Extracellular Vesicles for Therapeutic Nucleic Acid Delivery: Loading Strategies and Challenges
Abstract
1. Introduction
2. Classification of EVs
3. Challenges of EV Application as Drug Delivery Carriers
3.1. Interactions of EVs with Immune Cells
3.2. EVs in the Bloodstream
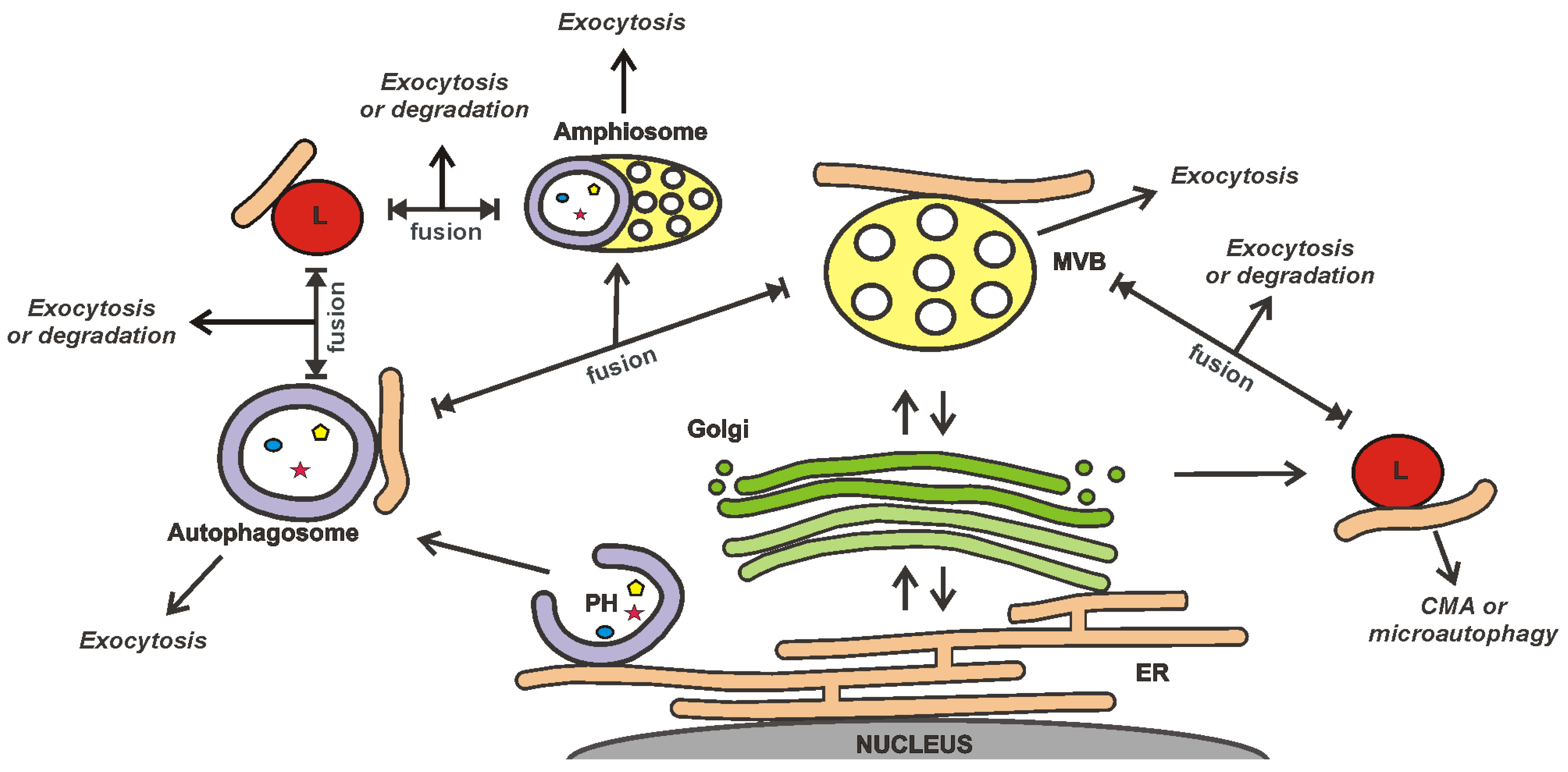
3.3. EVs as a Part of the Cellular Secretory Apparatus
3.4. Influence of the Non-Cellular Environment on EVs
3.5. Manufacture of EVs
4. Strategies of tNA Loading into EVs
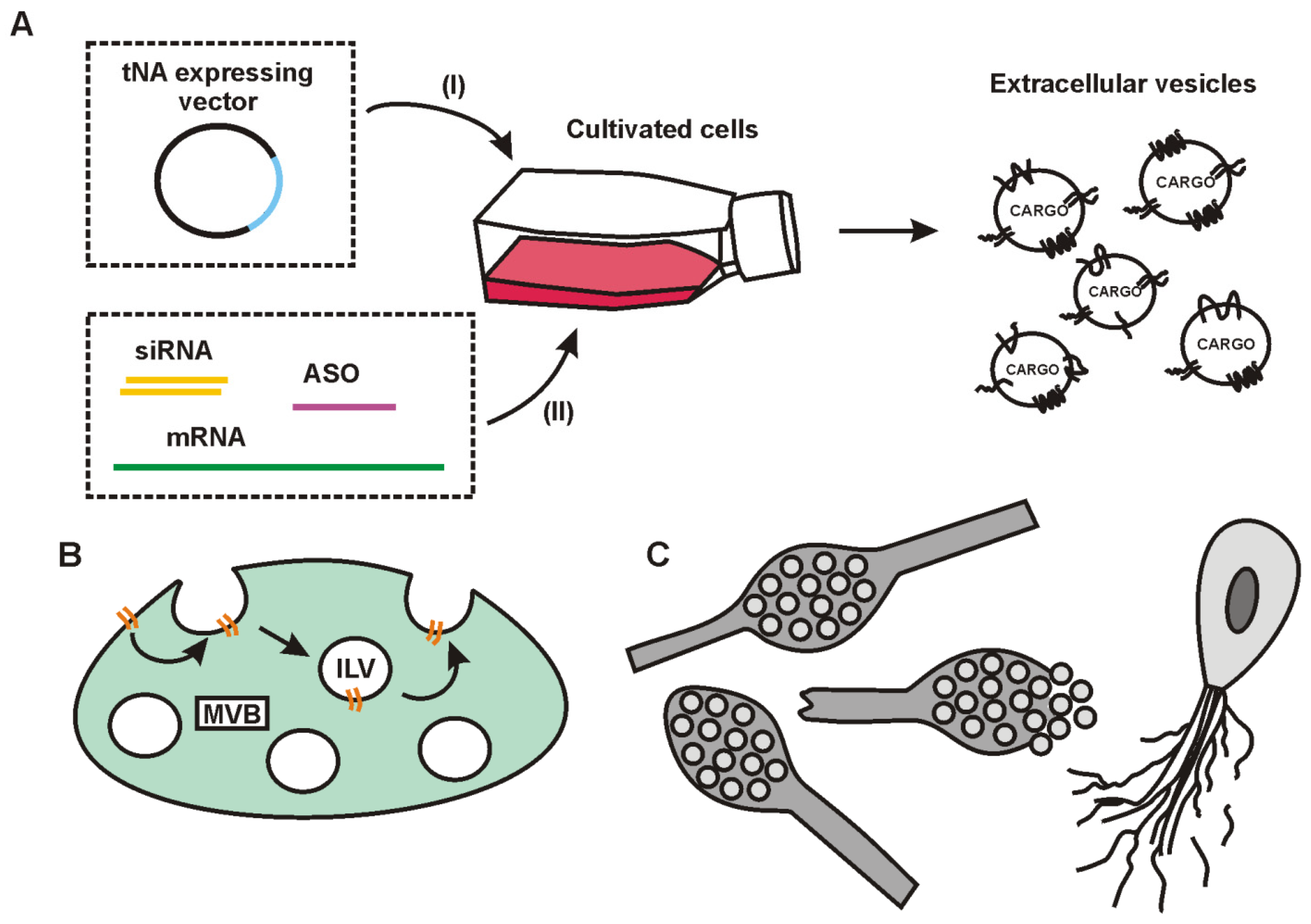
5. Exosome Loading with tNAs
5.1. Basic Principles of Cargo Sorting into Exosomes and Exosome Release from Cells
5.2. Involvement of RNA-Binding Proteins in tNA Sorting into Exosomes
5.3. Upstream ESCRTs (0-II)-Mediated Binding of RBPs and RNA
5.4. ALIX, Syntenin-1, and Syndecans Participate in tNA Sorting into Exosomes

5.5. Some Aspects of mRNA Loading into Exosomes
6. Microvesicle Loading with tNAs

7. Apoptotic Body Loading with tNAs
8. The Use of Surface Proteins of EVs for Endogenous Loading of tNAs
9. Direct (Exogenous) Strategies for tNA Loading into EVs
9.1. Binding of tNAs with the EV Membrane
9.2. The Loading Using Transient Permeabilization of the EV’s Membrane
10. Clinical Trials and Some Recent Patents Related to the EV Application for tNA Delivery
11. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
References
- Bost, J.P.; Barriga, H.; Holme, M.N.; Gallud, A.; Maugeri, M.; Gupta, D.; Lehto, T.; Valadi, H.; Esbjörner, E.K.; Stevens, M.M.; et al. Delivery of Oligonucleotide Therapeutics: Chemical Modifications, Lipid Nanoparticles, and Extracellular Vesicles. ACS Nano 2021, 15, 13993–14021. [Google Scholar] [CrossRef] [PubMed]
- Smith, C.I.E.; Zain, R. Therapeutic Oligonucleotides: State of the Art. Annu. Rev. Pharmacol. Toxicol. 2019, 59, 605–630. [Google Scholar] [CrossRef] [PubMed]
- Yin, H.; Xue, W.; Anderson, D.G. CRISPR-Cas: A tool for cancer research and therapeutics. Nature reviews. Clin. Oncol. 2019, 16, 281–295. [Google Scholar] [CrossRef]
- Adamus, T.; Kortylewski, M. The revival of CpG oligonucleotide-based cancer immunotherapies. Contemp. Oncol. 2018, 22, 56–60. [Google Scholar] [CrossRef]
- Lima, J.F.; Cerqueira, L.; Figueiredo, C.; Oliveira, C.; Azevedo, N.F. Anti-miRNA oligonucleotides: A comprehensive guide for design. RNA Biol. 2018, 15, 338–352. [Google Scholar] [CrossRef]
- Pardi, N.; Hogan, M.J.; Porter, F.W.; Weissman, D. mRNA vaccines—A new era in vaccinology. Nat. Rev. Drug Discov. 2018, 17, 261–279. [Google Scholar] [CrossRef]
- Ma, L.; Liu, J. Catalytic Nucleic Acids: Biochemistry, Chemical Biology, Biosensors, and Nanotechnology. iScience 2020, 23, 100815. [Google Scholar] [CrossRef]
- Kulkarni, J.A.; Witzigmann, D.; Thomson, S.B.; Chen, S.; Leavitt, B.R.; Cullis, P.R.; van der Meel, R. The current landscape of nucleic acid therapeutics. Nat. Nanotechnol. 2021, 16, 630–643. [Google Scholar] [CrossRef]
- Xiong, H.; Veedu, R.N.; Diermeier, S.D. Recent Advances in Oligonucleotide Therapeutics in Oncology. Int. J. Mol. Sci. 2021, 22, 3295. [Google Scholar] [CrossRef]
- Din, F.U.; Aman, W.; Ullah, I.; Qureshi, O.S.; Mustapha, O.; Shafique, S.; Zeb, A. Effective use of nanocarriers as drug delivery systems for the treatment of selected tumors. Int. J. Nanomed. 2017, 12, 7291–7309. [Google Scholar] [CrossRef]
- Khan, S.A.; Ali, H. Novel Drug Delivery Systems. In Essentials of Industrial Pharmacy; Khan, S.A., Ed.; Springer International Publishing: Cham, Switzerland, 2022; pp. 235–250. [Google Scholar] [CrossRef]
- Nikezić, A.V.V.; Bondžić, A.M.; Vasić, V.M. Drug delivery systems based on nanoparticles and related nanostructures. Eur. J. Pharm Sci. 2020, 151, 105412. [Google Scholar] [CrossRef] [PubMed]
- Tan, X.; Jia, F.; Wang, P.; Zhang, K. Nucleic acid-based drug delivery strategies. J. Control. Release Off. J. Control. Release Soc. 2020, 323, 240–252. [Google Scholar] [CrossRef]
- Xu, F.; Xia, Q.; Wang, P. Rationally Designed DNA Nanostructures for Drug Delivery. Front. Chem. 2020, 8, 751. [Google Scholar] [CrossRef] [PubMed]
- Rampado, R.; Caliceti, P.; Agostini, M. Latest Advances in Biomimetic Cell Membrane-Coated and Membrane-Derived Nanovectors for Biomedical Applications. Nanomaterials 2022, 12, 1543. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Zhao, X.; Zhong, Y.; Shen, J.; An, W. Biomimetic Exosomes: A New Generation of Drug Delivery System. Front. Bioeng. Biotechnol. 2022, 10, 865682. [Google Scholar] [CrossRef] [PubMed]
- Herrmann, I.K.; Wood, M.J.A.; Fuhrmann, G. Extracellular vesicles as a next-generation drug delivery platform. Nat. Nanotechnol. 2021, 16, 748–759. [Google Scholar] [CrossRef]
- Stephenson, M.L.; Zamecnik, P.C. Inhibition of Rous sarcoma viral RNA translation by a specific oligodeoxyribonucleotide. Proc. Natl. Acad. Sci. USA 1978, 75, 285–288. [Google Scholar] [CrossRef]
- Crooke, S.T.; Liang, X.H.; Baker, B.F.; Crooke, R.M. Antisense technology: A review. J. Biol. Chem. 2021, 296, 100416. [Google Scholar] [CrossRef]
- Hall, J. Future Directions for Medicinal Chemistry in the Field of Oligonucleotide Therapeutics; RNA: New York, NY, USA, 2023. [Google Scholar] [CrossRef]
- Zhang, C.; Zhang, B. RNA therapeutics: Updates and future potential. Science China. Life Sci. 2023, 66, 12–30. [Google Scholar] [CrossRef]
- Curreri, A.; Sankholkar, D.; Mitragotri, S.; Zhao, Z. RNA therapeutics in the clinic. Bioeng. Transl. Med. 2023, 8, e10374. [Google Scholar] [CrossRef]
- Gupta, A.; Andresen, J.L.; Manan, R.S.; Langer, R. Nucleic acid delivery for therapeutic applications. Adv. Drug Deliv. Rev. 2021, 178, 113834. [Google Scholar] [CrossRef] [PubMed]
- Duffy, K.; Arangundy-Franklin, S.; Holliger, P. Modified nucleic acids: Replication, evolution, and next-generation therapeutics. BMC Biol. 2020, 18, 112. [Google Scholar] [CrossRef] [PubMed]
- Liczner, C.; Duke, K.; Juneau, G.; Egli, M.; Wilds, C.J. Beyond ribose and phosphate: Selected nucleic acid modifications for structure-function investigations and therapeutic applications. Beilstein J. Org. Chem. 2021, 17, 908–931. [Google Scholar] [CrossRef] [PubMed]
