 0.97 | 3.65 × 10−3 |
| Vitellinogen, open beta-sheet | 3 | 0.73 | 3.8 × 10−3 |
| Vitellinogen, open beta-sheet, subdomain 1 | 3 | 0.73 | 3.8 × 10−3 |
| MD-2-related lipid-recognition (ML) domain | 3 | 0.73 | 3.85 × 10−3 |
| Low-density lipoprotein (LDL) receptor class A repeat | 6 | 1.45 | 4.4 × 10−3 |
| Carbonic anhydrase, alpha-class, conserved site | 3 | 0.73 | 4.91 × 10−3 |
| Actin-related protein | 4 | 0.97 | 6.04 × 10−3 |
| Actin, conserved site | 3 | 0.73 | 6.09 × 10−3 |
| Actinin-type, actin-binding, conserved site | 4 | 0.97 | 6.60 × 10−3 |
| Peptidase, metallopeptidase | 4 | 0.97 | 7.20 × 10−3 |
| Alpha carbonic anhydrase | 3 | 0.73 | 7.38 × 10−3 |
| Carbonic anhydrase, alpha-class | 3 | 0.73 | 7.38 × 10−3 |
| Glycoside hydrolase family 20, catalytic core | 3 | 0.73 | 7.38 × 10−3 |
| Beta-hexosaminidase subunit alpha/beta | 3 | 0.73 | 7.38 × 10−3 |
| Peptidase C14, caspase non-catalytic subunit p10 | 5 | 1.21 | 7.40 × 10−3 |
| Peptidase C14, caspase precursor p45, core | 5 | 1.21 | 7.82 × 10−3 |
| Peptidase C14, ICE, catalytic subunit p20 | 5 | 1.21 | 8.24 × 10−3 |
| Peptidase C14, caspase precursor p45 | 5 | 1.21 | 9.14 × 10−3 |
| EGF domain, merozoite surface protein 1-like | 3 | 0.73 | 1.19 × 10−2 |
| Translation elongation factor EFTu/EF1A, domain 2 | 3 | 0.73 | 1.19 × 10−2 |
| Low-density lipoprotein (LDL) receptor class A, conserved site | 5 | 1.21 | 1.28 × 10−2 |
| Cystine knot, C-terminal | 3 | 0.73 | 1.37 × 10−2 |
| Chitinase II | 4 | 0.97 | 1.68 × 10−2 |
| Villin/Gelsolin | 3 | 0.73 | 1.74 × 10−2 |
| Glycoside hydrolase, family 18, catalytic domain | 4 | 0.97 | 2.00 × 10−2 |
| Aldehyde dehydrogenase, C-terminal | 3 | 0.73 | 2.38 × 10−2 |
| Myosin tail | 3 | 0.73 | 2.38 × 10−2 |
| Gelsolin domain | 3 | 0.73 | 2.60 × 10−2 |
| Aldehyde dehydrogenase domain | 3 | 0.73 | 2.60 × 10−2 |
| Aldehyde/histidinol dehydrogenase | 3 | 0.73 | 2.60 × 10−2 |
| Aldehyde dehydrogenase, N-terminal | 3 | 0.73 | 2.60 × 10−2 |
| Uncharacterized domain, di-copper center | 3 | 0.73 | 2.84 × 10−2 |
| Tyrosinase | 3 | 0.73 | 2.84 × 10−2 |
| Spectrin repeat | 4 | 0.97 | 3.72 × 10−2 |
| Spectrin/alpha-actinin | 4 | 0.97 | 3.87 × 10−2 |
| Peptidase C14, ICE, catalytic subunit p20, active site | 3 | 0.73 | 3.88 × 10−2 |
| Epidermal growth factor-like domain | 10 | 2.42 | 3.95 × 10−2 |
| Epidermal growth factor-like domain | 10 | 2.42 | 3.95 × 10−2 |
| Elongation factor, GTP-binding domain | 3 | 0.73 | 4.15 × 10−2 |
| Cyclophilin-like peptidyl-prolyl cis-trans isomerase domain | 3 | 0.73 | 4.44 × 10−2 |
| Calponin homology domain | 6 | 1.45 | 4.55 × 10−2 |
| Glycoside hydrolase, chitinase active site | 3 | 0.73 | 7.29 × 10−2 |
| Translation elongation/initiation factor/Ribosomal, beta-barrel | 3 | 0.73 | 7.29 × 10−2 |
| EGF-like calcium-binding, conserved site | 5 | 1.21 | 7.54 × 10−2 |
