NLRC5-CIITA Fusion Protein as an Effective Inducer of MHC-I Expression and Antitumor Immunity
Abstract
1. Introduction
2. Results
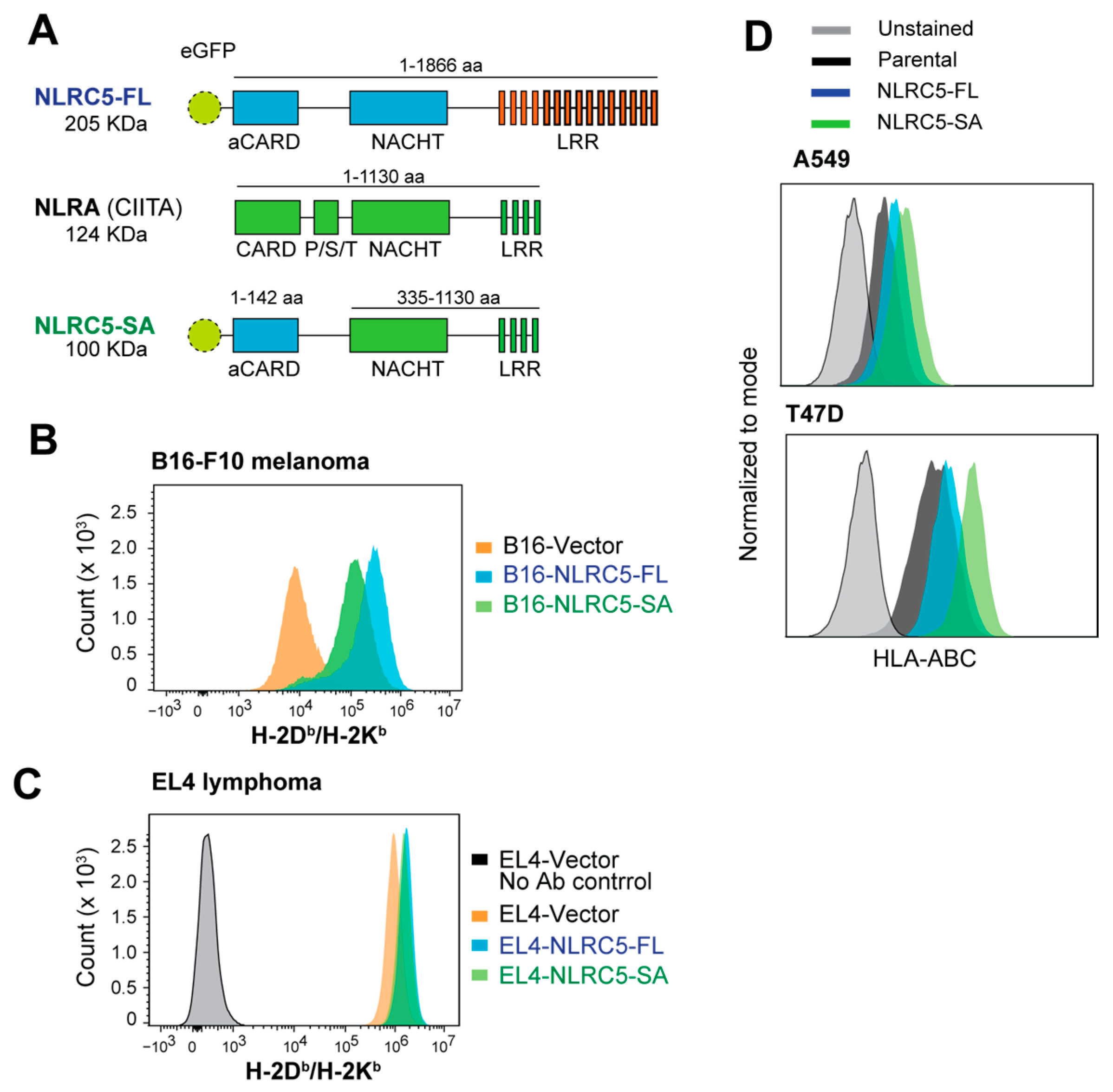
2.1. Induction of MHC-I Expression by NLRC5-SA in Mouse and Human Cancer Cell Lines
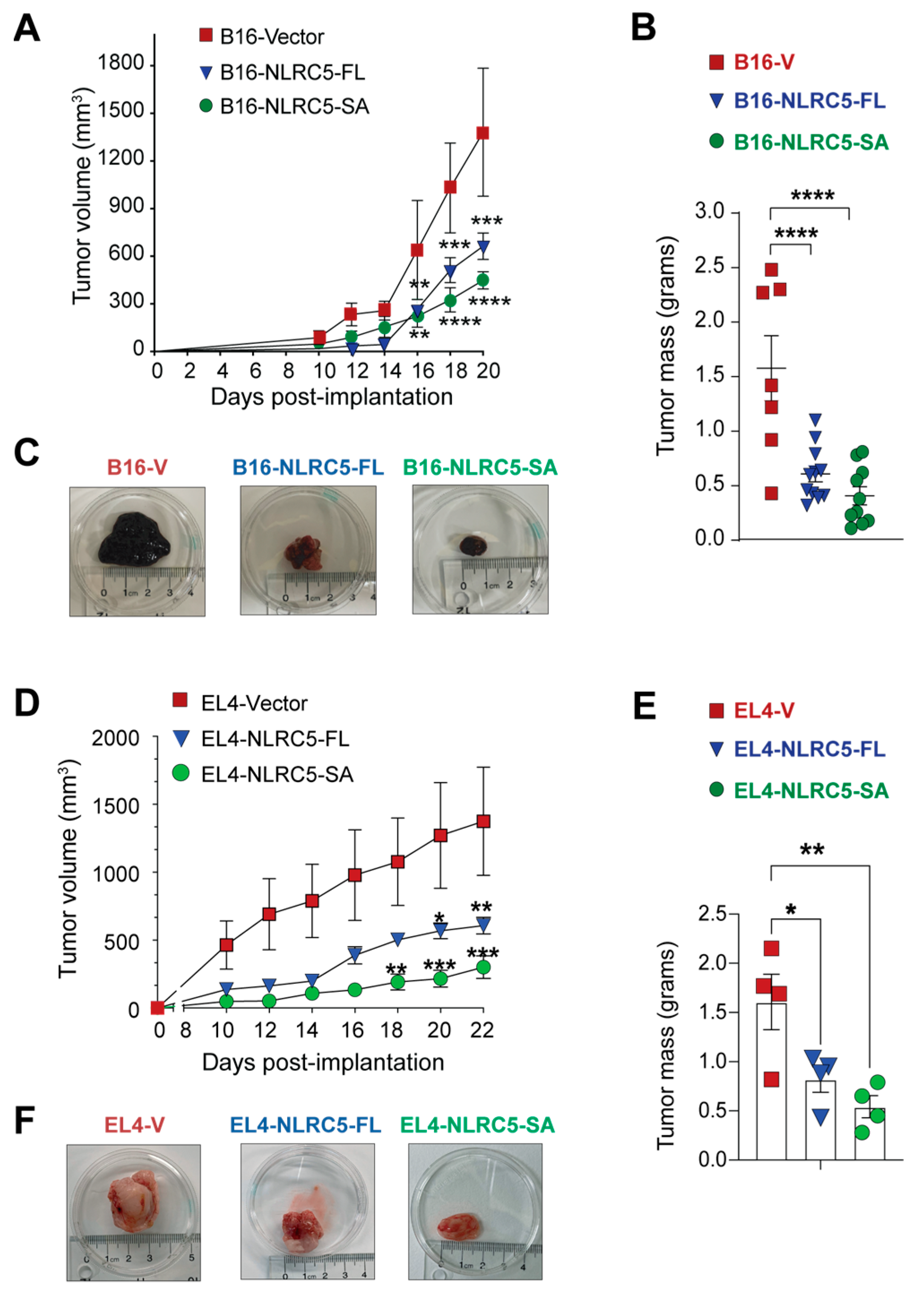
2.2. Attenuation of B16 Melanoma and EL4 Lymphoma Tumor Growth by NLRC5-SA
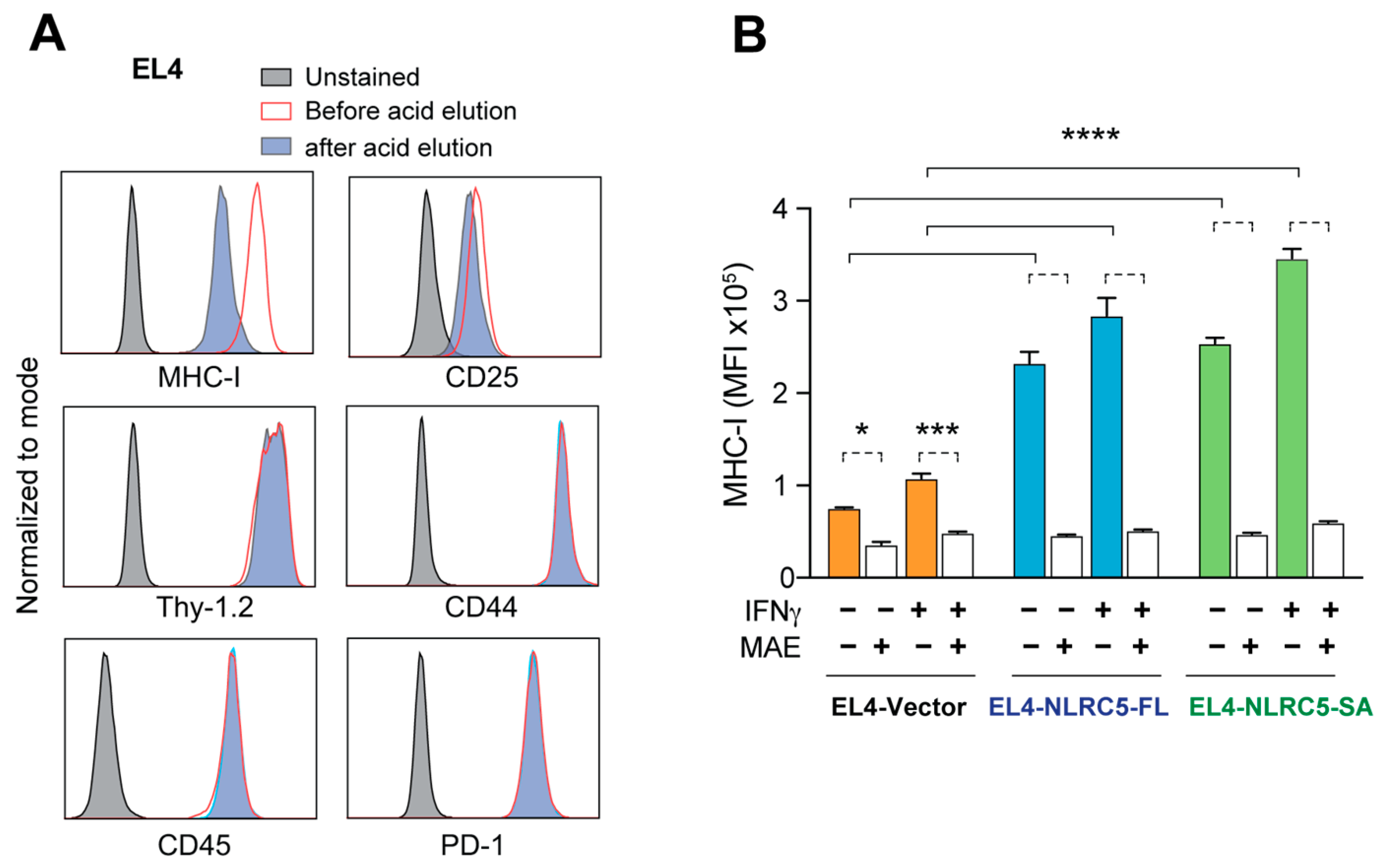
2.3. Acid Elution of MHC-I-Bound Peptides EL4-NLRC5-SA Cells
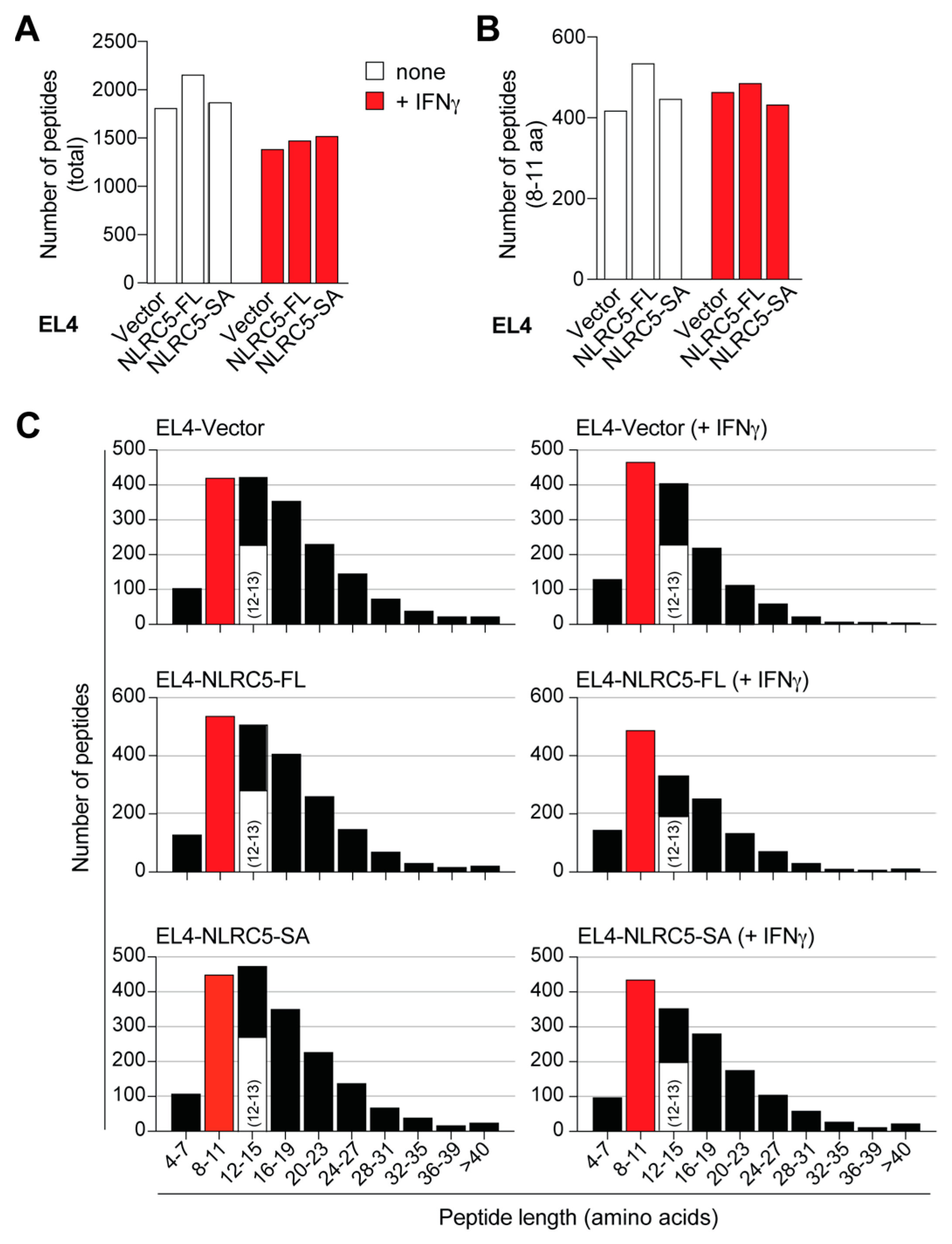
2.4. MAP Repertoire of NLRC5-Expressing EL4 Cells
2.5. NLRC5-SA and NLRC5-FL Differentially Modulate MAPs
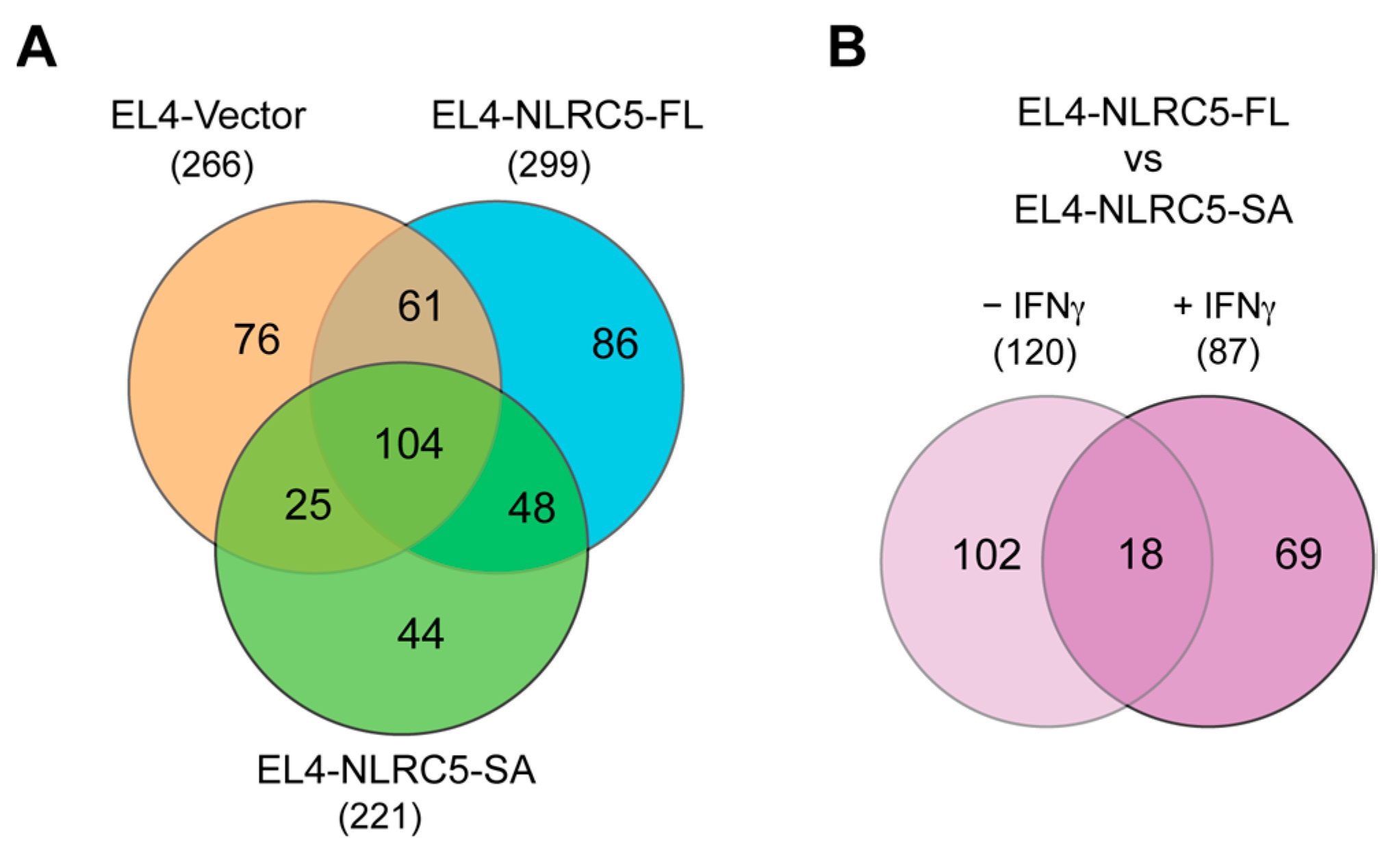
2.6. NLRC5-SA Substantially Differs from NLRC5-FL in Modulating MAPs
2.7. Potentially Immunogenic NLRC5-SA-Induced MAPs
3. Discussion
4. Materials and Methods
4.1. Cell Culture
4.2. Plasmids and Derivation of NLRC5-Expressing Lines
4.3. IFNγ Stimulation and Flow Cytometry
4.4. Tumor Growth
4.5. Mild Acid Elution and Purification of MHC-I-Associated Peptides
4.6. Elution of MAPs and Mass Spectrometry
4.7. Data Analysis and Mining
4.8. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Dunn, G.P.; Bruce, A.T.; Ikeda, H.; Old, L.J.; Schreiber, R.D. Cancer immunoediting: From immunosurveillance to tumor escape. Nat. Immunol. 2002, 3, 991–998. [Google Scholar] [CrossRef] [PubMed]
- Vyas, J.M.; Van der Veen, A.G.; Ploegh, H.L. The known unknowns of antigen processing and presentation. Nat. Rev. Immunol. 2008, 8, 607–618. [Google Scholar] [CrossRef] [PubMed]
- Neefjes, J.; Jongsma, M.L.; Paul, P.; Bakke, O. Towards a systems understanding of MHC class I and MHC class II antigen presentation. Nat. Rev. Immunol. 2011, 11, 823–836. [Google Scholar] [CrossRef] [PubMed]
- Chen, D.S.; Mellman, I. Oncology meets immunology: The cancer-immunity cycle. Immunity 2013, 39, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Coulie, P.G.; Van den Eynde, B.J.; van der Bruggen, P.; Boon, T. Tumour antigens recognized by T lymphocytes: At the core of cancer immunotherapy. Nat. Rev. Cancer 2014, 14, 135–146. [Google Scholar] [CrossRef]
- Schumacher, T.N.; Schreiber, R.D. Neoantigens in cancer immunotherapy. Science 2015, 348, 69–74. [Google Scholar] [CrossRef]
- Laumont, C.M.; Daouda, T.; Laverdure, J.P.; Bonneil, E.; Caron-Lizotte, O.; Hardy, M.P.; Granados, D.P.; Durette, C.; Lemieux, S.; Thibault, P.; et al. Global proteogenomic analysis of human MHC class I-associated peptides derived from non-canonical reading frames. Nat. Commun. 2016, 7, 10238. [Google Scholar] [CrossRef]
- Bassani-Sternberg, M.; Coukos, G. Mass spectrometry-based antigen discovery for cancer immunotherapy. Curr. Opin. Immunol. 2016, 41, 9–17. [Google Scholar] [CrossRef]
- Zhao, Q.; Laverdure, J.P.; Lanoix, J.; Durette, C.; Cote, C.; Bonneil, E.; Laumont, C.M.; Gendron, P.; Vincent, K.; Courcelles, M.; et al. Proteogenomics Uncovers a Vast Repertoire of Shared Tumor-Specific Antigens in Ovarian Cancer. Cancer Immunol. Res. 2020, 8, 544–555. [Google Scholar] [CrossRef]
- Haen, S.P.; Loffler, M.W.; Rammensee, H.G.; Brossart, P. Towards new horizons: Characterization, classification and implications of the tumour antigenic repertoire. Nat. Rev. Clin. Oncol. 2020, 17, 595–610. [Google Scholar] [CrossRef]
- Wang, Z.; Cao, Y.J. Adoptive Cell Therapy Targeting Neoantigens: A Frontier for Cancer Research. Front. Immunol. 2020, 11, 176. [Google Scholar] [CrossRef]