- Torres-Vanegas, J.D.; Cruz, J.C.; Reyes, L.H. Delivery Systems for Nucleic Acids and Proteins: Barriers, Cell Capture Pathways and Nanocarriers. Pharmaceutics 2021, 13, 428. [Google Scholar] [CrossRef]
- van den Berg, A.I.S.; Yun, C.O.; Schiffelers, R.M.; Hennink, W.E. Polymeric delivery systems for nucleic acid therapeutics: Approaching the clinic. J. Control. Release Off. J. Control. Release Soc. 2021, 331, 121–141. [Google Scholar] [CrossRef]
- Steffens, R.C.; Wagner, E. Directing the Way-Receptor and Chemical Targeting Strategies for Nucleic Acid Delivery. Pharm. Res. 2022, 40, 47–76. [Google Scholar] [CrossRef]
- Kuriyama, N.; Yoshioka, Y.; Kikuchi, S.; Okamura, A.; Azuma, N.; Ochiya, T. Challenges for the Development of Extracellular Vesicle-Based Nucleic Acid Medicines. Cancers 2021, 13, 6137. [Google Scholar] [CrossRef]
- Yao, X.; Lyu, P.; Yoo, K.; Yadav, M.K.; Singh, R.; Atala, A.; Lu, B. Engineered extracellular vesicles as versatile ribonucleoprotein delivery vehicles for efficient and safe CRISPR genome editing. J. Extracell. Vesicles 2021, 10, e12076. [Google Scholar] [CrossRef]
- Piffoux, M.; Volatron, J.; Silva, A.K.A.; Gazeau, F. Thinking Quantitatively of RNA-Based Information Transfer via Extracellular Vesicles: Lessons to Learn for the Design of RNA-Loaded EVs. Pharmaceutics 2021, 13, 1931. [Google Scholar] [CrossRef]
- Schulz-Siegmund, M.; Aigner, A. Nucleic acid delivery with extracellular vesicles. Adv. Drug Deliv. Rev. 2021, 173, 89–111. [Google Scholar] [CrossRef]
- Du, R.; Wang, C.; Zhu, L.; Yang, Y. Extracellular Vesicles as Delivery Vehicles for Therapeutic Nucleic Acids in Cancer Gene Therapy: Progress and Challenges. Pharmaceutics 2022, 14, 2236. [Google Scholar] [CrossRef] [PubMed]
- Lu, M.; Shao, W.; Xing, H.; Huang, Y. Extracellular Vesicle-Based Nucleic Acid Delivery. Interdiscip. Med. 2023, e20220007. [Google Scholar] [CrossRef]
- Jurgielewicz, B.; Stice, S.; Yao, Y. Therapeutic Potential of Nucleic Acids when Combined with Extracellular Vesicles. Aging Dis. 2021, 12, 1476–1493. [Google Scholar] [CrossRef] [PubMed]
- Jiang, L.; Vader, P.; Schiffelers, R.M. Extracellular vesicles for nucleic acid delivery: Progress and prospects for safe RNA-based gene therapy. Gene Ther. 2017, 24, 157–166. [Google Scholar] [CrossRef] [PubMed]
- Massaro, C.; Sgueglia, G.; Frattolillo, V.; Baglio, S.R.; Altucci, L.; Dell’Aversana, C. Extracellular Vesicle-Based Nucleic Acid Delivery: Current Advances and Future Perspectives in Cancer Therapeutic Strategies. Pharmaceutics 2020, 12, 980. [Google Scholar] [CrossRef] [PubMed]
- Han, Y.; Jones, T.W.; Dutta, S.; Zhu, Y.; Wang, X.; Narayanan, S.P.; Fagan, S.C.; Zhang, D. Overview and Update on Methods for Cargo Loading into Extracellular Vesicles. Processes 2021, 9, 356. [Google Scholar] [CrossRef]
- Lara, P.; Chan, A.B.; Cruz, L.J.; Quest, A.F.G.; Kogan, M.J. Exploiting the Natural Properties of Extracellular Vesicles in Targeted Delivery towards Specific Cells and Tissues. Pharmaceutics 2020, 12, 1022. [Google Scholar] [CrossRef]
- Biagiotti, S.; Abbas, F.; Montanari, M.; Barattini, C.; Rossi, L.; Magnani, M.; Papa, S.; Canonico, B. Extracellular Vesicles as New Players in Drug Delivery: A Focus on Red Blood Cells-Derived EVs. Pharmaceutics 2023, 15, 365. [Google Scholar] [CrossRef]
- Zhao, Y.; Li, X.; Zhang, W.; Yu, L.; Wang, Y.; Deng, Z.; Liu, M.; Mo, S.; Wang, R.; Zhao, J.; et al. Trends in the biological functions and medical applications of extracellular vesicles and analogues. Acta Pharm. Sinica. B 2021, 11, 2114–2135. [Google Scholar] [CrossRef]
- Lou, K.; Feng, S.; Luo, H.; Zou, J.; Zhang, G.; Zou, X. Extracellular vesicles derived from macrophages: Current applications and prospects in tumors. Front. Bioeng. Biotechnol. 2022, 10, 1097074. [Google Scholar] [CrossRef]
- Lesage, F.; Thébaud, B. Mesenchymal Stromal Cell-Derived Extracellular Vesicles for Neonatal Lung Disease: Tiny Particles, Major Promise, Rigorous Requirements for Clinical Translation. Cells 2022, 11, 1176. [Google Scholar] [CrossRef] [PubMed]
- Lee, I.; Choi, Y.; Shin, D.U.; Kwon, M.; Kim, S.; Jung, H.; Nam, G.H.; Kwon, M. Small Extracellular Vesicles as a New Class of Medicines. Pharmaceutics 2023, 15, 325. [Google Scholar] [CrossRef]
- Chitti, S.V.; Nedeva, C.; Manickam, R.; Fonseka, P.; Mathivanan, S. Extracellular Vesicles as Drug Targets and Delivery Vehicles for Cancer Therapy. Pharmaceutics 2022, 14, 2822. [Google Scholar] [CrossRef] [PubMed]
- Ozkocak, D.C.; Phan, T.K.; Poon, I.K.H. Translating extracellular vesicle packaging into therapeutic applications. Front. Immunol. 2022, 13, 946422. [Google Scholar] [CrossRef] [PubMed]
- Escudé Martinez de Castilla, P.; Tong, L.; Huang, C.; Sofias, A.M.; Pastorin, G.; Chen, X.; Storm, G.; Schiffelers, R.M.; Wang, J.W. Extracellular vesicles as a drug delivery system: A systematic review of preclinical studies. Adv. Drug Deliv. Rev. 2021, 175, 113801. [Google Scholar] [CrossRef] [PubMed]
- Clancy, J.W.; D’Souza-Schorey, C. Tumor-Derived Extracellular Vesicles: Multifunctional Entities in the Tumor Microenvironment. Annu. Rev. Pathol. 2023, 18, 205–229. [Google Scholar] [CrossRef]
- Kugeratski, F.G.; Kalluri, R. Exosomes as mediators of immune regulation and immunotherapy in cancer. FEBS J. 2021, 288, 10–35. [Google Scholar] [CrossRef]
- Rajput, A.; Varshney, A.; Bajaj, R.; Pokharkar, V. Exosomes as New Generation Vehicles for Drug Delivery: Biomedical Applications and Future Perspectives. Molecules 2022, 27, 7289. [Google Scholar] [CrossRef]
- Takahashi, Y.; Takakura, Y. Extracellular vesicle-based therapeutics: Extracellular vesicles as therapeutic targets and agents. Pharmacol. Ther. 2023, 242, 108352. [Google Scholar] [CrossRef]
- Teng, F.; Fussenegger, M. Shedding Light on Extracellular Vesicle Biogenesis and Bioengineering. Adv. Sci. 2020, 8, 2003505. [Google Scholar] [CrossRef]
