Function Investigations and Applications of Membrane Proteins on Artificial Lipid Membranes
Abstract
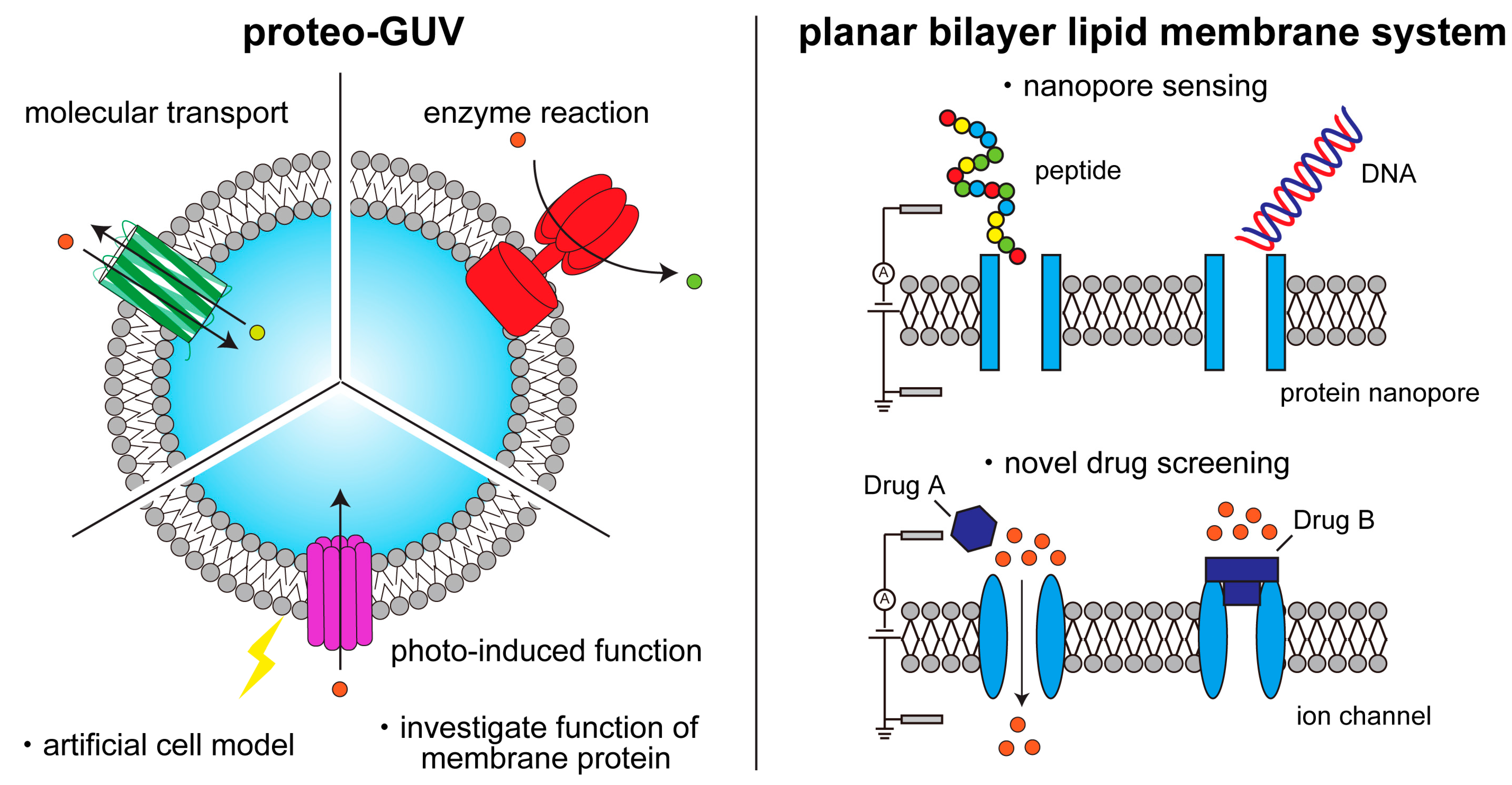
:1. Introduction
2. Formation of Cell-Sized Lipid Vesicles

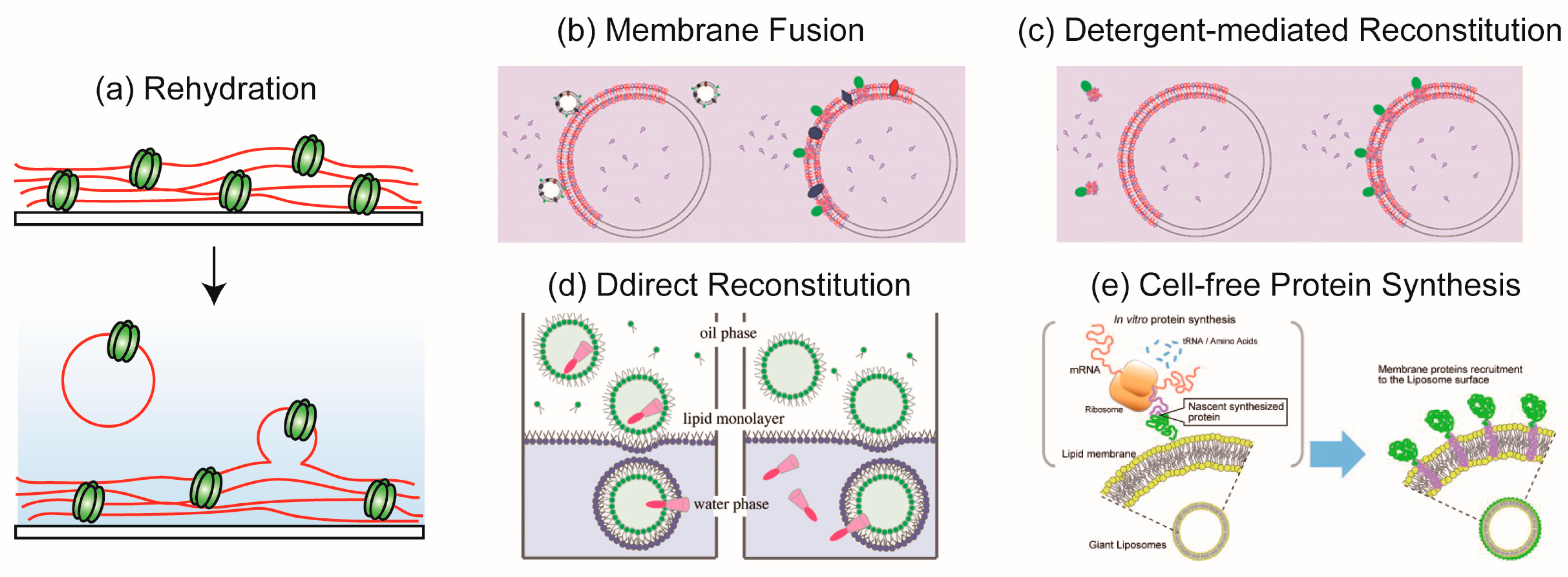
3. Methodology of Reconstitution of Membrane Proteins into Cell-Sized Artificial Membrane
3.1. Rehydration Method
3.2. Membrane Fusion Method
3.3. Detergent-Mediated Reconstitution Method
3.4. Direct Reconstitution Method
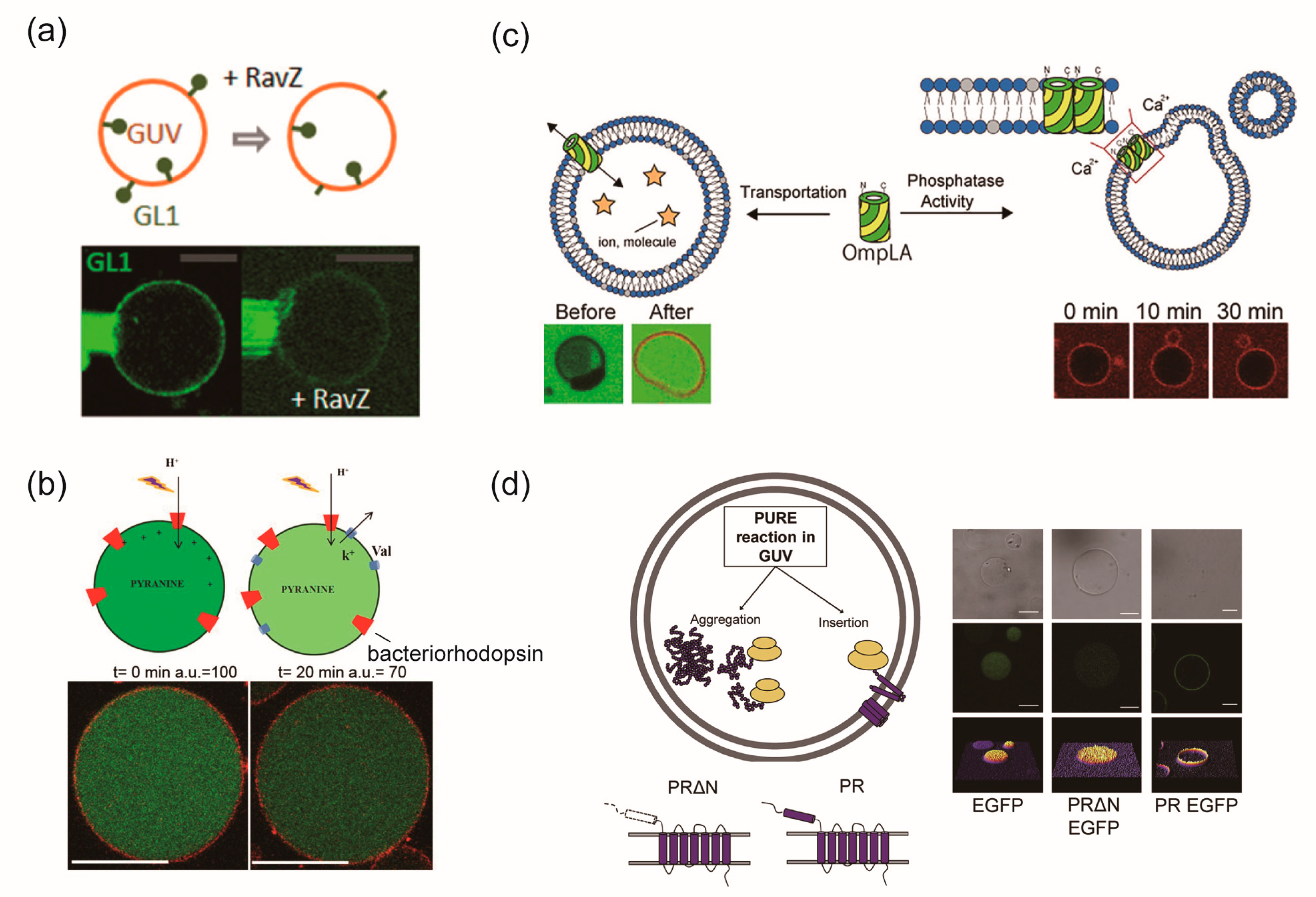
3.5. Reconstitution of Membrane Proteins Using the Cell-Free Protein Synthesis
3.6. Reconstitution of Membrane Proteins into Cell-Sized Vesicles Composed of Polymer or Proteins
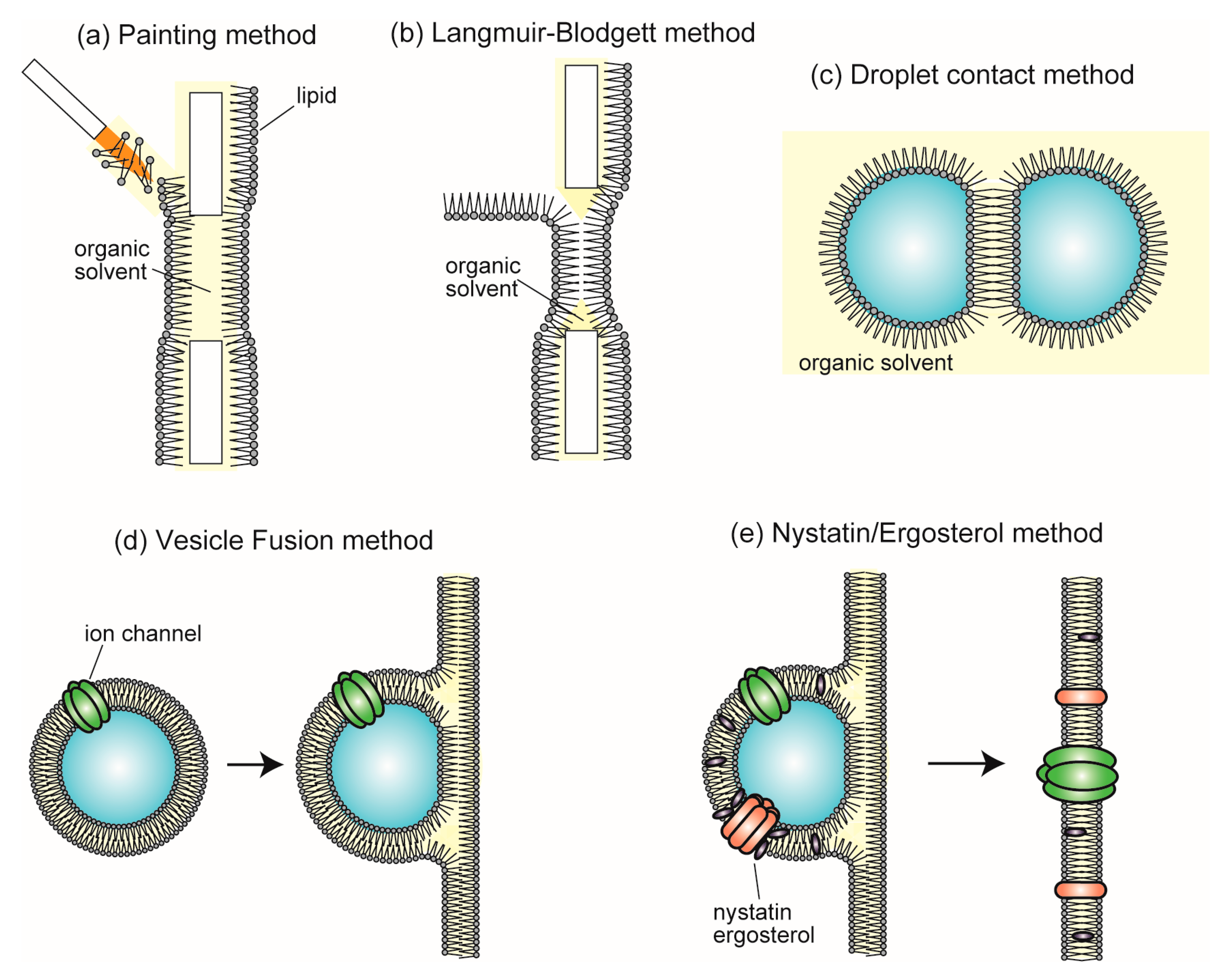
4. Single-Channel Recording Using Planar Bilayer Lipid Membrane Systems
4.1. Reconstitution of Ion Channel into BLMs
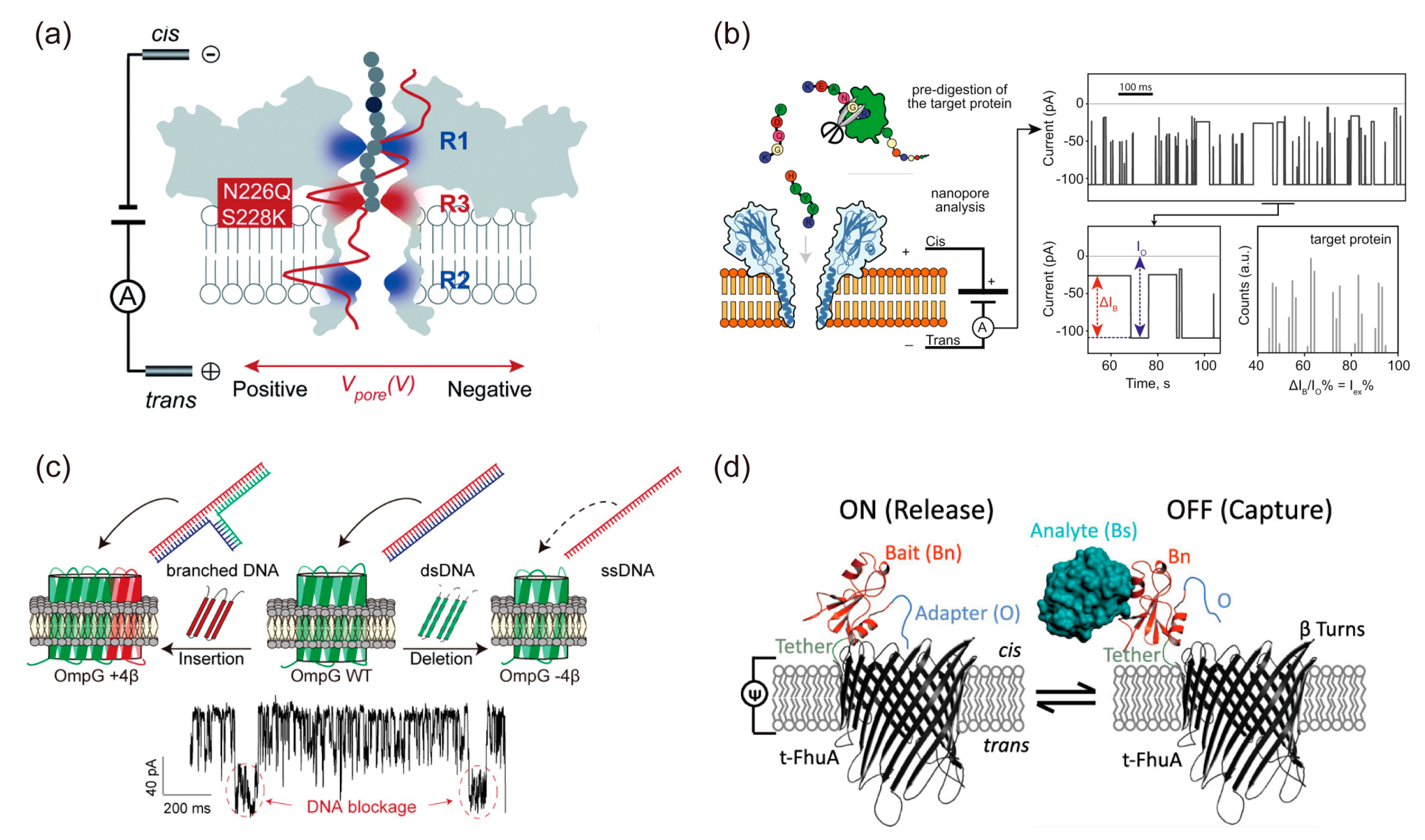
4.2. Protein Nanopore Analysis Using the BLM System
4.3. Ion Channel Analysis Using the BLM System
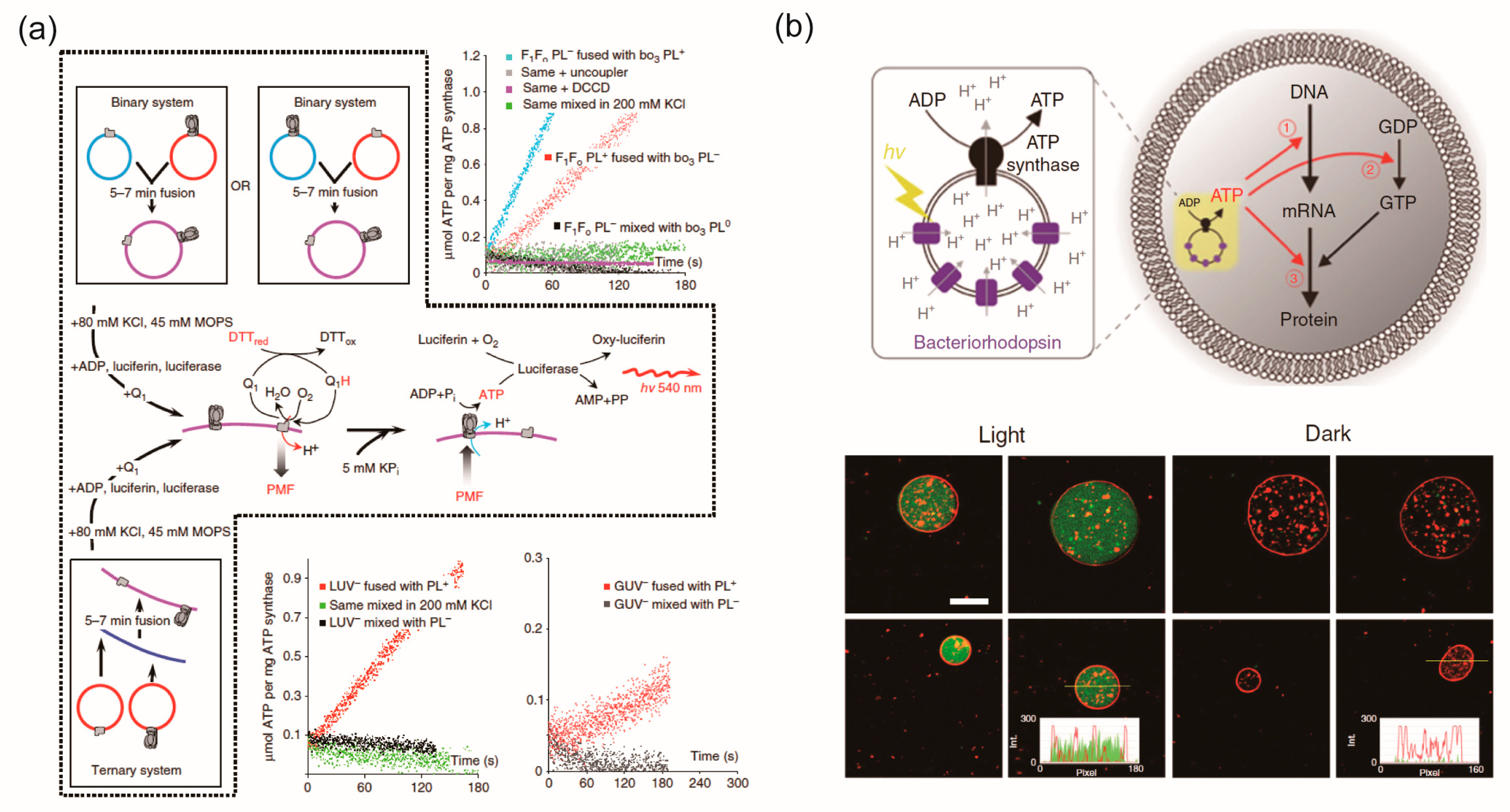
5. Lipid Vesicles Containing Some Types of Membrane Proteins for Creating Complex Artificial Cell Models
6. Conclusions and Future Direction
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Gilman, A.G. G proteins: Transducers of receptor-generated signals. Annu. Rev. Biochem. 1987, 56, 615–649. [Google Scholar] [CrossRef] [PubMed]
- Springer, T.A. Adhesion Receptors of the Immune System. Nature 1990, 346, 425–434. [Google Scholar] [CrossRef] [PubMed]
- Poitelon, Y.; Kopec, A.M.; Belin, S. Myelin Fat Facts: An Overview of Lipids and Fatty Acid Metabolism. Cells 2020, 9, 812. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schenk, P.W.; Snaar-Jagalska, B.E. Signal Perception and Transduction: The Role of Protein Kinases. Biochim. Biophys. Acta Mol. Cell Res. 1999, 1449, 1–24. [Google Scholar] [CrossRef] [Green Version]
- Martin, T.F.J. Phosphoinositide Lipids as Signaling Molecules: Common Themes for Signal Transduction, Cytoskeletal Regulation, and Membrane Trafficking. Annu. Rev. Cell Dev. Biol. 1998, 14, 231–264. [Google Scholar] [CrossRef]
- Qin, J.; Guo, Y.; Xue, B.; Shi, P.; Chen, Y.; Su, Q.P.; Hao, H.; Zhao, S.; Wu, C.; Yu, L.; et al. ER-Mitochondria Contacts Promote MtDNA Nucleoids Active Transportation via Mitochondrial Dynamic Tubulation. Nat. Commun. 2020, 11, 4471. [Google Scholar] [CrossRef]
- Li, L.; Conradson, D.M.; Bharat, V.; Kim, M.J.; Hsieh, C.H.; Minhas, P.S.; Papakyrikos, A.M.; Durairaj, A.S.; Ludlam, A.; Andreasson, K.I.; et al. A Mitochondrial Membrane-Bridging Machinery Mediates Signal Transduction of Intramitochondrial Oxidation. Nat. Metab. 2021, 3, 1242–1258. [Google Scholar] [CrossRef] [PubMed]
- Raff, M.C. Social Controls on Cell Survival and Cell Death. Nature 1992, 356, 397–400. [Google Scholar] [CrossRef]
- Zhang, Y.; Chen, X.; Gueydan, C.; Han, J. Plasma Membrane Changes during Programmed Cell Deaths. Cell Res. 2018, 28, 9–21. [Google Scholar] [CrossRef] [Green Version]
- Han, Y.; Liu, D.; Li, L. PD-1/PD-L1 Pathway: Current Researches in Cancer. Am. J. Cancer Res. 2020, 10, 727–742. [Google Scholar] [PubMed]
- Mizushima, N.; Klionsky, D.J. Protein Turnover via Autophagy: Implications for Metabolism. Annu. Rev. Nutr. 2007, 27, 19–40. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Congreve, M.; de Graaf, C.; Swain, N.A.; Tate, C.G. Impact of GPCR Structures on Drug Discovery. Cell 2020, 181, 81–91. [Google Scholar] [CrossRef] [PubMed]
- Davenport, A.P.; Scully, C.C.G.; de Graaf, C.; Brown, A.J.H.; Maguire, J.J. Advances in Therapeutic Peptides Targeting G Protein-Coupled Receptors. Nat. Rev. Drug Discov. 2020, 19, 389–413. [Google Scholar] [CrossRef] [PubMed]
- Lee, Y.; Warne, T.; Nehmé, R.; Pandey, S.; Dwivedi-Agnihotri, H.; Chaturvedi, M.; Edwards, P.C.; García-Nafría, J.; Leslie, A.G.W.; Shukla, A.K.; et al. Molecular Basis of β-Arrestin Coupling to Formoterol-Bound Β1-Adrenoceptor. Nature 2020, 583, 862–866. [Google Scholar] [CrossRef] [PubMed]
- Koivisto, A.P.; Belvisi, M.G.; Gaudet, R.; Szallasi, A. Advances in TRP Channel Drug Discovery: From Target Validation to Clinical Studies. Nat. Rev. Drug Discov. 2022, 21, 41–59. [Google Scholar] [CrossRef]
- Rosenbaum, M.I.; Clemmensen, L.S.; Bredt, D.S.; Bettler, B.; Strømgaard, K. Targeting Receptor Complexes: A New Dimension in Drug Discovery. Nat. Rev. Drug Discov. 2020, 19, 884–901. [Google Scholar] [CrossRef] [PubMed]
- He, S.; Moutaoufik, M.T.; Islam, S.; Persad, A.; Wu, A.; Aly, K.A.; Fonge, H.; Babu, M.; Cayabyab, F.S. HERG Channel and Cancer: A Mechanistic Review of Carcinogenic Processes and Therapeutic Potential. Biochim. Biophys. Acta Rev. Cancer 2020, 1873, 188355. [Google Scholar] [CrossRef] [PubMed]
- Pullman, M.E.; Penefsky, H.S.; Datta, A.; Racker, E. Partial Resolution of the Enzymes Catalyzing Oxidative Phosphorylation. J. Biol. Chem. 1960, 235, 3322–3329. [Google Scholar] [CrossRef] [PubMed]
- Racker, E. Reconstitution of a Calcium Pump with Phospholipids and a Purified Ca++—Adenosine Triphosphatase from Sacroplasmic Reticulum. J. Biol. Chem. 1972, 247, 8198–8200. [Google Scholar] [CrossRef]
- Keller, B.U.; Hedrich, R.; Vaz, W.L.C.; Criado, M. Single Channel Recordings of Reconstituted Ion Channel Proteins: An Improved Technique. Pflügers Arch. Eur. J. Physiol. 1988, 411, 94–100. [Google Scholar] [CrossRef]
- Ishmukhametov, R.R.; Russell, A.N.; Berry, R.M. A Modular Platform for One-Step Assembly of Multi-Component Membrane Systems by Fusion of Charged Proteoliposomes. Nat. Commun. 2016, 7, 13025. [Google Scholar] [CrossRef] [Green Version]
- Berhanu, S.; Ueda, T.; Kuruma, Y. Artificial Photosynthetic Cell Producing Energy for Protein Synthesis. Nat. Commun. 2019, 10, 1325. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lee, K.Y.; Park, S.J.; Lee, K.A.; Kim, S.H.; Kim, H.; Meroz, Y.; Mahadevan, L.; Jung, K.H.; Ahn, T.K.; Parker, K.K.; et al. Photosynthetic Artificial Organelles Sustain and Control ATP-Dependent Reactions in a Protocellular System. Nat. Biotechnol. 2018, 36, 530–535. [Google Scholar] [CrossRef] [PubMed]