- Zou, W. Immunosuppressive networks in the tumour environment and their therapeutic relevance. Nat. Rev. Cancer 2005, 5, 263–274. [Google Scholar] [CrossRef]
- Hicklin, D.J.; Marincola, F.M.; Ferrone, S. HLA class I antigen downregulation in human cancers: T-cell immunotherapy revives an old story. Mol. Med. Today 1999, 5, 178–186. [Google Scholar] [CrossRef]
- Seliger, B. Molecular mechanisms of MHC class I abnormalities and APM components in human tumors. Cancer Immunol. Immunother. CII 2008, 57, 1719–1726. [Google Scholar] [CrossRef] [PubMed]
- Garrido, F.; Cabrera, T.; Aptsiauri, N. “Hard” and “soft” lesions underlying the HLA class I alterations in cancer cells: Implications for immunotherapy. Int. J. Cancer. J. Int. Du Cancer 2010, 127, 249–256. [Google Scholar] [CrossRef]
- Seliger, B.; Maeurer, M.J.; Ferrone, S. TAP off—Tumors on. Immunol. Today 1997, 18, 292–299. [Google Scholar] [CrossRef] [PubMed]
- Benitez, R.; Godelaine, D.; Lopez-Nevot, M.A.; Brasseur, F.; Jimenez, P.; Marchand, M.; Oliva, M.R.; van Baren, N.; Cabrera, T.; Andry, G.; et al. Mutations of the beta2-microglobulin gene result in a lack of HLA class I molecules on melanoma cells of two patients immunized with MAGE peptides. Tissue Antigens 1998, 52, 520–529. [Google Scholar] [CrossRef] [PubMed]
- Koopman, L.A.; Corver, W.E.; van der Slik, A.R.; Giphart, M.J.; Fleuren, G.J. Multiple genetic alterations cause frequent and heterogeneous human histocompatibility leukocyte antigen class I loss in cervical cancer. J. Exp. Med. 2000, 191, 961–976. [Google Scholar] [CrossRef] [PubMed]
- Paschen, A.; Mendez, R.M.; Jimenez, P.; Sucker, A.; Ruiz-Cabello, F.; Song, M.; Garrido, F.; Schadendorf, D. Complete loss of HLA class I antigen expression on melanoma cells: A result of successive mutational events. Int. J. Cancer J. Int. Cancer 2003, 103, 759–767. [Google Scholar] [CrossRef]
- Leone, P.; Shin, E.C.; Perosa, F.; Vacca, A.; Dammacco, F.; Racanelli, V. MHC class I antigen processing and presenting machinery: Organization, function, and defects in tumor cells. J. Natl. Cancer Inst. 2013, 105, 1172–1187. [Google Scholar] [CrossRef]
- Nie, Y.; Yang, G.; Song, Y.; Zhao, X.; So, C.; Liao, J.; Wang, L.D.; Yang, C.S. DNA hypermethylation is a mechanism for loss of expression of the HLA class I genes in human esophageal squamous cell carcinomas. Carcinogenesis 2001, 22, 1615–1623. [Google Scholar] [CrossRef] [PubMed]
- Sigalotti, L.; Fratta, E.; Coral, S.; Tanzarella, S.; Danielli, R.; Colizzi, F.; Fonsatti, E.; Traversari, C.; Altomonte, M.; Maio, M. Intratumor heterogeneity of cancer/testis antigens expression in human cutaneous melanoma is methylation-regulated and functionally reverted by 5-aza-2′-deoxycytidine. Cancer Res. 2004, 64, 9167–9171. [Google Scholar] [CrossRef]
- Khan, A.N.; Gregorie, C.J.; Tomasi, T.B. Histone deacetylase inhibitors induce TAP, LMP, Tapasin genes and MHC class I antigen presentation by melanoma cells. Cancer Immunol. Immunother. CII 2008, 57, 647–654. [Google Scholar] [CrossRef]