- Poon, I.K.H.; Parkes, M.A.F.; Jiang, L.; Atkin-Smith, G.K.; Tixeira, R.; Gregory, C.D.; Ozkocak, D.C.; Rutter, S.F.; Caruso, S.; Santavanond, J.P.; et al. Moving beyond size and phosphatidylserine exposure: Evidence for a diversity of apoptotic cell-derived extracellular vesicles in vitro. J. Extracell. Vesicles 2019, 8, 1608786. [Google Scholar] [CrossRef] [PubMed]
- Théry, C.; Witwer, K.W.; Aikawa, E.; Alcaraz, M.J.; Anderson, J.D.; Andriantsitohaina, R.; Antoniou, A.; Arab, T.; Archer, F.; Atkin-Smith, G.K.; et al. Minimal information for studies of extracellular vesicles 2018 (MISEV2018): A position statement of the International Society for Extracellular Vesicles and update of the MISEV2014 guidelines. J. Extracell. Vesicles 2018, 7, 1535750. [Google Scholar] [CrossRef] [PubMed]
- Kowal, J.; Arras, G.; Colombo, M.; Jouve, M.; Morath, J.P.; Primdal-Bengtson, B.; Dingli, F.; Loew, D.; Tkach, M.; Théry, C. Proteomic comparison defines novel markers to characterize heterogeneous populations of extracellular vesicle subtypes. Proc. Natl. Acad. Sci. USA 2016, 113, E968–E977. [Google Scholar] [CrossRef]
- Yoshioka, Y.; Konishi, Y.; Kosaka, N.; Katsuda, T.; Kato, T.; Ochiya, T. Comparative marker analysis of extracellular vesicles in different human cancer types. J. Extracell. Vesicles 2013, 2, 20424. [Google Scholar] [CrossRef] [PubMed]
- Ji, Y.; Qi, D.; Li, L.; Su, H.; Li, X.; Luo, Y.; Sun, B.; Zhang, F.; Lin, B.; Liu, T.; et al. Multiplexed profiling of single-cell extracellular vesicles secretion. Proc. Natl. Acad. Sci. USA 2019, 116, 5979–5984. [Google Scholar] [CrossRef] [PubMed]
- Jackson, C.E.; Scruggs, B.S.; Schaffer, J.E.; Hanson, P.I. Effects of Inhibiting VPS4 Support a General Role for ESCRTs in Extracellular Vesicle Biogenesis. Biophys. J. 2017, 113, 1342–1352. [Google Scholar] [CrossRef] [PubMed]
- Mathieu, M.; Névo, N.; Jouve, M.; Valenzuela, J.I.; Maurin, M.; Verweij, F.J.; Palmulli, R.; Lankar, D.; Dingli, F.; Loew, D.; et al. Specificities of exosome versus small ectosome secretion revealed by live intracellular tracking of CD63 and CD9. Nat. Commun. 2021, 12, 4389. [Google Scholar] [CrossRef] [PubMed]
- Kooijmans, S.A.A.; de Jong, O.G.; Schiffelers, R.M. Exploring interactions between extracellular vesicles and cells for innovative drug delivery system design. Adv. Drug Deliv. Rev. 2021, 173, 252–278. [Google Scholar] [CrossRef]
- Gupta, D.; Zickler, A.M.; El Andaloussi, S. Dosing extracellular vesicles. Adv. Drug Deliv. Rev. 2021, 178, 113961. [Google Scholar] [CrossRef]
- Ferreira, D.; Moreira, J.N.; Rodrigues, L.R. New advances in exosome-based targeted drug delivery systems. Crit. Rev. Oncol./Hematol. 2022, 172, 103628. [Google Scholar] [CrossRef]
- Murphy, D.E.; de Jong, O.G.; Brouwer, M.; Wood, M.J.; Lavieu, G.; Schiffelers, R.M.; Vader, P. Extracellular vesicle-based therapeutics: Natural versus engineered targeting and trafficking. Exp. Mol. Med. 2019, 51, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Liang, Y.; Duan, L.; Lu, J.; Xia, J. Engineering exosomes for targeted drug delivery. Theranostics 2021, 11, 3183–3195. [Google Scholar] [CrossRef]
- Frolova, L.; Li, I.T.S. Targeting Capabilities of Native and Bioengineered Extracellular Vesicles for Drug Delivery. Bioengineering 2022, 9, 496. [Google Scholar] [CrossRef] [PubMed]
- Walker, S.; Busatto, S.; Pham, A.; Tian, M.; Suh, A.; Carson, K.; Quintero, A.; Lafrence, M.; Malik, H.; Santana, M.X.; et al. Extracellular vesicle-based drug delivery systems for cancer treatment. Theranostics 2019, 9, 8001–8017. [Google Scholar] [CrossRef] [PubMed]
- Vilanova-Perez, T.; Jones, C.; Balint, S.; Dragovic, R.; Dustin, M.L.; Yeste, M.; Coward, K. Exosomes derived from HEK293T cells interact in an efficient and noninvasive manner with mammalian sperm in vitro. Nanomedicine 2020, 15, 1965–1980. [Google Scholar] [CrossRef] [PubMed]
- Hovhannisyan, L.; Czechowska, E.; Gutowska-Owsiak, D. The Role of Non-Immune Cell-Derived Extracellular Vesicles in Allergy. Front. Immunol. 2021, 12, 702381. [Google Scholar] [CrossRef]
- Hezel, M.E.V.; Nieuwland, R.; Bruggen, R.V.; Juffermans, N.P. The Ability of Extracellular Vesicles to Induce a Pro-Inflammatory Host Response. Int. J. Mol. Sci. 2017, 18, 1285. [Google Scholar] [CrossRef]
- Xu, L.; Liang, Y.; Xu, X.; Xia, J.; Wen, C.; Zhang, P.; Duan, L. Blood cell-derived extracellular vesicles: Diagnostic biomarkers and smart delivery systems. Bioengineered 2021, 12, 7929–7940. [Google Scholar] [CrossRef]
- Kim, S.M.; Yang, Y.; Oh, S.J.; Hong, Y.; Seo, M.; Jang, M. Cancer-derived exosomes as a delivery platform of CRISPR/Cas9 confer cancer cell tropism-dependent targeting. J. Control. Release Off. J. Control. Release Soc. 2017, 266, 8–16. [Google Scholar] [CrossRef]
- Munagala, R.; Aqil, F.; Jeyabalan, J.; Gupta, R.C. Bovine milk-derived exosomes for drug delivery. Cancer Lett. 2016, 371, 48–61. [Google Scholar] [CrossRef]
- Busatto, S.; Yang, Y.; Walker, S.A.; Davidovich, I.; Lin, W.H.; Lewis-Tuffin, L.; Anastasiadis, P.Z.; Sarkaria, J.; Talmon, Y.; Wurtz, G.; et al. Brain metastases-derived extracellular vesicles induce binding and aggregation of low-density lipoprotein. J. Nanobiotechnol. 2020, 18, 162. [Google Scholar] [CrossRef] [PubMed]
- Menard, J.A.; Cerezo-Magaña, M.; Belting, M. Functional role of extracellular vesicles and lipoproteins in the tumour microenvironment. Philosophical transactions of the Royal Society of London. Ser. B Biol. Sci. 2018, 373, 20160480. [Google Scholar] [CrossRef] [PubMed]
- Busatto, S.; Yang, Y.; Iannotta, D.; Davidovich, I.; Talmon, Y.; Wolfram, J. Considerations for extracellular vesicle and lipoprotein interactions in cell culture assays. J. Extracell. Vesicles 2022, 11, e12202. [Google Scholar] [CrossRef] [PubMed]
- Xiao, C.; Stahel, P.; Lewis, G.F. Regulation of Chylomicron Secretion: Focus on Post-Assembly Mechanisms. Cell. Mol. Gastroenterol. Hepatol. 2019, 7, 487–501. [Google Scholar] [CrossRef]
- Cieślik, M.; Nazimek, K.; Bryniarski, K. Extracellular Vesicles-Oral Therapeutics of the Future. Int. J. Mol. Sci. 2022, 23, 7554. [Google Scholar] [CrossRef] [PubMed]