- Seddon, A.M.; Curnow, P.; Booth, P.J. Membrane Proteins, Lipids and Detergents: Not Just a Soap Opera. Biochim. Biophys. Acta Biomembr. 2004, 1666, 105–117. [Google Scholar] [CrossRef] [Green Version]
- Denisov, I.G.; Sligar, S.G. Nanodiscs in Membrane Biochemistry and Biophysics. Chem. Rev. 2017, 117, 4669–4713. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dürr, U.H.N.; Gildenberg, M.; Ramamoorthy, A. The Magic of Bicelles Lights up Membrane Protein Structure. Chem. Rev. 2012, 112, 6054–6074. [Google Scholar] [CrossRef]
- Kim, H.J.; Howell, S.C.; Van Horn, W.D.; Jeon, Y.H.; Sanders, C.R. Recent Advances in the Application of Solution NMR Spectroscopy to Multi-Span Integral Membrane Proteins. Prog. Nucl. Magn. Reson. Spectrosc. 2009, 55, 335–360. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hong, M.; Zhang, Y.; Hu, F. Membrane Protein Structure and Dynamics from NMR Spectroscopy. Annu. Rev. Phys. Chem. 2012, 63, 1–24. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Binder, D.; Drepper, T.; Jaeger, K.E.; Delvigne, F.; Wiechert, W.; Kohlheyer, D.; Grünberger, A. Homogenizing Bacterial Cell Factories: Analysis and Engineering of Phenotypic Heterogeneity. Metab. Eng. 2017, 42, 145–156. [Google Scholar] [CrossRef] [PubMed]
- Frank, J. Single-Particle Imaging of Macromolecules by Cryo-Electron Microscopy. Annu. Rev. Biophys. Biomol. Struct. 2002, 31, 303–319. [Google Scholar] [CrossRef] [Green Version]
- Fernandez-Leiro, R.; Scheres, S.H.W. Unravelling Biological Macromolecules with Cryo-Electron Microscopy. Nature 2016, 537, 339–346. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hilgemann, D.W. Cytoplasmic ATP-Dependent Regulation of Ion Transporters and Channels: Mechanisms and Messengers. Annu. Rev. Physiol. 1997, 59, 193–220. [Google Scholar] [CrossRef]
- Darszon, A.; Labarca, P.; Nishigaki, T.; Espinosa, F. Ion Channels in Sperm Physiology. Physiol. Rev. 1999, 79, 481–510. [Google Scholar] [CrossRef]
- Wan, Y.; Zong, C.; Li, X.; Wang, A.; Li, Y.; Yang, T.; Bao, Q.; Dubow, M.; Yang, M.; Rodrigo, L.A.; et al. New Insights for Biosensing: Lessons from Microbial Defense Systems. Chem. Rev. 2021, 122, 8126–8180. [Google Scholar] [CrossRef]
- Misawa, N.; Osaki, T.; Takeuchi, S. Membrane Protein-Based Biosensors. J. R. Soc. Interface 2018, 15, 20170952. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Misawa, N.; Fujii, S.; Kamiya, K.; Osaki, T.; Takaku, T.; Takahashi, Y.; Takeuchi, S. Construction of a Biohybrid Odorant Sensor Using Biological Olfactory Receptors Embedded into Bilayer Lipid Membrane on a Chip. ACS Sens. 2019, 4, 711–716. [Google Scholar] [CrossRef]
- Yamada, T.; Sugiura, H.; Mimura, H.; Kamiya, K.; Osaki, T.; Takeuchi, S. Highly Sensitive VOC Detectors Using Insect Olfactory Receptors Reconstituted into Lipid Bilayers. Sci. Adv. 2021, 7, 2013–2026. [Google Scholar] [CrossRef]
- Bangham, A.D.; Horne, R.W. Negative Staining of Phospholipids and Their Structural Modification by Surface-Active Agents as Observed in the Electron Microscope. J. Mol. Biol. 1964, 8, 660–668. [Google Scholar] [CrossRef]
- Akashi, K.I.; Miyata, H.; Itoh, H.; Kinosita, K. Preparation of Giant Liposomes in Physiological Conditions and Their Characterization under an Optical Microscope. Biophys. J. 1996, 71, 3242–3250. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Horger, K.S.; Estes, D.J.; Capone, R.; Mayer, M. Films of Agarose Enable Rapid Formation of Giant Liposomes in Solutions of Physiologic Ionic Strength. J. Am. Chem. Soc. 2009, 131, 1810–1819. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lira, R.B.; Dimova, R.; Riske, K.A. Giant Unilamellar Vesicles Formed by Hybrid Films of Agarose and Lipids Display Altered Mechanical Properties. Biophys. J. 2014, 107, 1609–1619. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Angelova, M.I.; Dimitrov, D.S. Liposome Electroformation. Faraday Discuss. Chem. Soc. 1986, 81, 303–311. [Google Scholar] [CrossRef]
- Dimitrov, D.S.; Angelova, M.I. Lipid Swelling and Liposome Formation Mediated by Electric Fields. Bioelectrochem. Bioenerg. 1988, 19, 323–336. [Google Scholar] [CrossRef]
- Pautot, S.; Frisken, B.J.; Weitz, D.A. Production of Unilamellar Vesicles Using an Inverted Emulsion. Langmuir 2003, 19, 2870–2879. [Google Scholar] [CrossRef]
- Pautot, S.; Frisken, B.J.; Weitz, D.A. Engineering Asymmetric Vesicles. Proc. Natl. Acad. Sci. USA 2003, 100, 10718–10721. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tan, Y.C.; Hettiarachchi, K.; Siu, M.; Pan, Y.R.; Lee, A.P. Controlled Microfluidic Encapsulation of Cells, Proteins, and Microbeads in Lipid Vesicles. J. Am. Chem. Soc. 2006, 128, 5656–5658. [Google Scholar] [CrossRef]
- Hu, P.C.; Li, S.; Malmstadt, N. Microfluidic Fabrication of Asymmetric Giant Lipid Vesicles. ACS Appl. Mater. Interfaces 2011, 3, 1434–1440. [Google Scholar] [CrossRef] [Green Version]
- Matosevic, S.; Paegel, B.M. Stepwise Synthesis of Giant Unilamellar Vesicles on a Microfluidic Assembly Line. J. Am. Chem. Soc. 2011, 133, 2798–2800. [Google Scholar] [CrossRef] [PubMed]
- Abkarian, M.; Loiseau, E.; Massiera, G. Continuous Droplet Interface Crossing Encapsulation (CDICE) for High Throughput Monodisperse Vesicle Design. Soft Matter 2011, 7, 4610–4614. [Google Scholar] [CrossRef]
- Karamdad, K.; Law, R.V.; Seddon, J.M.; Brooks, N.J.; Ces, O. Preparation and Mechanical Characterisation of Giant Unilamellar Vesicles by a Microfluidic Method. Lab Chip 2015, 15, 557–562. [Google Scholar] [CrossRef] [Green Version]