- Setiadi, A.F.; Omilusik, K.; David, M.D.; Seipp, R.P.; Hartikainen, J.; Gopaul, R.; Choi, K.B.; Jefferies, W.A. Epigenetic enhancement of antigen processing and presentation promotes immune recognition of tumors. Cancer Res. 2008, 68, 9601–9607. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Chiappinelli, K.B.; Guzzetta, A.A.; Easwaran, H.; Yen, R.W.; Vatapalli, R.; Topper, M.J.; Luo, J.; Connolly, R.M.; Azad, N.S.; et al. Immune regulation by low doses of the DNA methyltransferase inhibitor 5-azacitidine in common human epithelial cancers. Oncotarget 2014, 5, 587–598. [Google Scholar] [CrossRef] [PubMed]
- Vlkova, V.; Stepanek, I.; Hruskova, V.; Senigl, F.; Mayerova, V.; Sramek, M.; Simova, J.; Bieblova, J.; Indrova, M.; Hejhal, T.; et al. Epigenetic regulations in the IFNgamma signalling pathway: IFNgamma-mediated MHC class I upregulation on tumour cells is associated with DNA demethylation of antigen-presenting machinery genes. Oncotarget 2014, 5, 6923–6935. [Google Scholar] [CrossRef] [PubMed]
- Lampen, M.H.; van Hall, T. Strategies to counteract MHC-I defects in tumors. Curr. Opin. Immunol. 2011, 23, 293–298. [Google Scholar] [CrossRef]
- Garrido, F.; Aptsiauri, N.; Doorduijn, E.M.; Garcia Lora, A.M.; van Hall, T. The urgent need to recover MHC class I in cancers for effective immunotherapy. Curr. Opin. Immunol. 2016, 39, 44–51. [Google Scholar] [CrossRef] [PubMed]
- Meissner, T.B.; Li, A.; Biswas, A.; Lee, K.H.; Liu, Y.J.; Bayir, E.; Iliopoulos, D.; van den Elsen, P.J.; Kobayashi, K.S. NLR family member NLRC5 is a transcriptional regulator of MHC class I genes. Proc. Natl. Acad. Sci. USA 2010, 107, 13794–13799. [Google Scholar] [CrossRef] [PubMed]
- Kobayashi, K.S.; van den Elsen, P.J. NLRC5: A key regulator of MHC class I-dependent immune responses. Nat. Rev. Immunol. 2012, 12, 813–820. [Google Scholar] [CrossRef]
- Yoshihama, S.; Roszik, J.; Downs, I.; Meissner, T.B.; Vijayan, S.; Chapuy, B.; Sidiq, T.; Shipp, M.A.; Lizee, G.A.; Kobayashi, K.S. NLRC5/MHC class I transactivator is a target for immune evasion in cancer. Proc. Natl. Acad. Sci. USA 2016, 113, 5999–6004. [Google Scholar] [CrossRef]
- Zong, Z.; Song, Y.; Xue, Y.; Ruan, X.; Liu, X.; Yang, C.; Zheng, J.; Cao, S.; Li, Z.; Liu, Y. Knockdown of LncRNA SCAMP1 suppressed malignant biological behaviours of glioma cells via modulating miR-499a-5p/LMX1A/NLRC5 pathway. J. Cell. Mol. Med. 2019, 23, 5048–5062. [Google Scholar] [CrossRef] [PubMed]
- Zhan, L.; Zhang, J.; Zhang, J.; Liu, X.; Zhu, S.; Shi, Y.; He, Y.; Wang, W.; Fan, Y.; Tang, Z.; et al. LC3 and NLRC5 interaction inhibits NLRC5-mediated MHC class I antigen presentation pathway in endometrial cancer. Cancer Lett. 2022, 529, 37–52. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Guo, F.; Liu, Y.; Chen, H.J.; Wen, F.; Zou, B.; Li, D.; Qin, Q.; Liu, X.; Shen, Y.; et al. NLRC5 expression in tumors and its role as a negative prognostic indicator in stage III non-small-cell lung cancer patients. Oncol. Lett. 2015, 10, 1533–1540. [Google Scholar] [CrossRef]
- Wang, Q.; Ding, H.; He, Y.; Li, X.; Cheng, Y.; Xu, Q.; Yang, Y.; Liao, G.; Meng, X.; Huang, C.; et al. NLRC5 mediates cell proliferation, migration, and invasion by regulating the Wnt/beta-catenin signalling pathway in clear cell renal cell carcinoma. Cancer Lett. 2019, 444, 9–19. [Google Scholar] [CrossRef]
- Rodriguez, G.M.; Bobbala, D.; Serrano, D.; Mayhue, M.; Champagne, A.; Saucier, C.; Steimle, V.; Kufer, T.A.; Menendez, A.; Ramanathan, S.; et al. NLRC5 elicits antitumor immunity by enhancing processing and presentation of tumor antigens to CD8(+) T lymphocytes. Oncoimmunology 2016, 5, e1151593. [Google Scholar] [CrossRef] [PubMed]