- Shi, Y.; van der Meel, R.; Chen, X.; Lammers, T. The EPR effect and beyond: Strategies to improve tumor targeting and cancer nanomedicine treatment efficacy. Theranostics 2020, 10, 7921–7924. [Google Scholar] [CrossRef] [PubMed]
- Wu, J. The Enhanced Permeability and Retention (EPR) Effect: The Significance of the Concept and Methods to Enhance Its Application. J. Pers. Med. 2021, 11, 771. [Google Scholar] [CrossRef]
- Hyenne, V.; Ghoroghi, S.; Collot, M.; Bons, J.; Follain, G.; Harlepp, S.; Mary, B.; Bauer, J.; Mercier, L.; Busnelli, I.; et al. Studying the Fate of Tumor Extracellular Vesicles at High Spatiotemporal Resolution Using the Zebrafish Embryo. Dev. Cell 2019, 48, 554–572.e557. [Google Scholar] [CrossRef]
- Smyth, T.; Kullberg, M.; Malik, N.; Smith-Jones, P.; Graner, M.W.; Anchordoquy, T.J. Biodistribution and delivery efficiency of unmodified tumor-derived exosomes. J. Control. Release Off. J. Control. Release Soc. 2015, 199, 145–155. [Google Scholar] [CrossRef]
- Ponpuak, M.; Mandell, M.A.; Kimura, T.; Chauhan, S.; Cleyrat, C.; Deretic, V. Secretory autophagy. Curr. Opin. Cell Biol. 2015, 35, 106–116. [Google Scholar] [CrossRef]
- Gudbergsson, J.M.; Johnsen, K.B. Exosomes and autophagy: Rekindling the vesicular waste hypothesis. J. Cell Commun. Signal. 2019, 13, 443–450. [Google Scholar] [CrossRef] [PubMed]
- Buratta, S.; Tancini, B.; Sagini, K.; Delo, F.; Chiaradia, E.; Urbanelli, L.; Emiliani, C. Lysosomal Exocytosis, Exosome Release and Secretory Autophagy: The Autophagic- and Endo-Lysosomal Systems Go Extracellular. Int. J. Mol. Sci. 2020, 21, 2576. [Google Scholar] [CrossRef] [PubMed]
- Zhao, X.; Guan, Y.; Liu, F.; Yan, S.; Wang, Y.; Hu, M.; Li, Y.; Li, R.; Zhang, C.X. SNARE Proteins Mediate α-Synuclein Secretion via Multiple Vesicular Pathways. Mol. Neurobiol. 2021, 59, 405–419. [Google Scholar] [CrossRef] [PubMed]
- Sagini, K.; Buratta, S.; Delo, F.; Pellegrino, R.M.; Giovagnoli, S.; Urbanelli, L.; Emiliani, C. Drug-Induced Lysosomal Impairment Is Associated with the Release of Extracellular Vesicles Carrying Autophagy Markers. Int. J. Mol. Sci. 2021, 22, 2922. [Google Scholar] [CrossRef] [PubMed]
- Leidal, A.M.; Debnath, J. Emerging roles for the autophagy machinery in extracellular vesicle biogenesis and secretion. FASEB Bioadvances 2021, 3, 377–386. [Google Scholar] [CrossRef]
- Raiborg, C.; Wenzel, E.M.; Stenmark, H. ER-endosome contact sites: Molecular compositions and functions. EMBO J. 2015, 34, 1848–1858. [Google Scholar] [CrossRef]
- Kohler, V.; Aufschnaiter, A.; Büttner, S. Closing the Gap: Membrane Contact Sites in the Regulation of Autophagy. Cells 2020, 9, 1184. [Google Scholar] [CrossRef]
- Trivedi, P.C.; Bartlett, J.J.; Pulinilkunnil, T. Lysosomal Biology and Function: Modern View of Cellular Debris Bin. Cells 2020, 9, 1131. [Google Scholar] [CrossRef]
- Guo, H.; Chitiprolu, M.; Roncevic, L.; Javalet, C.; Hemming, F.J.; Trung, M.T.; Meng, L.; Latreille, E.; Tanese de Souza, C.; McCulloch, D.; et al. Atg5 Disassociates the V(1)V(0)-ATPase to Promote Exosome Production and Tumor Metastasis Independent of Canonical Macroautophagy. Dev. Cell 2017, 43, 716–730.e717. [Google Scholar] [CrossRef]
- Leidal, A.M.; Huang, H.H.; Marsh, T.; Solvik, T.; Zhang, D.; Ye, J.; Kai, F.; Goldsmith, J.; Liu, J.Y.; Huang, Y.H.; et al. The LC3-conjugation machinery specifies the loading of RNA-binding proteins into extracellular vesicles. Nat. Cell Biol. 2020, 22, 187–199. [Google Scholar] [CrossRef]
- Ferreira, J.V.; da Rosa Soares, A.; Ramalho, J.; Máximo Carvalho, C.; Cardoso, M.H.; Pintado, P.; Carvalho, A.S.; Beck, H.C.; Matthiesen, R.; Zuzarte, M.; et al. LAMP2A regulates the loading of proteins into exosomes. Sci. Adv. 2022, 8, eabm1140. [Google Scholar] [CrossRef] [PubMed]
- Stranford, D.M.; Hung, M.E.; Gargus, E.S.; Shah, R.N.; Leonard, J.N. A Systematic Evaluation of Factors Affecting Extracellular Vesicle Uptake by Breast Cancer Cells. Tissue Eng. Part A 2017, 23, 1274–1282. [Google Scholar] [CrossRef] [PubMed]
- Schwager, S.C.; Bordeleau, F.; Zhang, J.; Antonyak, M.A.; Cerione, R.A.; Reinhart-King, C.A. Matrix stiffness regulates microvesicle-induced fibroblast activation. Am. J. Physiol. Cell Physiol. 2019, 317, C82–C92. [Google Scholar] [CrossRef] [PubMed]
- Parolini, I.; Federici, C.; Raggi, C.; Lugini, L.; Palleschi, S.; De Milito, A.; Coscia, C.; Iessi, E.; Logozzi, M.; Molinari, A.; et al. Microenvironmental pH is a key factor for exosome traffic in tumor cells. J. Biol. Chem. 2009, 284, 34211–34222. [Google Scholar] [CrossRef] [PubMed]
- Endzeliņš, E.; Ābols, A.; Bušs, A.; Zandberga, E.; Palviainen, M.; Siljander, P.; Linē, A. Extracellular Vesicles Derived from Hypoxic Colorectal Cancer Cells Confer Metastatic Phenotype to Non-metastatic Cancer Cells. Anticancer. Res. 2018, 38, 5139–5147. [Google Scholar] [CrossRef] [PubMed]
- Jurgielewicz, B.J.; Yao, Y.; Stice, S.L. Kinetics and Specificity of HEK293T Extracellular Vesicle Uptake using Imaging Flow Cytometry. Nanoscale Res. Lett. 2020, 15, 170. [Google Scholar] [CrossRef] [PubMed]
- Qiao, L.; Hu, S.; Huang, K.; Su, T.; Li, Z.; Vandergriff, A.; Cores, J.; Dinh, P.U.; Allen, T.; Shen, D.; et al. Tumor cell-derived exosomes home to their cells of origin and can be used as Trojan horses to deliver cancer drugs. Theranostics 2020, 10, 3474–3487. [Google Scholar] [CrossRef]
- Sancho-Albero, M.; Navascués, N.; Mendoza, G.; Sebastián, V.; Arruebo, M.; Martín-Duque, P.; Santamaría, J. Exosome origin determines cell targeting and the transfer of therapeutic nanoparticles towards target cells. J. Nanobiotechnol. 2019, 17, 16. [Google Scholar] [CrossRef]
- Wang, X.; Cheng, K.; Zhang, G.; Jia, Z.; Yu, Y.; Guo, J.; Hua, Y.; Guo, F.; Li, X.; Zou, W.; et al. Enrichment of CD44 in Exosomes From Breast Cancer Cells Treated With Doxorubicin Promotes Chemoresistance. Front. Oncol. 2020, 10, 960. [Google Scholar] [CrossRef]