- Karamdad, K.; Law, R.V.; Seddon, J.M.; Brooks, N.J.; Ces, O. Studying the Effects of Asymmetry on the Bending Rigidity of Lipid Membranes Formed by Microfluidics. Chem. Commun. 2016, 52, 5277–5280. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Blosser, M.C.; Horst, B.G.; Keller, S.L. CDICE Method Produces Giant Lipid Vesicles under Physiological Conditions of Charged Lipids and Ionic Solutions. Soft Matter 2016, 12, 7364–7371. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Romanov, V.; McCullough, J.; Gale, B.K.; Frost, A. A Tunable Microfluidic Device Enables Cargo Encapsulation by Cell- or Organelle-Sized Lipid Vesicles Comprising Asymmetric Lipid Bilayers. Adv. Biosyst. 2019, 3, 1900010. [Google Scholar] [CrossRef] [PubMed]
- Shum, H.C.; Lee, D.; Yoon, I.; Kodger, T.; Weitz, D.A. Double Emulsion Templated Monodisperse Phospholipid Vesicles. Langmuir 2008, 24, 7651–7653. [Google Scholar] [CrossRef] [PubMed]
- Arriaga, L.R.; Datta, S.S.; Kim, S.H.; Amstad, E.; Kodger, T.E.; Monroy, F.; Weitz, D.A. Ultrathin Shell Double Emulsion Templated Giant Unilamellar Lipid Vesicles with Controlled Microdomain Formation. Small 2014, 10, 950–956. [Google Scholar] [CrossRef]
- Kim, S.H.; Kim, J.W.; Cho, J.C.; Weitz, D.A. Double-Emulsion Drops with Ultra-Thin Shells for Capsule Templates. Lab Chip 2011, 11, 3162–3166. [Google Scholar] [CrossRef]
- Caschera, F.; Lee, J.W.; Ho, K.K.Y.; Liu, A.P.; Jewett, M.C. Cell-Free Compartmentalized Protein Synthesis inside Double Emulsion Templated Liposomes with In Vitro Synthesized and Assembled Ribosomes. Chem. Commun. 2016, 52, 5467–5469. [Google Scholar] [CrossRef] [Green Version]
- Deshpande, S.; Caspi, Y.; Meijering, A.E.C.; Dekker, C. Octanol-Assisted Liposome Assembly on Chip. Nat. Commun. 2016, 7, 10447. [Google Scholar] [CrossRef] [Green Version]
- Deng, N.-N.; Yelleswarapu, M.; Huck, W.T.S. Monodisperse Uni- and Multicompartment Liposomes. J. Am. Chem. Soc. 2016, 138, 7584–7591. [Google Scholar] [CrossRef] [Green Version]
- Deng, N.; Huck, W.T.S. Microfluidic Formation of Monodisperse Coacervate Organelles in Liposomes. Angew. Chem. Int. Ed. Engl. 2017, 56, 9736–9740. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Michelon, M.; Huang, Y.; de la Torre, L.G.; Weitz, D.A.; Cunha, R.L. Single-Step Microfluidic Production of W/O/W Double Emulsions as Templates for β-Carotene-Loaded Giant Liposomes Formation. Chem. Eng. J. 2019, 366, 27–32. [Google Scholar] [CrossRef]
- Ushiyama, R.; Koiwai, K.; Suzuki, H. Plug-and-Play Microfluidic Production of Monodisperse Giant Unilamellar Vesicles Using Droplet Transfer across Water–Oil Interface. Sens. Actuators B Chem. 2022, 355, 131281. [Google Scholar] [CrossRef]
- Funakoshi, K.; Suzuki, H.; Takeuchi, S. Lipid Bilayer Formation by Contacting Monolayers in a Microfluidic Device for Membrane Protein Analysis. Anal. Chem. 2006, 78, 8169–8174. [Google Scholar] [CrossRef]
- Kamiya, K.; Kawano, R.; Osaki, T.; Akiyoshi, K.; Takeuchi, S. Cell-Sized Asymmetric Lipid Vesicles Facilitate the Investigation of Asymmetric Membranes. Nat. Chem. 2016, 8, 881–889. [Google Scholar] [CrossRef] [PubMed]
- Kamiya, K.; Arisaka, C.; Suzuki, M. Investigation of Fusion between Nanosized Lipid Vesicles and a Lipid Monolayer toward Formation of Giant Lipid Vesicles with Various Kinds of Biomolecules. Micromachines 2021, 12, 133. [Google Scholar] [CrossRef]
- Kamiya, K.; Osaki, T.; Takeuchi, S. Formation of Nano-Sized Lipid Vesicles with Asymmetric Lipid Components Using a Pulsed-Jet Flow Method. Sens. Actuators B Chem. 2021, 327, 128917. [Google Scholar] [CrossRef]
- Gotanda, M.; Kamiya, K.; Osaki, T.; Fujii, S.; Misawa, N.; Miki, N.; Takeuchi, S. Sequential Generation of Asymmetric Lipid Vesicles Using a Pulsed-Jetting Method in Rotational Wells. Sens. Actuators B Chem. 2018, 261, 392–397. [Google Scholar] [CrossRef]
- Gotanda, M.; Kamiya, K.; Osaki, T.; Miki, N.; Takeuchi, S. Automatic Generation System of Cell-Sized Liposomes. Sens. Actuators B Chem. 2019, 292, 57–63. [Google Scholar] [CrossRef]
- Kamiya, K.; Osaki, T.; Takeuchi, S. Formation of Vesicles-in-a-Vesicle with Asymmetric Lipid Components Using a Pulsed-Jet Flow Method. RSC Adv. 2019, 9, 30071–30075. [Google Scholar] [CrossRef] [Green Version]
- Elani, Y.; Law, R.V.; Ces, O. Vesicle-Based Artificial Cells as Chemical Microreactors with Spatially Segregated Reaction Pathways. Nat. Commun. 2014, 5, 5305. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Miwa, A.; Kamiya, K. Control of Enzyme Reaction Initiation inside Giant Unilamellar Vesicles by the Cell-Penetrating Peptide-Mediated Translocation of Cargo Proteins. ACS Synth. Biol. 2022, 11, 3836–3846. [Google Scholar] [CrossRef]
- Kamiya, K. Development of Artificial Cell Models Using Microfluidic Technology and Synthetic Biology. Micromachines 2020, 11, 559. [Google Scholar] [CrossRef] [PubMed]
- Kamiya, K.; Takeuchi, S. Giant Liposome Formation toward the Synthesis of Well-Defined Artificial Cells. J. Mater. Chem. B 2017, 5, 5911–5923. [Google Scholar] [CrossRef] [PubMed]
- Dezi, M.; Di Cicco, A.; Bassereau, P.; Lévy, D. Detergent-Mediated Incorporation of Transmembrane Proteins in Giant Unilamellar Vesicles with Controlled Physiological Contents. Proc. Natl. Acad. Sci. USA 2013, 110, 7276–7281. [Google Scholar] [CrossRef] [Green Version]
- Fenz, S.F.; Sachse, R.; Schmidt, T.; Kubick, S. Cell-Free Synthesis of Membrane Proteins: Tailored Cell Models out of Microsomes. Biochim. Biophys. Acta Biomembr. 2014, 1838, 1382–1388. [Google Scholar] [CrossRef] [PubMed]
- Suzuki, M.; Kamiya, K. Cell-Sized Asymmetric Phospholipid-Amphiphilic Protein Vesicles with Growth, Fission, and Molecule Transportation. iScience 2023, 26, 106086. [Google Scholar] [CrossRef]
- Ohnishi, S.; Kamiya, K. Formation of Giant Lipid Vesicle Containing Dual Functions Facilitates Outer Membrane Phospholipase. ACS Synth. Biol. 2021, 10, 1837–1846. [Google Scholar] [CrossRef] [PubMed]
- Eaglesfield, R.; Madsen, M.A.; Sanyal, S.; Reboud, J.; Amtmann, A. Cotranslational Recruitment of Ribosomes in Protocells Recreates a Translocon-Independent Mechanism of Proteorhodopsin Biogenesis. iScience 2021, 24, 102429. [Google Scholar] [CrossRef]