- Kalbasi, A.; Tariveranmoshabad, M.; Hakimi, K.; Kremer, S.; Campbell, K.M.; Funes, J.M.; Vega-Crespo, A.; Parisi, G.; Champekar, A.; Nguyen, C.; et al. Uncoupling interferon signaling and antigen presentation to overcome immunotherapy resistance due to JAK1 loss in melanoma. Sci. Transl. Med. 2020, 12, eabb0152. [Google Scholar] [CrossRef]
- Kim, H.; Kim, H.; Feng, Y.; Li, Y.; Tamiya, H.; Tocci, S.; Ronai, Z.A. PRMT5 control of cGAS/STING and NLRC5 pathways defines melanoma response to antitumor immunity. Sci. Transl. Med. 2020, 12, eaaz5683. [Google Scholar] [CrossRef]
- Yoshihama, S.; Cho, S.X.; Yeung, J.; Pan, X.; Lizee, G.; Konganti, K.; Johnson, V.E.; Kobayashi, K.S. NLRC5/CITA expression correlates with efficient response to checkpoint blockade immunotherapy. Sci. Rep. 2021, 11, 3258. [Google Scholar] [CrossRef]
- Kungulovski, G.; Jeltsch, A. Epigenome Editing: State of the Art, Concepts, and Perspectives. Trends Genet. 2016, 32, 101–113. [Google Scholar] [CrossRef]
- Nebbioso, A.; Tambaro, F.P.; Dell’Aversana, C.; Altucci, L. Cancer epigenetics: Moving forward. PLoS Genet. 2018, 14, e1007362. [Google Scholar] [CrossRef]
- Cui, J.; Zhu, L.; Xia, X.; Wang, H.Y.; Legras, X.; Hong, J.; Ji, J.; Shen, P.; Zheng, S.; Chen, Z.J.; et al. NLRC5 negatively regulates the NF-kappaB and type I interferon signaling pathways. Cell 2010, 141, 483–496. [Google Scholar] [CrossRef]
- Neerincx, A.; Jakobshagen, K.; Utermohlen, O.; Buning, H.; Steimle, V.; Kufer, T.A. The N-terminal domain of NLRC5 confers transcriptional activity for MHC class I and II gene expression. J. Immunol. 2014, 193, 3090–3100. [Google Scholar] [CrossRef] [PubMed]
- Fortier, M.H.; Caron, E.; Hardy, M.P.; Voisin, G.; Lemieux, S.; Perreault, C.; Thibault, P. The MHC class I peptide repertoire is molded by the transcriptome. J. Exp. Med. 2008, 205, 595–610. [Google Scholar] [CrossRef]
- Goldwich, A.; Hahn, S.S.; Schreiber, S.; Meier, S.; Kampgen, E.; Wagner, R.; Lutz, M.B.; Schubert, U. Targeting HIV-1 Gag into the defective ribosomal product pathway enhances MHC class I antigen presentation and CD8+ T cell activation. J. Immunol. 2008, 180, 372–382. [Google Scholar] [CrossRef] [PubMed]
- Granados, D.P.; Tanguay, P.L.; Hardy, M.P.; Caron, E.; de Verteuil, D.; Meloche, S.; Perreault, C. ER stress affects processing of MHC class I-associated peptides. BMC Immunol. 2009, 10, 10. [Google Scholar] [CrossRef] [PubMed]
- Palmer, A.L.; Dolan, B.P. MHC class I antigen presentation of DRiP-derived peptides from a model antigen is not dependent on the AAA ATPase p97. PLoS ONE 2013, 8, e67796. [Google Scholar] [CrossRef]
- Storkus, W.J.; Zeh, H.J., 3rd; Salter, R.D.; Lotze, M.T. Identification of T-cell epitopes: Rapid isolation of class I-presented peptides from viable cells by mild acid elution. J. Immunother. Emphas. Tumor Immunol. 1993, 14, 94–103. [Google Scholar] [CrossRef]
- Zhang, J.; Xin, L.; Shan, B.; Chen, W.; Xie, M.; Yuen, D.; Zhang, W.; Zhang, Z.; Lajoie, G.A.; Ma, B. PEAKS DB: De novo sequencing assisted database search for sensitive and accurate peptide identification. Mol. Cell. Proteom. 2012, 11, M111.010587. [Google Scholar] [CrossRef]
- Yewdell, J.W.; Reits, E.; Neefjes, J. Making sense of mass destruction: Quantitating MHC class I antigen presentation. Nat. Rev. Immunol. 2003, 3, 952–961. [Google Scholar] [CrossRef]