- Raab-Traub, N.; Dittmer, D.P. Viral effects on the content and function of extracellular vesicles. Nature reviews. Microbiology 2017, 15, 559–572. [Google Scholar] [CrossRef]
- Vallhov, H.; Gutzeit, C.; Johansson, S.M.; Nagy, N.; Paul, M.; Li, Q.; Friend, S.; George, T.C.; Klein, E.; Scheynius, A.; et al. Exosomes containing glycoprotein 350 released by EBV-transformed B cells selectively target B cells through CD21 and block EBV infection in vitro. J. Immunol. 2011, 186, 73–82. [Google Scholar] [CrossRef]
- Raposo, G.; Stahl, P.D. Extracellular vesicles: A new communication paradigm? Nature reviews. Mol. Cell Biol. 2019, 20, 509–510. [Google Scholar] [CrossRef]
- Wang, S.; Li, C.; Yuan, Y.; Xiong, Y.; Xu, H.; Pan, W.; Pan, H.; Zhu, Z. Microvesicles as drug delivery systems: A new frontier for bionic therapeutics in cancer. J. Drug Deliv. Sci. Technol. 2023, 79, 104088. [Google Scholar] [CrossRef]
- Böröczky, T.; Dobra, G.; Bukva, M.; Gyukity-Sebestyén, E.; Hunyadi-Gulyás, É.; Darula, Z.; Horváth, P.; Buzás, K.; Harmati, M. Impact of Experimental Conditions on Extracellular Vesicles’ Proteome: A Comparative Study. Life 2023, 13, 206. [Google Scholar] [CrossRef] [PubMed]
- Dixson, A.C.; Dawson, T.R.; Di Vizio, D.; Weaver, A.M. Context-Specific regulation of extracellular vesicle biogenesis and cargo selection. Nat. Rev. Mol. Cell Biol. 2023, in press. [Google Scholar] [CrossRef]
- Kojima, R.; Bojar, D.; Rizzi, G.; Hamri, G.C.; El-Baba, M.D.; Saxena, P.; Ausländer, S.; Tan, K.R.; Fussenegger, M. Designer exosomes produced by implanted cells intracerebrally deliver therapeutic cargo for Parkinson’s disease treatment. Nat. Commun. 2018, 9, 1305. [Google Scholar] [CrossRef]
- Chen, J.; Li, P.; Zhang, T.; Xu, Z.; Huang, X.; Wang, R.; Du, L. Review on Strategies and Technologies for Exosome Isolation and Purification. Front. Bioeng. Biotechnol. 2021, 9, 811971. [Google Scholar] [CrossRef]
- Akbar, A.; Malekian, F.; Baghban, N.; Kodam, S.P.; Ullah, M. Methodologies to Isolate and Purify Clinical Grade Extracellular Vesicles for Medical Applications. Cells 2022, 11, 186. [Google Scholar] [CrossRef]
- Syromiatnikova, V.; Prokopeva, A.; Gomzikova, M. Methods of the Large-Scale Production of Extracellular Vesicles. Int. J. Mol. Sci. 2022, 23, 522. [Google Scholar] [CrossRef]
- Clos-Sansalvador, M.; Monguió-Tortajada, M.; Roura, S.; Franquesa, M.; Borràs, F.E. Commonly used methods for extracellular vesicles’ enrichment: Implications in downstream analyses and use. Eur. J. Cell Biol. 2022, 101, 151227. [Google Scholar] [CrossRef]
- Di Bella, M.A. Overview and Update on Extracellular Vesicles: Considerations on Exosomes and Their Application in Modern Medicine. Biology 2022, 11, 804. [Google Scholar] [CrossRef] [PubMed]
- Davidson, S.M.; Boulanger, C.M.; Aikawa, E.; Badimon, L.; Barile, L.; Binder, C.J.; Brisson, A.; Buzas, E.; Emanueli, C.; Jansen, F.; et al. Methods for the identification and characterization of extracellular vesicles in cardiovascular studies: From exosomes to microvesicles. Cardiovasc. Res. 2023, 119, 45–63. [Google Scholar] [CrossRef] [PubMed]
- Bağcı, C.; Sever-Bahcekapili, M.; Belder, N.; Bennett, A.P.S.; Erdener, Ş.E.; Dalkara, T. Overview of extracellular vesicle characterization techniques and introduction to combined reflectance and fluorescence confocal microscopy to distinguish extracellular vesicle subpopulations. Neurophotonics 2022, 9, 021903. [Google Scholar] [CrossRef] [PubMed]
- Driedonks, T.; Jiang, L.; Carlson, B.; Han, Z.; Liu, G.; Queen, S.E.; Shirk, E.N.; Gololobova, O.; Liao, Z.; Nyberg, L.H.; et al. Pharmacokinetics and biodistribution of extracellular vesicles administered intravenously and intranasally to Macaca nemestrina. J. Extracell. Biol. 2022, 1, e59. [Google Scholar] [CrossRef] [PubMed]
- Sork, H.; Conceicao, M.; Corso, G.; Nordin, J.; Lee, Y.X.F.; Krjutskov, K.; Orzechowski Westholm, J.; Vader, P.; Pauwels, M.; Vandenbroucke, R.E.; et al. Profiling of Extracellular Small RNAs Highlights a Strong Bias towards Non-Vesicular Secretion. Cells 2021, 10, 1543. [Google Scholar] [CrossRef]
- Möbius, W.; Ohno-Iwashita, Y.; van Donselaar, E.G.; Oorschot, V.M.; Shimada, Y.; Fujimoto, T.; Heijnen, H.F.; Geuze, H.J.; Slot, J.W. Immunoelectron microscopic localization of cholesterol using biotinylated and non-cytolytic perfringolysin O. J. Histochem. Cytochem. Off. J. Histochem. Soc. 2002, 50, 43–55. [Google Scholar] [CrossRef]
- Choezom, D.; Gross, J.C. Neutral sphingomyelinase 2 controls exosome secretion by counteracting V-ATPase-mediated endosome acidification. J. Cell Sci. 2022, 135. [Google Scholar] [CrossRef]
- Ghossoub, R.; Chéry, M.; Audebert, S.; Leblanc, R.; Egea-Jimenez, A.L.; Lembo, F.; Mammar, S.; Le Dez, F.; Camoin, L.; Borg, J.P.; et al. Tetraspanin-6 negatively regulates exosome production. Proc. Natl. Acad. Sci. USA 2020, 117, 5913–5922. [Google Scholar] [CrossRef]
- Villarroya-Beltri, C.; Baixauli, F.; Mittelbrunn, M.; Fernández-Delgado, I.; Torralba, D.; Moreno-Gonzalo, O.; Baldanta, S.; Enrich, C.; Guerra, S.; Sánchez-Madrid, F. ISGylation controls exosome secretion by promoting lysosomal degradation of MVB proteins. Nat. Commun. 2016, 7, 13588. [Google Scholar] [CrossRef]
- Perrin, P.; Janssen, L.; Janssen, H.; van den Broek, B.; Voortman, L.M.; van Elsland, D.; Berlin, I.; Neefjes, J. Retrofusion of intralumenal MVB membranes parallels viral infection and coexists with exosome release. Curr. Biol. CB 2021, 31, 3884–3893.e3884. [Google Scholar] [CrossRef]
- Valcz, G.; Buzás, E.I.; Kittel, Á.; Krenács, T.; Visnovitz, T.; Spisák, S.; Török, G.; Homolya, L.; Zsigrai, S.; Kiszler, G.; et al. En bloc release of MVB-like small extracellular vesicle clusters by colorectal carcinoma cells. J. Extracell. Vesicles 2019, 8, 1596668. [Google Scholar] [CrossRef] [PubMed]
- Yu, S.; Yu, L. Migrasome biogenesis and functions. FEBS J. 2021, 289, 7246–7254. [Google Scholar] [CrossRef] [PubMed]
- Ma, L.; Li, Y.; Peng, J.; Wu, D.; Zhao, X.; Cui, Y.; Chen, L.; Yan, X.; Du, Y.; Yu, L. Discovery of the migrasome, an organelle mediating release of cytoplasmic contents during cell migration. Cell Res. 2015, 25, 24–38. [Google Scholar] [CrossRef] [PubMed]