- Altamura, E.; Milano, F.; Tangorra, R.R.; Trotta, M.; Omar, O.H.; Stano, P.; Mavelli, F. Highly Oriented Photosynthetic Reaction Centers Generate a Proton Gradient in Synthetic Protocells. Proc. Natl. Acad. Sci. USA 2017, 114, 3837–3842. [Google Scholar] [CrossRef] [Green Version]
- Kajii, K.; Shimomura, A.; Higashide, M.T.; Oki, M.; Tsuji, G. Effects of Sugars on Giant Unilamellar Vesicle Preparation, Fusion, PCR in Liposomes, and Pore Formation. Langmuir 2022, 38, 8871–8880. [Google Scholar] [CrossRef] [PubMed]
- Diederichs, T.; Tampé, R. Single Cell-like Systems Reveal Active Unidirectional and Light-Controlled Transport by Nanomachineries. ACS Nano 2021, 15, 6747–6755. [Google Scholar] [CrossRef] [PubMed]
- Varnier, A.; Kermarrec, F.; Blesneac, I.; Moreau, C.; Liguori, L.; Lenormand, J.L.; Picollet-D’Hahan, N. A Simple Method for the Reconstitution of Membrane Proteins into Giant Unilamellar Vesicles. J. Membr. Biol. 2010, 233, 85–92. [Google Scholar] [CrossRef] [PubMed]
- Soga, H.; Fujii, S.; Yomo, T.; Kato, Y.; Watanabe, H.; Matsuura, T. In Vitro Membrane Protein Synthesis inside Cell-Sized Vesicles Reveals the Dependence of Membrane Protein Integration on Vesicle Volume. ACS Synth. Biol. 2014, 3, 372–379. [Google Scholar] [CrossRef] [PubMed]
- Almendro-Vedia, V.G.; Natale, P.; Mell, M.; Bonneau, S.; Monroy, F.; Joubert, F.; López-Montero, I. Nonequilibrium Fluctuations of Lipid Membranes by the Rotating Motor Protein F1F0-ATP Synthase. Proc. Natl. Acad. Sci. USA 2017, 114, 11291–11296. [Google Scholar] [CrossRef] [Green Version]
- Motta, I.; Gohlke, A.; Adrien, V.; Li, F.; Gardavot, H.; Rothman, J.E.; Pincet, F. Formation of Giant Unilamellar Proteo-Liposomes by Osmotic Shock. Langmuir 2015, 31, 7091–7099. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jang, H.S.; Cho, Y.K.; Granick, S. Biologically-Active Unilamellar Vesicles from Red Blood Cells. Biomater. Sci. 2019, 7, 1393–1398. [Google Scholar] [CrossRef] [PubMed]
- Guo, X.; Steinkühler, J.; Marin, M.; Li, X.; Lu, W.; Dimova, R.; Melikyan, G.B. Interferon-Induced Transmembrane Protein 3 Blocks Fusion of Diverse Enveloped Viruses by Altering Mechanical Properties of Cell Membranes. ACS Nano 2021, 15, 8155–8170. [Google Scholar] [CrossRef] [PubMed]
- Yanagisawa, M.; Iwamoto, M.; Kato, A.; Yoshikawa, K.; Oiki, S. Oriented Reconstitution of a Membrane Protein in a Giant Unilamellar Vesicle: Experimental Verification with the Potassium Channel KcsA. J. Am. Chem. Soc. 2011, 133, 11774–11779. [Google Scholar] [CrossRef]
- Aimon, S.; Manzi, J.; Schmidt, D.; Larrosa, J.A.P.; Bassereau, P.; Toombes, G.E.S. Functional Reconstitution of a Voltage-Gated Potassium Channel in Giant Unilamellar Vesicles. PLoS ONE 2011, 6, 25529. [Google Scholar] [CrossRef]
- dos Santos, E.C.; Belluati, A.; Necula, D.; Scherrer, D.; Meyer, C.E.; Wehr, R.P.; Lörtscher, E.; Palivan, C.G.; Meier, W. Combinatorial Strategy for Studying Biochemical Pathways in Double Emulsion Templated Cell-Sized Compartments. Adv. Mater. 2020, 32, 2004804. [Google Scholar] [CrossRef]
- Darszon, A.; Vandenberg, C.A.; Schönfeld, M.; Ellisman, M.H.; Spitzer, N.C.; Montal, M. Reassembly of Protein-Lipid Complexes into Large Bilayer Vesicles: Perspectives for Membrane Reconstitution. Proc. Natl. Acad. Sci. USA 1980, 77, 239–243. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Girard, P.; Pécréaux, J.; Lenoir, G.; Falson, P.; Rigaud, J.L.; Bassereau, P. A New Method for the Reconstitution of Membrane Proteins into Giant Unilamellar Vesicles. Biophys. J. 2004, 87, 419–429. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dolder, N.; Müller, P.; von Ballmoos, C. Experimental Platform for the Functional Investigation of Membrane Proteins in Giant Unilamellar Vesicles. Soft Matter 2022, 18, 5877–5893. [Google Scholar] [CrossRef]
- Kahya, N.; Pécheur, E.-I.; de Boeij, W.P.; Wiersma, D.A.; Hoekstra, D. Reconstitution of Membrane Proteins into Giant Unilamellar Vesicles via Peptide-Induced Fusion. Biophys. J. 2001, 81, 1464–1474. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pécheur, E.I.; Hoekstra, D.; Sainte-Marie, J.; Maurin, L.; Bienvenüe, A.; Philippot, J.R. Membrane Anchorage Brings about Fusogenic Properties in a Short Synthetic Peptide. Biochemistry 1997, 36, 3773–3781. [Google Scholar] [CrossRef]
- Martin, I.; Pécheur, E.I.; Ruysschaert, J.M.; Hoekstra, D. Membrane Fusion Induced by a Short Fusogenic Peptide Is Assessed by Its Insertion and Orientation into Target Bilayers. Biochemistry 1999, 38, 9337–9347. [Google Scholar] [CrossRef]
- Kragh-Hansen, U.; le Maire, M.; Møller, J.V. The Mechanism of Detergent Solubilization of Liposomes and Protein-Containing Membranes. Biophys. J. 1998, 75, 2932–2946. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Paternostre, M.-T.; Roux, M.; Rigaud, J. louis Mechanisms of Membrane Protein Insertion into Liposomes during Reconstitution Procedures Involving the Use of Detergents. 1. Solubilization of Large Unilamellar Liposomes (Prepared by Reverse-Phase Evaporation) by Triton X-100, Octyl Glucoside, and Sodium. Biochemistry 1988, 27, 2668–2677. [Google Scholar] [CrossRef] [PubMed]
- Rigaud, J.L.; Paternostre, M.T.; Bluzat, A. Mechanisms of Membrane Protein Insertion into Liposomes during Reconstitution Procedures Involving the Use of Detergents. 2. Incorporation of the Light-Driven Proton Pump Bacteriorhodopsint. Biochemistry 1988, 27, 2677–2688. [Google Scholar] [CrossRef]
- Urbaneja, M.-A.; Goñi, F.M.; Alonso, A. Structural Changes Induced by Triton X-100 on Sonicated Phosphatidylcholine Liposomes. Eur. J. Biochem. 1988, 173, 585–588. [Google Scholar] [CrossRef] [PubMed]
- Kreir, M.; Farre, C.; Beckler, M.; George, M.; Fertig, N. Rapid Screening of Membrane Protein Activity: Electrophysiological Analysis of OmpF Reconstituted in Proteoliposomes. Lab Chip 2008, 8, 587–595. [Google Scholar] [CrossRef] [PubMed]