- Sturm, T.; Sautter, B.; Worner, T.P.; Stevanovic, S.; Rammensee, H.G.; Planz, O.; Heck, A.J.R.; Aebersold, R. Mild Acid Elution and MHC Immunoaffinity Chromatography Reveal Similar Albeit Not Identical Profiles of the HLA Class I Immunopeptidome. J. Proteome Res. 2021, 20, 289–304. [Google Scholar] [CrossRef] [PubMed]
- Biswas, A.; Meissner, T.B.; Kawai, T.; Kobayashi, K.S. Cutting edge: Impaired MHC class I expression in mice deficient for Nlrc5/class I transactivator. J. Immunol. 2012, 189, 516–520. [Google Scholar] [CrossRef] [PubMed]
- Yao, Y.; Wang, Y.; Chen, F.; Huang, Y.; Zhu, S.; Leng, Q.; Wang, H.; Shi, Y.; Qian, Y. NLRC5 regulates MHC class I antigen presentation in host defense against intracellular pathogens. Cell Res. 2012, 22, 836–847. [Google Scholar] [CrossRef] [PubMed]
- Dhatchinamoorthy, K.; Colbert, J.D.; Rock, K.L. Cancer Immune Evasion Through Loss of MHC Class I Antigen Presentation. Front. Immunol. 2021, 12, 636568. [Google Scholar] [CrossRef]
- Gettinger, S.; Choi, J.; Hastings, K.; Truini, A.; Datar, I.; Sowell, R.; Wurtz, A.; Dong, W.; Cai, G.; Melnick, M.A.; et al. Impaired HLA Class I Antigen Processing and Presentation as a Mechanism of Acquired Resistance to Immune Checkpoint Inhibitors in Lung Cancer. Cancer Discov. 2017, 7, 1420–1435. [Google Scholar] [CrossRef]
- Taylor, B.C.; Balko, J.M. Mechanisms of MHC-I Downregulation and Role in Immunotherapy Response. Front. Immunol. 2022, 13, 844866. [Google Scholar] [CrossRef]
- Castro, F.; Cardoso, A.P.; Goncalves, R.M.; Serre, K.; Oliveira, M.J. Interferon-Gamma at the Crossroads of Tumor Immune Surveillance or Evasion. Front. Immunol. 2018, 9, 847. [Google Scholar] [CrossRef] [PubMed]
- Shukla, A.; Cloutier, M.; Appiya Santharam, M.; Ramanathan, S.; Ilangumaran, S. The MHC Class-I Transactivator NLRC5: Implications to Cancer Immunology and Potential Applications to Cancer Immunotherapy. Int. J. Mol. Sci. 2021, 22, 1964. [Google Scholar] [CrossRef]
- Dersh, D.; Holly, J.; Yewdell, J.W. A few good peptides: MHC class I-based cancer immunosurveillance and immunoevasion. Nat. Rev. Immunol. 2020, 21, 116–128. [Google Scholar] [CrossRef]
- Castelli, C.; Storkus, W.J.; Maeurer, M.J.; Martin, D.M.; Huang, E.C.; Pramanik, B.N.; Nagabhushan, T.L.; Parmiani, G.; Lotze, M.T. Mass spectrometric identification of a naturally processed melanoma peptide recognized by CD8+ cytotoxic T lymphocytes. J. Exp. Med. 1995, 181, 363–368. [Google Scholar] [CrossRef]
- Caron, E.; Kowalewski, D.J.; Chiek Koh, C.; Sturm, T.; Schuster, H.; Aebersold, R. Analysis of Major Histocompatibility Complex (MHC) Immunopeptidomes Using Mass Spectrometry. Mol. Cell. Proteom. 2015, 14, 3105–3117. [Google Scholar] [CrossRef] [PubMed]
- Purcell, A.W.; Ramarathinam, S.H.; Ternette, N. Mass spectrometry-based identification of MHC-bound peptides for immunopeptidomics. Nat. Protoc. 2019, 14, 1687–1707. [Google Scholar] [CrossRef] [PubMed]
- Stryhn, A.; Pedersen, L.O.; Holm, A.; Buus, S. Longer peptide can be accommodated in the MHC class I binding site by a protrusion mechanism. Eur. J. Immunol. 2000, 30, 3089–3099. [Google Scholar] [CrossRef] [PubMed]
- Koopmann, J.O.; Post, M.; Neefjes, J.J.; Hammerling, G.J.; Momburg, F. Translocation of long peptides by transporters associated with antigen processing (TAP). Eur. J. Immunol. 1996, 26, 1720–1728. [Google Scholar] [CrossRef]