| Peptidase S1, trypsin family, active site | 4 | 0.97 | 7.64 × 10−2 |
| Peptidase S1 | 4 | 0.97 | 9.46 × 10−2 |
| Trypsin-like cysteine/serine peptidase domain | 4 | 0.97 | 1.02 × 10−1 |
| EGF-like calcium-binding | 6 | 1.45 | 1.45 × 10−1 |
| Insulin-like growth factor binding protein, N-terminal | 5 | 1.21 | 1.58 × 10−1 |
| EGF-type aspartate/asparagine hydroxylation site | 5 | 1.21 | 1.72 × 10−1 |
| WD40 repeat, conserved site | 5 | 1.21 | 2.11 × 10−1 |
| Leucine-rich repeat, typical subtype | 6 | 1.45 | 2.11 × 10−1 |
| Sushi/SCR/CCP | 3 | 0.73 | 2.19 × 10−1 |
| Myosin head, motor domain | 3 | 0.73 | 2.55 × 10−1 |
| Apple-like | 3 | 0.73 | 3.39 × 10−1 |
| IQ motif, EF-hand binding site | 3 | 0.73 | 4.41 × 10−1 |
| Leucine-rich repeat | 6 | 1.45 | 4.64 × 10−1 |
| Immunoglobulin-like domain | 5 | 1.21 | 5.68 × 10−1 |
| WD40 repeat | 6 | 1.45 | 6.24 × 10−1 |
| Fibronectin, type III | 3 | 0.73 | 7.21 × 10−1 |
| WD40/YVTN repeat-like-containing domain | 6 | 1.45 | 7.23 × 10−1 |
| Immunoglobulin I-set | 3 | 0.73 | 7.90 × 10−1 |
| Death domain | 3 | 0.73 | 8.03 × 10−1 |
| Death-like domain | 4 | 0.97 | 8.12 × 10−1 |
| Src homology-3 domain | 4 | 0.97 | 8.60 × 10−1 |
| Immunoglobulin subtype | 3 | 0.73 | 8.67 × 10−1 |
| Ankyrin repeat-containing domain | 3 | 0.73 | 9.96 × 10−1 |
| Ankyrin repeat | 3 | 0.73 | 9.99 × 10−1 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Pérez-Polo, S.; Imran, M.A.S.; Dios, S.; Pérez, J.; Barros, L.; Carrera, M.; Gestal, C. Identifying Natural Bioactive Peptides from the Common Octopus (Octopus vulgaris Cuvier, 1797) Skin Mucus By-Products Using Proteogenomic Analysis. Int. J. Mol. Sci. 2023, 24, 7145. https://doi.org/10.3390/ijms24087145
Pérez-Polo S, Imran MAS, Dios S, Pérez J, Barros L, Carrera M, Gestal C. Identifying Natural Bioactive Peptides from the Common Octopus (Octopus vulgaris Cuvier, 1797) Skin Mucus By-Products Using Proteogenomic Analysis. International Journal of Molecular Sciences. 2023; 24(8):7145. https://doi.org/10.3390/ijms24087145
Chicago/Turabian StylePérez-Polo, Sara, Md Abdus Shukur Imran, Sonia Dios, Jaime Pérez, Lorena Barros, Mónica Carrera, and Camino Gestal. 2023. "Identifying Natural Bioactive Peptides from the Common Octopus (Octopus vulgaris Cuvier, 1797) Skin Mucus By-Products Using Proteogenomic Analysis" International Journal of Molecular Sciences 24, no. 8: 7145. https://doi.org/10.3390/ijms24087145
APA StylePérez-Polo, S., Imran, M. A. S., Dios, S., Pérez, J., Barros, L., Carrera, M., & Gestal, C. (2023). Identifying Natural Bioactive Peptides from the Common Octopus (Octopus vulgaris Cuvier, 1797) Skin Mucus By-Products Using Proteogenomic Analysis. International Journal of Molecular Sciences, 24(8), 7145. https://doi.org/10.3390/ijms24087145