- Battle, A.R.; Petrov, E.; Pal, P.; Martinac, B. Rapid and Improved Reconstitution of Bacterial Mechanosensitive Ion Channel Proteins MscS and MscL into Liposomes Using a Modified Sucrose Method. FEBS Lett. 2009, 583, 407–412. [Google Scholar] [CrossRef]
- Jørgensen, I.L.; Kemmer, G.C.; Pomorski, T.G. Membrane Protein Reconstitution into Giant Unilamellar Vesicles: A Review on Current Techniques. Eur. Biophys. J. 2017, 46, 103–119. [Google Scholar] [CrossRef] [PubMed]
- Berrier, C.; Park, K.H.; Abes, S.; Bibonne, A.; Betton, J.M.; Ghazi, A. Cell-Free Synthesis of a Functional Ion Channel in the Absence of a Membrane and in the Presence of Detergent. Biochemistry 2004, 43, 12585–12591. [Google Scholar] [CrossRef] [PubMed]
- Nomura, S.-i.M.; Kondoh, S.; Asayama, W.; Asada, A.; Nishikawa, S.; Akiyoshi, K. Direct Preparation of Giant Proteo-Liposomes by In Vitro Membrane Protein Synthesis. J. Biotechnol. 2008, 133, 190–195. [Google Scholar] [CrossRef]
- Spirin, A.S.; Baranov, V.I.; Ryabova, L.A.; Ovodov, S.Y.; Alakhov, Y.B. A Continuous Cell-Free Translation System Capable of Producing Polypeptides in High Yield. Science 1988, 242, 1162–1164. [Google Scholar] [CrossRef]
- Betton, J. Rapid Translation System (RTS): A Promising Alternative for Recombinant Protein Production. Curr. Protein Pept. Sci. 2005, 4, 73–80. [Google Scholar] [CrossRef]
- Berrier, C.; Besnard, M.; Ajouz, B.; Coulombe, A.; Ghazi, A. Multiple Mechanosensitive Ion Channels from Escherichia Coli, Activated at Different Thresholds of Applied Pressure. J. Membr. Biol. 1996, 151, 175–187. [Google Scholar] [CrossRef]
- Sukharev, S.I.; Sigurdson, W.J.; Kung, C.; Sachs, F. Energetic and Spatial Parameters for Gating of the Bacterial Large Conductance Mechanosensitive Channel, MscL. J. Gen. Physiol. 1999, 113, 525–539. [Google Scholar] [CrossRef] [Green Version]
- Sawasaki, T.; Hasegawa, Y.; Tsuchimochi, M.; Kamura, N.; Ogasawara, T.; Kuroita, T.; Endo, Y. A Bilayer Cell-Free Protein Synthesis System for High-Throughput Screening of Gene Products. In Proceedings of the FEBS Letters; John Wiley & Sons, Ltd.: Hoboken, NJ, USA, 6 March 2002; Volume 514, pp. 102–105. [Google Scholar]
- Shimizu, Y.; Inoue, A.; Tomari, Y.; Suzuki, T.; Yokogawa, T.; Nishikawa, K.; Ueda, T. Cell-Free Translation Reconstituted with Purified Components. Nat. Biotechnol. 2001, 19, 751–755. [Google Scholar] [CrossRef] [PubMed]
- Garni, M.; Einfalt, T.; Goers, R.; Palivan, C.G.; Meier, W. Live Follow-Up of Enzymatic Reactions Inside the Cavities of Synthetic Giant Unilamellar Vesicles Equipped with Membrane Proteins Mimicking Cell Architecture. ACS Synth. Biol. 2018, 7, 2116–2125. [Google Scholar] [CrossRef]
- Marušič, N.; Otrin, L.; Zhao, Z.; Lira, R.B.; Kyrilis, F.L.; Hamdi, F.; Kastritis, P.L.; Vidakovic-Koch, T.; Ivanov, I.; Sundmacher, K.; et al. Constructing Artificial Respiratory Chain in Polymer Compartments: Insights into the Interplay between Bo3 Oxidase and the Membrane. Proc. Natl. Acad. Sci. USA 2020, 117, 15006–15017. [Google Scholar] [CrossRef]
- Neher, E.; Sakmann, B. Single-Channel Currents Recorded from Membrane of Denervated Frog Muscle Fibres. Nature 1976, 260, 799–802. [Google Scholar] [CrossRef]
- Hirano-Iwata, A.; Ishinari, Y.; Yamamoto, H.; Niwano, M. Micro- and Nano-Technologies for Lipid Bilayer-Based Ion-Channel Functional Assays. Chem. Asian J. 2015, 10, 1266–1274. [Google Scholar] [CrossRef] [PubMed]
- Mueller, P.; Rudin, D.O.; Ti Tien, H.; Wescott, W.C. Reconstitution of Cell Membrane Structure In Vitro and Its Transformation into an Excitable System. Nature 1962, 194, 979–980. [Google Scholar] [CrossRef]
- Montal, M.; Mueller, P. Formation of Bimolecular Membranes from Lipid Monolayers and a Study of Their Electrical Properties. Proc. Natl. Acad. Sci. USA 1972, 69, 3561–3566. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rokitskaya, T.I.; Sorochkina, A.I.; Kovalchuk, S.I.; Egorova, N.S.; Kotova, E.A.; Sychev, S.V.; Antonenko, Y.N. The PH-Dependent Induction of Lipid Membrane Ionic Permeability by N-Terminally Lysine-Substituted Analogs of Gramicidin A. Eur. Biophys. J. 2012, 41, 129–138. [Google Scholar] [CrossRef]
- Harriss, L.M.; Cronin, B.; Thompson, J.R.; Wallace, M.I. Imaging Multiple Conductance States in an Alamethicin Pore. J. Am. Chem. Soc. 2011, 133, 14507–14509. [Google Scholar] [CrossRef]
- Butler, T.Z.; Gundlach, J.H.; Troll, M. Ionic Current Blockades from DNA and RNA Molecules in the α-Hemolysin Nanopore. Biophys. J. 2007, 93, 3229–3240. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Baslé, A.; Iyer, R.; Delcour, A.H. Subconductance States in OmpF Gating. Biochim. Biophys. Acta Biomembr. 2004, 1664, 100–107. [Google Scholar] [CrossRef] [Green Version]
- Vaisey, G.; Long, S.B. An Allosteric Mechanism of Inactivation in the Calcium-Dependent Chloride Channel BEST1. J. Gen. Physiol. 2018, 150, 1484–1497. [Google Scholar] [CrossRef] [Green Version]
- Maher, J.; Allen, M. Planar Lipid Bilayers in Recombinant Ion Channel Research. Methods 2018, 147, 206–212. [Google Scholar] [CrossRef] [PubMed]
- Montal, M. Reconstitution of Channel Proteins from Excitable Cells in Planar Lipid Bilayer Membranes. J. Membr. Biol. 1987, 98, 101–115. [Google Scholar] [CrossRef] [PubMed]
- Cohen, F.S.; Niles, W.D. [5] Reconstituting Channels into Planar Membranes: A Conceptual Framework and Methods for Fusing Vesicles to Planar Bilayer Phospholipid Membranes. Methods Enzymol. 1993, 220, 50–68. [Google Scholar] [CrossRef] [PubMed]
- Cohen, F.S.; Akabas, M.H.; Zimmerberg, J.; Finkelstein, A. Parameters Affecting the Fusion of Unilamellar Phospholipid Vesicles within Planar Bilayer Membranes. J. Cell Biol. 1984, 98, 1054–1062. [Google Scholar] [CrossRef] [Green Version]
- Woodbury, D.J.; Miller, C. Nystatin-Induced Liposome Fusion. A Versatile Approach to Ion Channel Reconstitution into Planar Bilayers. Biophys. J. 1990, 58, 833–839. [Google Scholar] [CrossRef] [Green Version]
- Woodbury, D.J.; Hall, J.E. Role of Channels in the Fusion of Vesicles with a Planar Bilayer. Biophys. J. 1988, 54, 1053–1063. [Google Scholar] [CrossRef] [Green Version]