- Milner, E.; Barnea, E.; Beer, I.; Admon, A. The turnover kinetics of major histocompatibility complex peptides of human cancer cells. Mol. Cell. Proteom. 2006, 5, 357–365. [Google Scholar] [CrossRef] [PubMed]
- Qian, S.B.; Princiotta, M.F.; Bennink, J.R.; Yewdell, J.W. Characterization of rapidly degraded polypeptides in mammalian cells reveals a novel layer of nascent protein quality control. J. Biol. Chem. 2006, 281, 392–400. [Google Scholar] [CrossRef]
- Shastri, N.; Nguyen, V.; Gonzalez, F. Major histocompatibility class I molecules can present cryptic translation products to T-cells. J. Biol. Chem. 1995, 270, 1088–1091. [Google Scholar] [CrossRef]
- Malarkannan, S.; Horng, T.; Shih, P.P.; Schwab, S.; Shastri, N. Presentation of out-of-frame peptide/MHC class I complexes by a novel translation initiation mechanism. Immunity 1999, 10, 681–690. [Google Scholar] [CrossRef]
- Schwab, S.R.; Li, K.C.; Kang, C.; Shastri, N. Constitutive display of cryptic translation products by MHC class I molecules. Science 2003, 301, 1367–1371. [Google Scholar] [CrossRef]
- Yewdell, J.W. Plumbing the sources of endogenous MHC class I peptide ligands. Curr. Opin. Immunol. 2007, 19, 79–86. [Google Scholar] [CrossRef]
- Anton, L.C.; Yewdell, J.W. Translating DRiPs: MHC class I immunosurveillance of pathogens and tumors. J. Leukoc. Biol. 2014, 95, 551–562. [Google Scholar] [CrossRef] [PubMed]
- Granados, D.P.; Laumont, C.M.; Thibault, P.; Perreault, C. The nature of self for T cells-a systems-level perspective. Curr. Opin. Immunol. 2015, 34, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Starck, S.R.; Shastri, N. Nowhere to hide: Unconventional translation yields cryptic peptides for immune surveillance. Immunol. Rev. 2016, 272, 8–16. [Google Scholar] [CrossRef] [PubMed]
- Ebstein, F.; Textoris-Taube, K.; Keller, C.; Golnik, R.; Vigneron, N.; Van den Eynde, B.J.; Schuler-Thurner, B.; Schadendorf, D.; Lorenz, F.K.; Uckert, W.; et al. Proteasomes generate spliced epitopes by two different mechanisms and as efficiently as non-spliced epitopes. Sci. Rep. 2016, 6, 24032. [Google Scholar] [CrossRef] [PubMed]
- Liepe, J.; Marino, F.; Sidney, J.; Jeko, A.; Bunting, D.E.; Sette, A.; Kloetzel, P.M.; Stumpf, M.P.; Heck, A.J.; Mishto, M. A large fraction of HLA class I ligands are proteasome-generated spliced peptides. Science 2016, 354, 354–358. [Google Scholar] [CrossRef] [PubMed]
- Platteel, A.C.M.; Liepe, J.; van Eden, W.; Mishto, M.; Sijts, A. An Unexpected Major Role for Proteasome-Catalyzed Peptide Splicing in Generation of T Cell Epitopes: Is There Relevance for Vaccine Development? Front. Immunol. 2017, 8, 1441. [Google Scholar] [CrossRef]
- Rolfs, Z.; Solntsev, S.K.; Shortreed, M.R.; Frey, B.L.; Smith, L.M. Global Identification of Post-Translationally Spliced Peptides with Neo-Fusion. J. Proteome Res. 2019, 18, 349–358. [Google Scholar] [CrossRef]
- Perkins, D.N.; Pappin, D.J.; Creasy, D.M.; Cottrell, J.S. Probability-based protein identification by searching sequence databases using mass spectrometry data. Electrophoresis 1999, 20, 3551–3567. [Google Scholar] [CrossRef]
- Cox, J.; Mann, M. MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat. Biotechnol. 2008, 26, 1367–1372. [Google Scholar] [CrossRef]