- Frankel, E.B.; Audhya, A. ESCRT-dependent cargo sorting at multivesicular endosomes. Semin. Cell Dev. Biol. 2018, 74, 4–10. [Google Scholar] [CrossRef] [PubMed]
- McGough, I.J.; Vincent, J.P. Exosomes in developmental signalling. Development 2016, 143, 2482–2493. [Google Scholar] [CrossRef]
- Kim, Y.S.; Ahn, J.S.; Kim, S.; Kim, H.J.; Kim, S.H.; Kang, J.S. The potential theragnostic (diagnostic+therapeutic) application of exosomes in diverse biomedical fields. Korean J. Physiol. Pharmacol. Off. J. Korean Physiol. Soc. Korean Soc. Pharmacol. 2018, 22, 113–125. [Google Scholar] [CrossRef]
- D’Souza-Schorey, C.; Schorey, J.S. Regulation and mechanisms of extracellular vesicle biogenesis and secretion. Essays Biochem. 2018, 62, 125–133. [Google Scholar] [CrossRef]
- Juan, T.; Fürthauer, M. Biogenesis and function of ESCRT-dependent extracellular vesicles. Semin. Cell Dev. Biol. 2018, 74, 66–77. [Google Scholar] [CrossRef]
- Kalra, H.; Drummen, G.P.; Mathivanan, S. Focus on Extracellular Vesicles: Introducing the Next Small Big Thing. Int. J. Mol. Sci. 2016, 17, 170. [Google Scholar] [CrossRef]
- van Niel, G.; D’Angelo, G.; Raposo, G. Shedding light on the cell biology of extracellular vesicles. Nature reviews. Mol. Cell Biol. 2018, 19, 213–228. [Google Scholar] [CrossRef]
- Schuh, A.L.; Audhya, A. The ESCRT machinery: From the plasma membrane to endosomes and back again. Crit. Rev. Biochem. Mol. Biol. 2014, 49, 242–261. [Google Scholar] [CrossRef] [PubMed]
- Vietri, M.; Radulovic, M.; Stenmark, H. The many functions of ESCRTs. Nature reviews. Mol. Cell Biol. 2020, 21, 25–42. [Google Scholar] [CrossRef]
- Migliano, S.M.; Wenzel, E.M.; Stenmark, H. Biophysical and molecular mechanisms of ESCRT functions, and their implications for disease. Curr. Opin. Cell Biol. 2022, 75, 102062. [Google Scholar] [CrossRef] [PubMed]
- Mathieu, M.; Martin-Jaular, L.; Lavieu, G.; Théry, C. Specificities of secretion and uptake of exosomes and other extracellular vesicles for cell-to-cell communication. Nat. Cell Biol. 2019, 21, 9–17. [Google Scholar] [CrossRef]
- Skotland, T.; Sagini, K.; Sandvig, K.; Llorente, A. An emerging focus on lipids in extracellular vesicles. Adv. Drug Deliv. Rev. 2020, 159, 308–321. [Google Scholar] [CrossRef]
- Verderio, C.; Gabrielli, M.; Giussani, P. Role of sphingolipids in the biogenesis and biological activity of extracellular vesicles. J. Lipid Res. 2018, 59, 1325–1340. [Google Scholar] [CrossRef]
- Skryabin, G.O.; Komelkov, A.V.; Savelyeva, E.E.; Tchevkina, E.M. Lipid Rafts in Exosome Biogenesis. Biochemistry. Biokhimiia 2020, 85, 177–191. [Google Scholar] [CrossRef]
- Egea-Jimenez, A.L.; Zimmermann, P. Lipids in Exosome Biology. Handb Exp. Pharm. 2020, 259, 309–336. [Google Scholar] [CrossRef]
- Egea-Jimenez, A.L.; Zimmermann, P. Phospholipase D and phosphatidic acid in the biogenesis and cargo loading of extracellular vesicles. J. Lipid Res. 2018, 59, 1554–1560. [Google Scholar] [CrossRef]
- Friand, V.; David, G.; Zimmermann, P. Syntenin and syndecan in the biogenesis of exosomes. Biol. Cell 2015, 107, 331–341. [Google Scholar] [CrossRef]
- Wenzel, E.M.; Schultz, S.W.; Schink, K.O.; Pedersen, N.M.; Nähse, V.; Carlson, A.; Brech, A.; Stenmark, H.; Raiborg, C. Concerted ESCRT and clathrin recruitment waves define the timing and morphology of intraluminal vesicle formation. Nat. Commun. 2018, 9, 2932. [Google Scholar] [CrossRef] [PubMed]
- Quinney, K.B.; Frankel, E.B.; Shankar, R.; Kasberg, W.; Luong, P.; Audhya, A. Growth factor stimulation promotes multivesicular endosome biogenesis by prolonging recruitment of the late-acting ESCRT machinery. Proc. Natl. Acad. Sci. USA 2019, 116, 6858–6867. [Google Scholar] [CrossRef] [PubMed]
- Wollert, T.; Hurley, J.H. Molecular mechanism of multivesicular body biogenesis by ESCRT complexes. Nature 2010, 464, 864–869. [Google Scholar] [CrossRef]
- Liese, S.; Wenzel, E.M.; Kjos, I.; Rojas Molina, R.; Schultz, S.W.; Brech, A.; Stenmark, H.; Raiborg, C.; Carlson, A. Protein crowding mediates membrane remodeling in upstream ESCRT-induced formation of intraluminal vesicles. Proc. Natl. Acad. Sci. USA 2020, 117, 28614–28624. [Google Scholar] [CrossRef] [PubMed]
- Adell, M.A.Y.; Migliano, S.M.; Upadhyayula, S.; Bykov, Y.S.; Sprenger, S.; Pakdel, M.; Vogel, G.F.; Jih, G.; Skillern, W.; Behrouzi, R.; et al. Recruitment dynamics of ESCRT-III and Vps4 to endosomes and implications for reverse membrane budding. eLife 2017, 6, e31652. [Google Scholar] [CrossRef]
- Frankel, E.B.; Shankar, R.; Moresco, J.J.; Yates, J.R., 3rd; Volkmann, N.; Audhya, A. Ist1 regulates ESCRT-III assembly and function during multivesicular endosome biogenesis in Caenorhabditis elegans embryos. Nat. Commun. 2017, 8, 1439. [Google Scholar] [CrossRef] [PubMed]
- Blanc, L.; Vidal, M. New insights into the function of Rab GTPases in the context of exosomal secretion. Small GTPases 2018, 9, 95–106. [Google Scholar] [CrossRef]
- Hinger, S.A.; Abner, J.J.; Franklin, J.L.; Jeppesen, D.K.; Coffey, R.J.; Patton, J.G. Rab13 regulates sEV secretion in mutant KRAS colorectal cancer cells. Sci. Rep. 2020, 10, 15804. [Google Scholar] [CrossRef]
- Matsui, T.; Sakamaki, Y.; Nakashima, S.; Fukuda, M. Rab39 and its effector UACA regulate basolateral exosome release from polarized epithelial cells. Cell Rep. 2022, 39, 110875. [Google Scholar] [CrossRef]
- Hyenne, V.; Apaydin, A.; Rodriguez, D.; Spiegelhalter, C.; Hoff-Yoessle, S.; Diem, M.; Tak, S.; Lefebvre, O.; Schwab, Y.; Goetz, J.G.; et al. RAL-1 controls multivesicular body biogenesis and exosome secretion. J. Cell Biol. 2015, 211, 27–37. [Google Scholar] [CrossRef]
- Wandinger-Ness, A.; Zerial, M. Rab proteins and the compartmentalization of the endosomal system. Cold Spring Harb. Perspect. Biol. 2014, 6, a022616. [Google Scholar] [CrossRef]