- Woodbury, D.J. Nystatin/Ergosterol Method for Reconstituting Ion Channels into Planar Lipid Bilayers. Methods Enzymol. 1999, 294, 319–339. [Google Scholar] [CrossRef]
- Pham, B.; Chisholm, C.M.; Foster, J.; Friis, E.; Fahie, M.A.; Chen, M. A PH-Independent Quiet OmpG Pore with Enhanced Electrostatic Repulsion among the Extracellular Loops. Biochim. Biophys. Acta Biomembr. 2021, 1863, 183485. [Google Scholar] [CrossRef] [PubMed]
- Perez-Rathke, A.; Fahie, M.A.; Chisholm, C.; Liang, J.; Chen, M. Mechanism of OmpG PH-Dependent Gating from Loop Ensemble and Single Channel Studies. J. Am. Chem. Soc. 2018, 140, 1105–1115. [Google Scholar] [CrossRef]
- Schmitt, C.; Bafna, J.A.; Schmid, B.; Klingl, S.; Baier, S.; Hemmis, B.; Wagner, R.; Winterhalter, M.; Voll, L.M. Manipulation of Charge Distribution in the Arginine and Glutamate Clusters of the OmpG Pore Alters Sugar Specificity and Ion Selectivity. Biochim. Biophys. Acta Biomembr. 2019, 1861, 183021. [Google Scholar] [CrossRef]
- Liang, B.; Tamm, L.K. Structure of Outer Membrane Protein G by Solution NMR Spectroscopy. Proc. Natl. Acad. Sci. USA 2007, 104, 16140–16145. [Google Scholar] [CrossRef] [Green Version]
- Subbarao, G.V.; van den Berg, B. Crystal Structure of the Monomeric Porin OmpG. J. Mol. Biol. 2006, 360, 750–759. [Google Scholar] [CrossRef]
- Sanganna Gari, R.R.; Seelheim, P.; Liang, B.; Tamm, L.K. Quiet Outer Membrane Protein G (OmpG) Nanopore for Biosensing. ACS Sens. 2019, 4, 1230–1235. [Google Scholar] [CrossRef] [PubMed]
- Yildiz, Ö.; Vinothkumar, K.R.; Goswami, P.; Kühlbrandt, W. Structure of the Monomeric Outer-Membrane Porin OmpG in the Open and Closed Conformation. EMBO J. 2006, 25, 3702–3713. [Google Scholar] [CrossRef]
- Chen, M.; Khalid, S.; Sansom, M.S.P.; Bayley, H. Outer Membrane Protein G: Engineering a Quiet Pore for Biosensing. Proc. Natl. Acad. Sci. USA 2008, 105, 6272–6277. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kamiya, K. Formation and Function of OmpG or OmpA-Incorporated Liposomes Using an In Vitro Translation System. Sci. Rep. 2022, 12, 2376. [Google Scholar] [CrossRef] [PubMed]
- Charbit, A.; Andersen, C.; Wang, J.; Schiffler, B.; Michel, V.; Benz, R.; Hofnung, M. In Vivo and In Vitro Studies of Transmembrane Beta-Strand Deletion, Insertion or Substitution Mutants of the Escherichia Coli K-12 Maltoporin. Mol. Microbiol. 2000, 35, 777–790. [Google Scholar] [CrossRef] [Green Version]
- Liu, Z.; Ghai, I.; Winterhalter, M.; Schwaneberg, U. Engineering Enhanced Pore Sizes Using FhuA Δ1-160 from E. Coli Outer Membrane as Template. ACS Sens. 2017, 2, 1619–1626. [Google Scholar] [CrossRef]
- Arnold, T.; Poynor, M.; Nussberger, S.; Lupas, A.N.; Linke, D. Gene Duplication of the Eight-Stranded β-Barrel OmpX Produces a Functional Pore: A Scenario for the Evolution of Transmembrane β-Barrels. J. Mol. Biol. 2007, 366, 1174–1184. [Google Scholar] [CrossRef] [PubMed]
- Tosaka, T.; Kamiya, K. Modified Outer Membrane Protein-G Nanopores with Expanded and Truncated β-Hairpins for Recognition of Double-Stranded DNA. ACS Appl. Nano Mater. 2022, 5, 6149–6158. [Google Scholar] [CrossRef]
- Shimizu, K.; Mijiddorj, B.; Usami, M.; Mizoguchi, I.; Yoshida, S.; Akayama, S.; Hamada, Y.; Ohyama, A.; Usui, K.; Kawamura, I.; et al. De Novo Design of a Nanopore for Single-Molecule Detection That Incorporates a β-Hairpin Peptide. Nat. Nanotechnol. 2022, 17, 67–75. [Google Scholar] [CrossRef]
- Kasianowicz, J.J.; Brandin, E.; Branton, D.; Deamer, D.W. Characterization of Individual Polynucleotide Molecules Using a Membrane Channel. Proc. Natl. Acad. Sci. USA 1996, 93, 13770–13773. [Google Scholar] [CrossRef] [Green Version]
- Niu, H.; Li, M.Y.; Ying, Y.L.; Long, Y.T. An Engineered Third Electrostatic Constriction of Aerolysin to Manipulate Heterogeneously Charged Peptide Transport. Chem. Sci. 2022, 13, 2456–2461. [Google Scholar] [CrossRef]
- Yuan, B.; Li, S.; Ying, Y.L.; Long, Y.T. The Analysis of Single Cysteine Molecules with an Aerolysin Nanopore. Analyst 2020, 145, 1179–1183. [Google Scholar] [CrossRef] [PubMed]
- Piguet, F.; Ouldali, H.; Pastoriza-Gallego, M.; Manivet, P.; Pelta, J.; Oukhaled, A. Identification of Single Amino Acid Differences in Uniformly Charged Homopolymeric Peptides with Aerolysin Nanopore. Nat. Commun. 2018, 9, 966. [Google Scholar] [CrossRef] [PubMed]
- Lucas, F.L.R.; Versloot, R.C.A.; Yakovlieva, L.; Walvoort, M.T.C.; Maglia, G. Protein Identification by Nanopore Peptide Profiling. Nat. Commun. 2021, 12, 5795. [Google Scholar] [CrossRef] [PubMed]
- Huang, G.; Willems, K.; Soskine, M.; Wloka, C.; Maglia, G. Electro-Osmotic Capture and Ionic Discrimination of Peptide and Protein Biomarkers with FraC Nanopores. Nat. Commun. 2017, 8, 935. [Google Scholar] [CrossRef] [Green Version]
- Fahie, M.A.; Chen, M. Electrostatic Interactions between OmpG Nanopore and Analyte Protein Surface Can Distinguish between Glycosylated Isoforms. J. Phys. Chem. B 2015, 119, 10198–10206. [Google Scholar] [CrossRef]
- Thakur, A.K.; Movileanu, L. Single-Molecule Protein Detection in a Biofluid Using a Quantitative Nanopore Sensor. ACS Sens. 2019, 4, 2320–2326. [Google Scholar] [CrossRef]
- Thakur, A.K.; Movileanu, L. Real-Time Measurement of Protein–Protein Interactions at Single-Molecule Resolution Using a Biological Nanopore. Nat. Biotechnol. 2019, 37, 96–104. [Google Scholar] [CrossRef] [PubMed]
- Afshar Bakshloo, M.; Kasianowicz, J.J.; Pastoriza-Gallego, M.; Mathé, J.; Daniel, R.; Piguet, F.; Oukhaled, A. Nanopore-Based Protein Identification. J. Am. Chem. Soc. 2022, 144, 2716–2725. [Google Scholar] [CrossRef]
- Brinkerhoff, H.; Kang, A.S.W.; Liu, J.; Aksimentiev, A.; Dekker, C. Multiple Rereads of Single Proteins at Single-Amino Acid Resolution Using Nanopores. Science 2021, 374, 1509–1513. [Google Scholar] [CrossRef]