- Latouche, J.B.; Sadelain, M. Induction of human cytotoxic T lymphocytes by artificial antigen-presenting cells. Nat. Biotechnol. 2000, 18, 405–409. [Google Scholar] [CrossRef]
- Harari, A.; Graciotti, M.; Bassani-Sternberg, M.; Kandalaft, L.E. Antitumour dendritic cell vaccination in a priming and boosting approach. Nat. Rev. Drug Discov. 2020, 19, 635–652. [Google Scholar] [CrossRef] [PubMed]
- Abusarah, J.; Khodayarian, F.; El-Hachem, N.; Salame, N.; Olivier, M.; Balood, M.; Roversi, K.; Talbot, S.; Bikorimana, J.P.; Chen, J.; et al. Engineering immunoproteasome-expressing mesenchymal stromal cells: A potent cellular vaccine for lymphoma and melanoma in mice. Cell. Rep. Med. 2021, 2, 100455. [Google Scholar] [CrossRef] [PubMed]
- Lee-Chang, C.; Lesniak, M.S. Next-generation antigen-presenting cell immune therapeutics for gliomas. J. Clin. Investig. 2023, 133, e163449. [Google Scholar] [CrossRef] [PubMed]
- Neerincx, A.; Rodriguez, G.M.; Steimle, V.; Kufer, T.A. NLRC5 controls basal MHC class I gene expression in an MHC enhanceosome-dependent manner. J. Immunol. 2012, 188, 4940–4950. [Google Scholar] [CrossRef]
- Dubois, M.L.; Meller, A.; Samandi, S.; Brunelle, M.; Frion, J.; Brunet, M.A.; Toupin, A.; Beaudoin, M.C.; Jacques, J.F.; Levesque, D.; et al. UBB pseudogene 4 encodes functional ubiquitin variants. Nat. Commun. 2020, 11, 1306. [Google Scholar] [CrossRef]
- Heberle, H.; Meirelles, G.V.; da Silva, F.R.; Telles, G.P.; Minghim, R. InteractiVenn: A web-based tool for the analysis of sets through Venn diagrams. BMC Bioinform. 2015, 16, 169. [Google Scholar] [CrossRef] [PubMed]
- Shankavaram, U.T.; Varma, S.; Kane, D.; Sunshine, M.; Chary, K.K.; Reinhold, W.C.; Pommier, Y.; Weinstein, J.N. CellMiner: A relational database and query tool for the NCI-60 cancer cell lines. BMC Genom. 2009, 10, 277. [Google Scholar] [CrossRef]
- Reinhold, W.C.; Sunshine, M.; Liu, H.; Varma, S.; Kohn, K.W.; Morris, J.; Doroshow, J.; Pommier, Y. CellMiner: A web-based suite of genomic and pharmacologic tools to explore transcript and drug patterns in the NCI-60 cell line set. Cancer Res. 2012, 72, 3499–3511. [Google Scholar] [CrossRef]
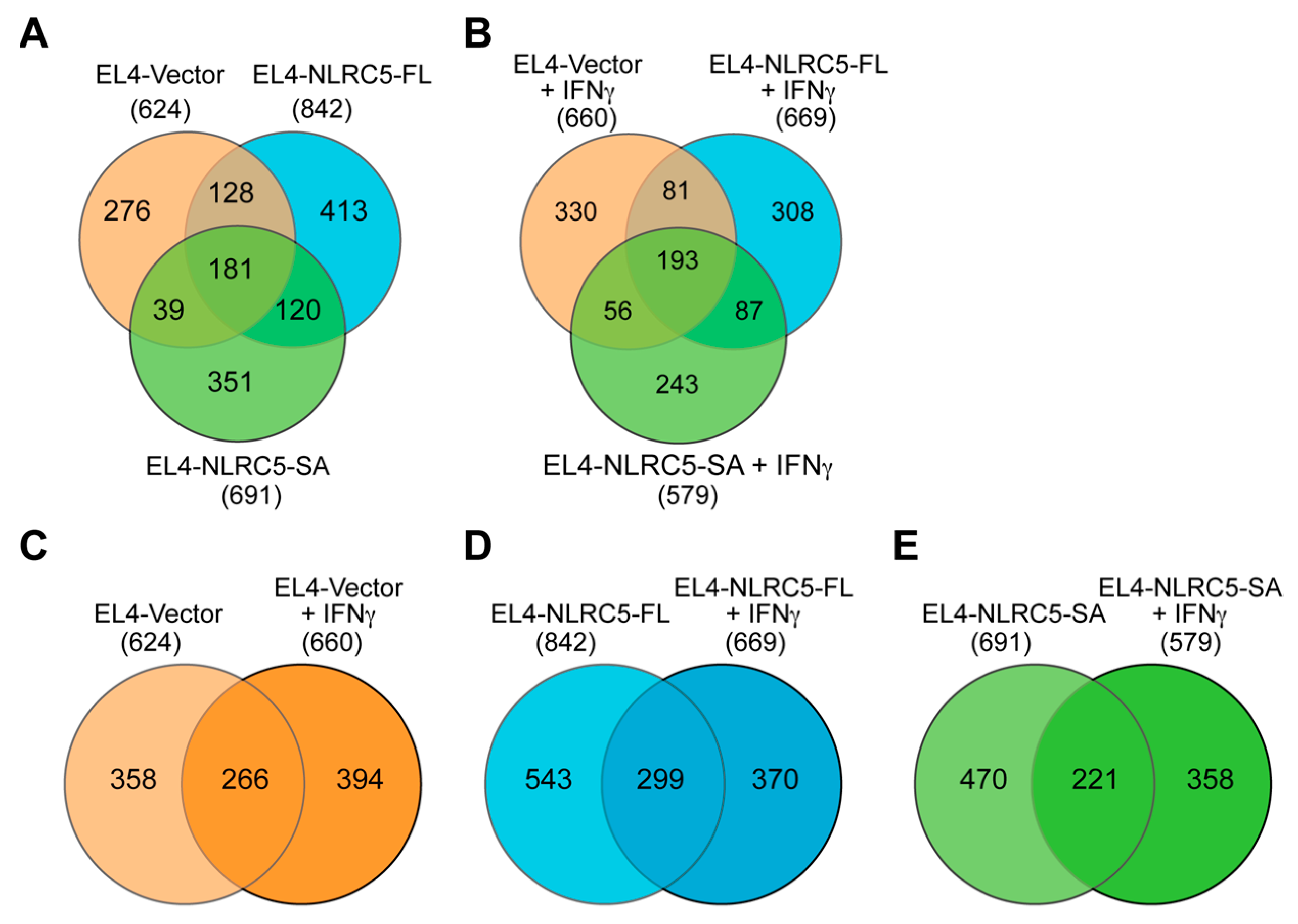

| Cell Population | Total Peptides | Known | Unknown | % Unknown |
|---|---|---|---|---|
| EL4-Vector | 624 | 374 | 250 | 40% |
| EL4-NLRC5-FL | 842 | 543 | 299 | 35% |
| EL4-NLRC5-SA | 691 | 381 | 310 | 45% |
| EL4-Vector (IFNγ) | 660 | 367 | 293 | 44% |
| EL4-NLRC5-FL (IFNγ) | 669 | 357 | 312 | 46% |
| EL4-NLRC5-SA (IFNγ) | 579 | 324 | 255 | 44% |
| Peptide | Length | Accession Name | Protein Functionality in | −10lgP | m/z |
|---|---|---|---|---|---|
| KVFLENVIRDA | 11 | Unknown | - | 57.42 | 435.2499 |
| KVFLENVIRD | 10 | Unknown | - | 57.26 | 411.5716 |
| VVLRNPLIAGK | 11 | SMD2 | Pre-mRNA splicing | 56.32 | 393.9244 |
| S(+42.01)GGLLKALRSDSY | 13 | NCBP2 | RNA processing | 54.98 | 704.8751 |
| AKALANVNIGSL | 12 | RLA1 | Protein synthesis | 49.39 | 585.8455 |
| AVRLLLPGEL | 10 | Unknown | - | 45.61 | 540.8425 |
| AVRLILPGEL | 10 | Unknown | - | 45.61 | 540.8425 |
| RFQSSAVM(+15.99)AL | 10 | H3.1 | Chromatin stability | 45.21 | 563.2897 |
| SVEDIHSRFQSL | 12 | ILEUA | Innate immune response | 42.6 | 473.2405 |
| LIRKLPFQRL | 10 | H3C | Chromatin stability | 37.55 | 428.616 |
| LLRKLPFQRL | 10 | Unknown | - | 37.55 | 428.616 |
| KQQIATFDT | 9 | Unknown | - | 31.22 | 526.2757 |