- Sinha, S.; Hoshino, D.; Hong, N.H.; Kirkbride, K.C.; Grega-Larson, N.E.; Seiki, M.; Tyska, M.J.; Weaver, A.M. Cortactin promotes exosome secretion by controlling branched actin dynamics. J. Cell Biol. 2016, 214, 197–213. [Google Scholar] [CrossRef] [PubMed]
- Beghein, E.; Devriese, D.; Van Hoey, E.; Gettemans, J. Cortactin and fascin-1 regulate extracellular vesicle release by controlling endosomal trafficking or invadopodia formation and function. Sci. Rep. 2018, 8, 15606. [Google Scholar] [CrossRef] [PubMed]
- Matsui, T.; Osaki, F.; Hiragi, S.; Sakamaki, Y.; Fukuda, M. ALIX and ceramide differentially control polarized small extracellular vesicle release from epithelial cells. EMBO Rep. 2021, 22, e51475. [Google Scholar] [CrossRef]
- Davies, B.A.; Morton, L.O.; Jefferson, J.R.; Rozeveld, C.N.; Doskey, L.C.; LaRusso, N.F.; Katzmann, D.J. Polarized human cholangiocytes release distinct populations of apical and basolateral small extracellular vesicles. Mol. Biol. Cell 2020, 31, 2463–2474. [Google Scholar] [CrossRef]
- Fabbiano, F.; Corsi, J.; Gurrieri, E.; Trevisan, C.; Notarangelo, M.; D’Agostino, V.G. RNA packaging into extracellular vesicles: An orchestra of RNA-binding proteins? J. Extracell. Vesicles 2020, 10, e12043. [Google Scholar] [CrossRef]
- Groot, M.; Lee, H. Sorting Mechanisms for MicroRNAs into Extracellular Vesicles and Their Associated Diseases. Cells 2020, 9, 1044. [Google Scholar] [CrossRef] [PubMed]
- Janas, T.; Janas, P.; Sapoń, K.; Janas, T. Binding of RNA Aptamers to Membrane Lipid Rafts: Implications for Exosomal miRNAs Transfer from Cancer to Immune Cells. Int. J. Mol. Sci. 2020, 21, 8503. [Google Scholar] [CrossRef]
- Mańka, R.; Janas, P.; Sapoń, K.; Janas, T.; Janas, T. Role of RNA Motifs in RNA Interaction with Membrane Lipid Rafts: Implications for Therapeutic Applications of Exosomal RNAs. Int. J. Mol. Sci. 2021, 22, 9416. [Google Scholar] [CrossRef]
- Mordovkina, D.; Lyabin, D.N.; Smolin, E.A.; Sogorina, E.M.; Ovchinnikov, L.P.; Eliseeva, I. Y-Box Binding Proteins in mRNP Assembly, Translation, and Stability Control. Biomolecules 2020, 10, 591. [Google Scholar] [CrossRef]
- Liu, X.M.; Ma, L.; Schekman, R. Selective sorting of microRNAs into exosomes by phase-separated YBX1 condensates. eLife 2021, 10, e71982. [Google Scholar] [CrossRef] [PubMed]
- Shurtleff, M.J.; Temoche-Diaz, M.M.; Karfilis, K.V.; Ri, S.; Schekman, R. Y-box protein 1 is required to sort microRNAs into exosomes in cells and in a cell-free reaction. eLife 2016, 5, e19276. [Google Scholar] [CrossRef] [PubMed]
- Kossinova, O.A.; Gopanenko, A.V.; Tamkovich, S.N.; Krasheninina, O.A.; Tupikin, A.E.; Kiseleva, E.; Yanshina, D.D.; Malygin, A.A.; Ven’yaminova, A.G.; Kabilov, M.R.; et al. Cytosolic YB-1 and NSUN2 are the only proteins recognizing specific motifs present in mRNAs enriched in exosomes. Biochim. Biophys. Acta Proteins Proteom. 2017, 1865, 664–673. [Google Scholar] [CrossRef] [PubMed]
- Yanshina, D.D.; Kossinova, O.A.; Gopanenko, A.V.; Krasheninina, O.A.; Malygin, A.A.; Venyaminova, A.G.; Karpova, G.G. Structural features of the interaction of the 3′-untranslated region of mRNA containing exosomal RNA-specific motifs with YB-1, a potential mediator of mRNA sorting. Biochimie 2018, 144, 134–143. [Google Scholar] [CrossRef]
- Shin, Y.; Brangwynne, C.P. Liquid phase condensation in cell physiology and disease. Science 2017, 357. [Google Scholar] [CrossRef] [PubMed]
- Barman, B.; Sung, B.H.; Krystofiak, E.; Ping, J.; Ramirez, M.; Millis, B.; Allen, R.; Prasad, N.; Chetyrkin, S.; Calcutt, M.W.; et al. VAP-A and its binding partner CERT drive biogenesis of RNA-containing extracellular vesicles at ER membrane contact sites. Dev. Cell 2022, 57, 974–994.e8. [Google Scholar] [CrossRef]
- Takahashi, H.; Mayers, J.R.; Wang, L.; Edwardson, J.M.; Audhya, A. Hrs and STAM function synergistically to bind ubiquitin-modified cargoes in vitro. Biophys. J. 2015, 108, 76–84. [Google Scholar] [CrossRef]
- Raiborg, C.; Bremnes, B.; Mehlum, A.; Gillooly, D.J.; D’Arrigo, A.; Stang, E.; Stenmark, H. FYVE and coiled-coil domains determine the specific localisation of Hrs to early endosomes. J. Cell Sci. 2001, 114, 2255–2263. [Google Scholar] [CrossRef]
- Kobayashi, H.; Tanaka, N.; Asao, H.; Miura, S.; Kyuuma, M.; Semura, K.; Ishii, N.; Sugamura, K. Hrs, a mammalian master molecule in vesicular transport and protein sorting, suppresses the degradation of ESCRT proteins signal transducing adaptor molecule 1 and 2. J. Biol. Chem. 2005, 280, 10468–10477. [Google Scholar] [CrossRef]
- Wozniak, A.L.; Adams, A.; King, K.E.; Dunn, W.; Christenson, L.K.; Hung, W.T.; Weinman, S.A. The RNA binding protein FMR1 controls selective exosomal miRNA cargo loading during inflammation. J. Cell Biol. 2020, 219. [Google Scholar] [CrossRef]
- Emerman, A.B.; Blower, M.D. The RNA-binding complex ESCRT-II in Xenopus laevis eggs recognizes purine-rich sequences through its subunit, Vps25. J. Biol. Chem. 2018, 293, 12593–12605. [Google Scholar] [CrossRef] [PubMed]
- Irion, U.; St Johnston, D. bicoid RNA localization requires specific binding of an endosomal sorting complex. Nature 2007, 445, 554–558. [Google Scholar] [CrossRef] [PubMed]
- Bissig, C.; Gruenberg, J. ALIX and the multivesicular endosome: ALIX in Wonderland. Trends Cell Biol. 2014, 24, 19–25. [Google Scholar] [CrossRef] [PubMed]
- Tseng, C.C.; Piper, R.C.; Katzmann, D.J. Bro1 family proteins harmonize cargo sorting with vesicle formation. BioEssays News Rev. Mol. Cell. Dev. Biol. 2022, 44, e2100276. [Google Scholar] [CrossRef] [PubMed]
- Jeppesen, D.K.; Fenix, A.M.; Franklin, J.L.; Higginbotham, J.N.; Zhang, Q.; Zimmerman, L.J.; Liebler, D.C.; Ping, J.; Liu, Q.; Evans, R.; et al. Reassessment of Exosome Composition. Cell 2019, 177, 428–445.e418. [Google Scholar] [CrossRef] [PubMed]
- David, G.; Zimmermann, P. Heparanase Involvement in Exosome Formation. Adv. Exp. Med. Biol. 2020, 1221, 285–307. [Google Scholar] [CrossRef]
- Shimada, T.; Yasuda, S.; Sugiura, H.; Yamagata, K. Syntenin: PDZ Protein Regulating Signaling Pathways and Cellular Functions. Int. J. Mol. Sci. 2019, 20, 4171. [Google Scholar] [CrossRef]