- Zhang, S.; Huang, G.; Versloot, R.C.A.; Bruininks, B.M.H.; de Souza, P.C.T.; Marrink, S.J.; Maglia, G. Bottom-up Fabrication of a Proteasome–Nanopore That Unravels and Processes Single Proteins. Nat. Chem. 2021, 13, 1192–1199. [Google Scholar] [CrossRef] [PubMed]
- Chakrapani, S.; Cordero-Morales, J.F.; Perozo, E. A Quantitative Description of KcsA Gating I: Macroscopic Currents. J. Gen. Physiol. 2007, 130, 465–478. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kiya, T.; Takeshita, K.; Kawanabe, A.; Fujiwara, Y. Intermolecular Functional Coupling between Phosphoinositides and the Potassium Channel KcsA. J. Biol. Chem. 2022, 298, 102257. [Google Scholar] [CrossRef] [PubMed]
- Lü, W.; Du, J.; Schwarzer, N.J.; Gerbig-Smentek, E.; Einsle, O.; Andrade, S.L.A. The Formate Channel FocA Exports the Products of Mixed-Acid Fermentation. Proc. Natl. Acad. Sci. USA 2012, 109, 13254–13259. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Focke, P.J.; Hein, C.; Hoffmann, B.; Matulef, K.; Bernhard, F.; Dötsch, V.; Valiyaveetil, F.I. Combining In Vitro Folding with Cell Free Protein Synthesis for Membrane Protein Expression. Biochemistry 2016, 55, 4212–4219. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Malik, C.; Ghosh, S. S6 Peptide Derived from KvAP Channel Shows Cooperativity in Gating on Bilayer Lipid Membrane. PLoS ONE 2013, 8, e78845. [Google Scholar] [CrossRef]
- Rosholm, K.R.; Baker, M.A.B.; Ridone, P.; Nakayama, Y.; Rohde, P.R.; Cuello, L.G.; Lee, L.K.; Martinac, B. Activation of the Mechanosensitive Ion Channel MscL by Mechanical Stimulation of Supported Droplet-Hydrogel Bilayers. Sci. Rep. 2017, 7, 45180. [Google Scholar] [CrossRef] [Green Version]
- Zhao, C.; Li, K.; Mou, X.; Zhu, Y.; Chen, C.; Zhang, M.; Wang, Y.; Zhou, K.; Sheng, Y.; Liu, H.; et al. High-Fidelity Biosensing of DNTPs and Nucleic Acids by Controllable Subnanometer Channel PaMscS. Biosens. Bioelectron. 2022, 200, 113894. [Google Scholar] [CrossRef]
- Jo, A.; Hoi, H.; Zhou, H.; Gupta, M.; Montemagno, C.D. Single-Molecule Study of Full-Length NaChBac by Planar Lipid Bilayer Recording. PLoS ONE 2017, 12, e0188861. [Google Scholar] [CrossRef] [Green Version]
- Lü, W.; Schwarzer, N.J.; Du, J.; Gerbig-Smentek, E.; Andrade, S.L.A.; Einsle, O. Structural and Functional Characterization of the Nitrite Channel NirC from Salmonella Typhimurium. Proc. Natl. Acad. Sci. USA 2012, 109, 18395–18400. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bagdany, M.; Veit, G.; Fukuda, R.; Avramescu, R.G.; Okiyoneda, T.; Baaklini, I.; Singh, J.; Sovak, G.; Xu, H.; Apaja, P.M.; et al. Chaperones Rescue the Energetic Landscape of Mutant CFTR at Single Molecule and in Cell. Nat. Commun. 2017, 8, 398. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Carnarius, C.; Kreir, M.; Krick, M.; Methfessel, C.; Moehrle, V.; Valerius, O.; Brüggemann, A.; Steinem, C.; Fertig, N. Green Fluorescent Protein Changes the Conductance of Connexin 43 (Cx43) Hemichannels Reconstituted in Planar Lipid Bilayers. J. Biol. Chem. 2012, 287, 2877–2886. [Google Scholar] [CrossRef] [Green Version]
- Corradi, V.; Bukiya, A.N.; Miranda, W.E.; Cui, M.; Plant, L.D.; Logothetis, D.E.; Tieleman, D.P.; Noskov, S.Y.; Rosenhouse-Dantsker, A. A Molecular Switch Controls the Impact of Cholesterol on a Kir Channel. Proc. Natl. Acad. Sci. USA 2022, 119, e2109431119. [Google Scholar] [CrossRef]
- De Souza, C.A.M.; Teixeira, P.C.N.; Faria, R.X.; Krylova, O.; Pohl, P.; Alves, L.A. A Consensus Segment in the M2 Domain of the HP2X 7 Receptor Shows Ion Channel Activity in Planar Lipid Bilayers and in Biological Membranes. Biochim. Biophys. Acta Biomembr. 2012, 1818, 64–71. [Google Scholar] [CrossRef] [Green Version]
- Gaburjakova, J.; Gaburjakova, M. Coupled Gating Modifies the Regulation of Cardiac Ryanodine Receptors by Luminal Ca2+. Biochim. Biophys. Acta Biomembr. 2014, 1838, 867–873. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lang, S.; Erdmann, F.; Jung, M.; Wagner, R.; Cavalié, A.; Zimmermann, R. Sec61 Complexes Form Ubiquitous ER Ca2+ Leak Channels. Channels 2011, 5, 228–235. [Google Scholar] [CrossRef] [Green Version]
- Lal, S.; Scarinci, N.; Perez, P.L.; del Rocío Cantero, M.; Cantiello, H.F. Lipid Bilayer-Atomic Force Microscopy Combined Platform Records Simultaneous Electrical and Topological Changes of the TRP Channel Polycystin-2 (TRPP2). PLoS ONE 2018, 13, e0202029. [Google Scholar] [CrossRef] [Green Version]
- Vandenberg, J.I.; Perry, M.D.; Perrin, M.J.; Mann, S.A.; Ke, Y.; Hill, A.P. HERG K(+) Channels: Structure, Function, and Clinical Significance. Physiol. Rev. 2012, 92, 1393–1478. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sanguinetti, M.C.; Tristani-Firouzi, M. HERG Potassium Channels and Cardiac Arrhythmia. Nature 2006, 440, 463–469. [Google Scholar] [CrossRef] [PubMed]
- Neubert, H.J. Patch Clamping Moves to Chips. Anal. Chem. 2004, 76, 327 A–330 A. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Oshima, A.; Hirano-Iwata, A.; Mozumi, H.; Ishinari, Y.; Kimura, Y.; Niwano, M. Reconstitution of Human Ether-a-Go-Go-Related Gene Channels in Microfabricated Silicon Chips. Anal. Chem. 2013, 85, 4363–4369. [Google Scholar] [CrossRef] [PubMed]
- Kawano, R.; Tsuji, Y.; Sato, K.; Osaki, T.; Kamiya, K.; Hirano, M.; Ide, T.; Miki, N.; Takeuchi, S. Automated Parallel Recordings of Topologically Identified Single Ion Channels. Sci. Rep. 2013, 3, 1995. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kamiya, K.; Osaki, T.; Nakao, K.; Kawano, R.; Fujii, S.; Misawa, N.; Hayakawa, M.; Takeuchi, S. Electrophysiological Measurement of Ion Channels on Plasma/Organelle Membranes Using an on-Chip Lipid Bilayer System. Sci. Rep. 2018, 8, 2–10. [Google Scholar] [CrossRef] [PubMed] [Green Version]
| Protein | Organism | Type | TM Region | Complex | TM Number | Method | Membrane Composition | Ref. |
|---|---|---|---|---|---|---|---|---|
| BmrC/BmrD | Bacillus subtilis | ABC transporter | α−helix | BmrC/BmrD | 12 | detergent mediate reconstitution | DPhPC + DOPC/DOPE or DOPC/Sph/chol | [74] |