| FAKALANVN | 9 | RLA1 | Protein synthesis | 30.9 | 474.2691 |
| DTLSEESYKDSTL | 13 | 1433Z | Adapter protein in regulation of multiple signaling pathways | 28.1 | 744.3399 |
| LVLPVPAF | 8 | Unknown | - | 28.06 | 428.2725 |
| IVLPVPAF | 8 | Unknown | - | 28.06 | 428.2725 |
| LAGTEAVVEAI | 11 | Unknown | - | 27.65 | 536.7983 |
| LEVQGRDSRLVL | 12 | ATPB | Mitochondrial electron transport complex | 25.71 | 462.2681 |
| Peptide | Length | Accession Name | Protein Functionality in | −10lgP | m/z |
|---|---|---|---|---|---|
| VREIAQDFKTDL | 12 | H3C | Chromatin stability | 56.04 | 478.9242 |
| DRLHISPDRVY | 11 | MIF | Innate immune response | 54.61 | 457.5773 |
| A(+42.01)EDIKTKIKNYK | 12 | CX6B1 | Mitochondrial electron transport complex | 53.05 | 498.2838 |
| SIRGNNIRY | 9 | SMD1 | Pre-mRNA splicing | 52.53 | 364.8692 |
| SLRGNNIRY | 9 | Unknown | - | 52.53 | 364.8692 |
| VYIKHPVSLEQYL | 13 | PSMD8 | Proteasomal processing | 47.32 | 530.296 |
| PRKIEEIKDFL | 11 | RL38 | Protein synthesis | 46.74 | 463.2697 |
| AAVLEYLTAE | 10 | H2A1 | Chromatin stability | 45.46 | 540.2855 |
| FASPTQVF | 8 | ATPD | Mitochondrial electron transport complex | 44.68 | 896.4516 |
| IEDDKSRLVL | 10 | MA2B2 | Spermatogenesis | 44.44 | 396.5593 |
| SLVYPFPGPIPN | 12 | Unknown | - | 40.72 | 650.8497 |
| LAEEAVTLD | 9 | ATPD | Mitochondrial electron transport complex | 39.51 | 960.4878 |
| TGPSNVDKL | 9 | CHK1 | Cell cycle regulation | 37.12 | 465.7493 |
| PLRAQQLAAEL | 11 | TIM10 | Mitochondrial intermembrane chaperone | 35.53 | 605.351 |
| T(+42.01)RDFKPGDLIFA | 12 | PSIP1 | Stress-induced apoptosis | 27.11 | 711.3745 |
| Marker | Antibody Clone | Conjugate | Source | Catalog Number |
|---|---|---|---|---|
| H-2Kb/Db | 28-8-6 | Alexa 647 | Biolegend | 114612 |
| CD45 | 30-F11 | BV605 | Biolegend | 103140 |
| CD44 | IM7 | PE Cy7 | eBiosciences | 25-0441-82 |
| CD25 | 3C7 | FITC | Biolegend | 101908 |
| Thy 1.2 | 30-H12 | APC | Biolegend | 105312 |
| PD-1 | J43 | PE | BD | 551892 |
| HLA-ABC | W6/32 | PE | eBiosciences | 12-9983-42 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Santharam, M.A.; Shukla, A.; Levesque, D.; Kufer, T.A.; Boisvert, F.-M.; Ramanathan, S.; Ilangumaran, S. NLRC5-CIITA Fusion Protein as an Effective Inducer of MHC-I Expression and Antitumor Immunity. Int. J. Mol. Sci. 2023, 24, 7206. https://doi.org/10.3390/ijms24087206
Santharam MA, Shukla A, Levesque D, Kufer TA, Boisvert F-M, Ramanathan S, Ilangumaran S. NLRC5-CIITA Fusion Protein as an Effective Inducer of MHC-I Expression and Antitumor Immunity. International Journal of Molecular Sciences. 2023; 24(8):7206. https://doi.org/10.3390/ijms24087206
Chicago/Turabian StyleSantharam, Madanraj Appiya, Akhil Shukla, Dominique Levesque, Thomas A. Kufer, François-Michel Boisvert, Sheela Ramanathan, and Subburaj Ilangumaran. 2023. "NLRC5-CIITA Fusion Protein as an Effective Inducer of MHC-I Expression and Antitumor Immunity" International Journal of Molecular Sciences 24, no. 8: 7206. https://doi.org/10.3390/ijms24087206
APA StyleSantharam, M. A., Shukla, A., Levesque, D., Kufer, T. A., Boisvert, F.-M., Ramanathan, S., & Ilangumaran, S. (2023). NLRC5-CIITA Fusion Protein as an Effective Inducer of MHC-I Expression and Antitumor Immunity. International Journal of Molecular Sciences, 24(8), 7206. https://doi.org/10.3390/ijms24087206