| BR | Halophilic archaea | proton pump | α−helix | − | 7 | fusion | EPC:EPA (9:1 [mol]) | [74] |
| CXCR4 | Homo sapiens | GPCR signaling protein | α−helix | homodimer | 14 (7 × 2) | fusion | DOPC | [75] |
| EmrE | Escherichia coli | multidrug transporter | α−helix | homodimer | 8 (4 × 2) | direct reconstitution | POPC | [83] |
| ETB | Homo sapiens | GPCR signaling protein | α−helix | − | 7 | fusion | DOPC | [75] |
| F1F0-ATP synthase | Escherichia coli | ATP production | α−helix | F1–F0 | 28 | rehydration | E. coli total lipid extract | [84] |
| FhuA | Escherichia coli | ferrichrome-iron receptor | β−strand | − | 22 | fusion | EPC:Tx-DHPE (99.5:0.5) | [74] |
| GL1 | Homo sapiens | γ-aminobutyric acid receptor | α−helix | − | anchored | rehydration | DOPE:DOPC:DOPE-Atto647 (30:69.5:0.5) | [85] |
| GLUT1 | Homo sapiens | glucose transporter | α−helix | − | 12 | fusion | DOPC:DMPE-RhB:Biotylated PE (99.7:0.2:0.1) | [86] |
| IFITM3 | Homo sapiens | enveloped virus inhibitor | α−helix | − | 1 | detergent mediate reconstitution | POPC:cholesterol:Liss-Rho-PE (99:0.5:0.5 [mol]) | [87] |
| KcsA | Streptomyces lividans | potassium channels | α−helix | homotetramer | 8 (2 × 4) | direct reconstitution | DOPG or DOPE | [88] |
| KvAP | Aeropyrum pernix | voltage-gated potassium channels | α−helix | heteromer or homotetramer | 8 (2 × 4) | rehydration | DPhPC or EPC:EPA (9:1) | [89] |
| OmpF | Escherichia coli | porin | β−strand | − | 16 | direct reconstitution | PDMS26-b-PMOXA9 | [90] |
| OmpG | Escherichia coli | porin | β−strand | − | 14 | direct reconstitution | Outer membrane: DOPC Inner membrane: oleosin | [76] |
| OmpLA | Escherichia coli | phospholipase | β−strand | homodimer | 24 (12 × 2) | direct reconstitution | DOPC:DOPG (1:3) | [77] |
| PR | SAR86 group of marine γ-proteobacteria | proton transport | α−helix | − | 7 | direct reconstitution | POPC | [78] |
| RC | Rhodobacter sphaeroides | electron transport | α−helix | − | 10 | detergent mediate reconstitution | POPC:POPG (9:1 [mol]) | [79] |
| SLO | Streptococcus pyogenes | toxin | α−helix | homo 36~40 mer | 36~40 | rehydration | POPC, DOPC, SOPC, POPG | [80] |
| TmrAB | thermus thermophilus | ABC transporter | α−helix | − | 6 × 2 | rehydration | POPC:POPG:POPE: biotinylated-DOPE (40:30:29:1 [mol]) | [81] |
| TolC | Escherichia coli | expulsion of diverse molecules from the cell | β−strand | homotrimer | 12 (4 × 3) | rehydration | DOPC:DOPS:Atto647-DOPE (91.2:8:0.8) | [85] |
| t-SNARE | Homo sapiens | endocytosis/exocytosis | α−helix | Syntaxin -SNAP25 | 1 | rehydration | DOPC:DOPS:Atto647-DOPE (91.2:8:0.8) | [85] |
| VDAC1 | Homo sapiens | voltage dependent anion channel | β−strand | − | 19 | fusion | soybean polar extract: cholesterol (9:1) | [82] |
| Ion Channels | Organism | Type | TM Region | Complex | TM Number | Membrane Composition | Ref. |
|---|---|---|---|---|---|---|---|
| FocA | Salmonella typhimurium | formate channel | α−helix | homohexamer | 30 (5 × 6) | E. coli polar lipid extract | [158] |
| KcsA | Streptomyces lividans | K+ channel | α−helix | homotetramer | 8 (2 × 4) | L-α-lecithin | [159] |
| KvAP | Aeropyrum pernix | K+ channel | α−helix | homotetrameric | 8 (2 × 4) | cardiolipin:cholesterol (6:1) | [160] |
| MscL | Escherichia coli | mechanosensitive channel | α−helix | homopentamer | 10 (2 × 5) | DPhPC | [161] |
| MscS | Pseudomonas aeruginosa | mechanosensitive channel | α−helix | homoheptamer | 31 (3 × 7) | E. coli polar lipid extract | [162] |
| MVP | Methanococcus jannaschii | voltage-gated K+ channel | α−helix | homotetramer | 24 (6 × 4) | POPE:POPG (3:1) | [159] |
| Nav | Bacilllus halodurans | voltage-gated Na+ channel | α−helix | homotetramer | 24 (6 × 4) | POPE:POPG (3:1 [wt]/[wt]) | [163] |
| NirC | Salmonella typhimurium | potential nitrite transporter | α−helix | homohexamer | 30 (5 × 6) | E. coli polar lipid extract | [164] |
| BEST1 | Homo sapiens | Ca2+ activated chloride channel | α−helix | homopentamer | 20 (4 × 5) | POPE:POPG (3:1 [wt/wt]) | [122] |
| BK | Homo sapiens | Ca2+ -and voltage-dependent potassium channel | α−helix | Homotetramer | 8 (2 × 4) | POPE:POPS (1:1) | [123] |
| CFTR | Homo sapiens | chloride channel | α−helix | − | 12 | POPE:POPS (2:1) | [165] |
| connexin43 | Homo sapiens | gap junction channel | α−helix | homodimer | 24 (4 × 6) | POPC | [166] |
| Kir3.4 | Mus musculus | cholesterol sensitive potassium channel | α−helix | homotetramer | 8 (2 × 4) | PE:POPS (1:1) | [167] |
| P2X7R | Homo sapiens | ATP-gated cation-selective channel | α−helix | homotrimer | 6 (2 × 3) | E. coli polar lipid extract | [168] |
| RYR2 | Homo sapiens | Ca2+ release channel | α−helix | homotetramer | 24 (6 × 4) | DOPE:DOPS (3:1) | [169] |
| Sec 61 | Homo sapiens | dynamic polypeptide-conducting channel | α−helix | 3-domain α/β/γ | 12 | L-α-lecithin | [170] |
| TRPP2 | Homo sapiens | Ca2+ permeable nonselective cation channel | α−helix | homotetramer | 24 (6 × 4) | POPC:POPE (7:3) | [171] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tosaka, T.; Kamiya, K. Function Investigations and Applications of Membrane Proteins on Artificial Lipid Membranes. Int. J. Mol. Sci. 2023, 24, 7231. https://doi.org/10.3390/ijms24087231
Tosaka T, Kamiya K. Function Investigations and Applications of Membrane Proteins on Artificial Lipid Membranes. International Journal of Molecular Sciences. 2023; 24(8):7231. https://doi.org/10.3390/ijms24087231
Chicago/Turabian StyleTosaka, Toshiyuki, and Koki Kamiya. 2023. "Function Investigations and Applications of Membrane Proteins on Artificial Lipid Membranes" International Journal of Molecular Sciences 24, no. 8: 7231. https://doi.org/10.3390/ijms24087231
APA StyleTosaka, T., & Kamiya, K. (2023). Function Investigations and Applications of Membrane Proteins on Artificial Lipid Membranes. International Journal of Molecular Sciences, 24(8), 7231. https://doi.org/10.3390/ijms24087231