- Latysheva, N.; Muratov, G.; Rajesh, S.; Padgett, M.; Hotchin, N.A.; Overduin, M.; Berditchevski, F. Syntenin-1 is a new component of tetraspanin-enriched microdomains: Mechanisms and consequences of the interaction of syntenin-1 with CD63. Mol. Cell. Biol. 2006, 26, 7707–7718. [Google Scholar] [CrossRef]
- Gräßel, L.; Fast, L.A.; Scheffer, K.D.; Boukhallouk, F.; Spoden, G.A.; Tenzer, S.; Boller, K.; Bago, R.; Rajesh, S.; Overduin, M.; et al. The CD63-Syntenin-1 Complex Controls Post-Endocytic Trafficking of Oncogenic Human Papillomaviruses. Sci. Rep. 2016, 6, 32337. [Google Scholar] [CrossRef]
- Ghossoub, R.; Lembo, F.; Rubio, A.; Gaillard, C.B.; Bouchet, J.; Vitale, N.; Slavík, J.; Machala, M.; Zimmermann, P. Syntenin-ALIX exosome biogenesis and budding into multivesicular bodies are controlled by ARF6 and PLD2. Nat. Commun. 2014, 5, 3477. [Google Scholar] [CrossRef]
- Leonova, E.I.; Galzitskaya, O.V. Structure and functions of syndecans in vertebrates. Biochemistry. Biokhimiia 2013, 78, 1071–1085. [Google Scholar] [CrossRef] [PubMed]
- Stepp, M.A.; Pal-Ghosh, S.; Tadvalkar, G.; Pajoohesh-Ganji, A. Syndecan-1 and Its Expanding List of Contacts. Adv. Wound Care 2015, 4, 235–249. [Google Scholar] [CrossRef] [PubMed]
- Ito, K.; Matsuda, Y.; Mine, A.; Shikida, N.; Takahashi, K.; Miyairi, K.; Shimbo, K.; Kikuchi, Y.; Konishi, A. Single-chain tandem macrocyclic peptides as a scaffold for growth factor and cytokine mimetics. Commun. Biol. 2022, 5, 56. [Google Scholar] [CrossRef] [PubMed]
- Ueki, R.; Atsuta, S.; Ueki, A.; Hoshiyama, J.; Li, J.; Hayashi, Y.; Sando, S. DNA aptamer assemblies as fibroblast growth factor mimics and their application in stem cell culture. Chem. Commun. 2019, 55, 2672–2675. [Google Scholar] [CrossRef] [PubMed]
- Bishani, A.; Chernolovskaya, E.L. Activation of Innate Immunity by Therapeutic Nucleic Acids. Int. J. Mol. Sci. 2021, 22, 3360. [Google Scholar] [CrossRef] [PubMed]
- Bolukbasi, M.F.; Mizrak, A.; Ozdener, G.B.; Madlener, S.; Ströbel, T.; Erkan, E.P.; Fan, J.B.; Breakefield, X.O.; Saydam, O. miR-1289 and “Zipcode”-like Sequence Enrich mRNAs in Microvesicles. Mol. Nucleic. Acids 2012, 1, e10. [Google Scholar] [CrossRef]
- Wang, J.-H.; Forterre, A.V.; Zhao, J.; Frimannsson, D.O.; Delcayre, A.; Antes, T.J.; Efron, B.; Jeffrey, S.S.; Pegram, M.D.; Matin, A.C. Anti-HER2 scFv-Directed Extracellular Vesicle-Mediated mRNA-Based Gene Delivery Inhibits Growth of HER2-Positive Human Breast Tumor Xenografts by Prodrug Activation. Mol. Cancer Ther. 2018, 17, 1133–1142. [Google Scholar] [CrossRef]
- Forterre, A.V.; Wang, J.H.; Delcayre, A.; Kim, K.; Green, C.; Pegram, M.D.; Jeffrey, S.S.; Matin, A.C. Extracellular Vesicle-Mediated In Vitro Transcribed mRNA Delivery for Treatment of HER2(+) Breast Cancer Xenografts in Mice by Prodrug CB1954 without General Toxicity. Mol. Cancer Ther. 2020, 19, 858–867. [Google Scholar] [CrossRef]
- Ikenouchi, J.; Aoki, K. A Clockwork Bleb: Cytoskeleton, calcium, and cytoplasmic fluidity. FEBS J. 2021, 289, 7907–7917. [Google Scholar] [CrossRef]
- Schick, J.; Raz, E. Blebs-Formation, Regulation, Positioning, and Role in Amoeboid Cell Migration. Front. Cell Dev. Biol. 2022, 10, 926394. [Google Scholar] [CrossRef]
- Ebstrup, M.L.; Dias, C.; Heitmann, A.S.B.; Sønder, S.L.; Nylandsted, J. Actin Cytoskeletal Dynamics in Single-Cell Wound Repair. Int. J. Mol. Sci. 2021, 22, 886. [Google Scholar] [CrossRef] [PubMed]
- Leow, R.S.; Wan, J.M.; Yu, A.C. Membrane blebbing as a recovery manoeuvre in site-specific sonoporation mediated by targeted microbubbles. J. R. Soc. Interface 2015, 12, 20150029. [Google Scholar] [CrossRef] [PubMed]
- Mageswaran, S.K.; Yang, W.Y.; Chakrabarty, Y.; Oikonomou, C.M.; Jensen, G.J. A cryo-electron tomography workflow reveals protrusion-mediated shedding on injured plasma membrane. Sci. Adv. 2021, 7, eabc6345. [Google Scholar] [CrossRef] [PubMed]
- Colombo, F.; Bastoni, M.; Nigro, A.; Podini, P.; Finardi, A.; Casella, G.; Ramesh, M.; Farina, C.; Verderio, C.; Furlan, R. Cytokines Stimulate the Release of Microvesicles from Myeloid Cells Independently from the P2X7 Receptor/Acid Sphingomyelinase Pathway. Front. Immunol. 2018, 9, 204. [Google Scholar] [CrossRef]
- Turola, E.; Furlan, R.; Bianco, F.; Matteoli, M.; Verderio, C. Microglial microvesicle secretion and intercellular signaling. Front. Physiol. 2012, 3, 149. [Google Scholar] [CrossRef]
- Rilla, K. Diverse plasma membrane protrusions act as platforms for extracellular vesicle shedding. J. Extracell. Vesicles 2021, 10, e12148. [Google Scholar] [CrossRef]
- Perez, G.I.; Bernard, M.P.; Vocelle, D.; Zarea, A.A.; Saleh, N.A.; Gagea, M.A.; Schneider, D.; Bauzon, M.; Hermiston, T.; Kanada, M. Phosphatidylserine-Exposing Annexin A1-Positive Extracellular Vesicles: Potential Cancer Biomarkers. Vaccines 2023, 11, 639. [Google Scholar] [CrossRef]
- Gabrielli, M.; Prada, I.; Joshi, P.; Falcicchia, C.; D’Arrigo, G.; Rutigliano, G.; Battocchio, E.; Zenatelli, R.; Tozzi, F.; Radeghieri, A.; et al. Microglial large extracellular vesicles propagate early synaptic dysfunction in Alzheimer’s disease. Brain A J. Neurol. 2022, 145, 2849–2868. [Google Scholar] [CrossRef]
- Antonucci, F.; Turola, E.; Riganti, L.; Caleo, M.; Gabrielli, M.; Perrotta, C.; Novellino, L.; Clementi, E.; Giussani, P.; Viani, P.; et al. Microvesicles released from microglia stimulate synaptic activity via enhanced sphingolipid metabolism. EMBO J. 2012, 31, 1231–1240. [Google Scholar] [CrossRef]
- Dar, G.H.; Mendes, C.C.; Kuan, W.L.; Speciale, A.A.; Conceição, M.; Görgens, A.; Uliyakina, I.; Lobo, M.J.; Lim, W.F.; El Andaloussi, S.; et al. GAPDH controls extracellular vesicle biogenesis and enhances the therapeutic potential of EV mediated siRNA delivery to the brain. Nat. Commun. 2021, 12, 6666. [Google Scholar] [CrossRef]
- Taylor, J.; Bebawy, M. Proteins Regulating Microvesicle Biogenesis and Multidrug Resistance in Cancer. Proteomics 2019, 19, e1800165. [Google Scholar] [CrossRef] [PubMed]
- Roseblade, A.; Luk, F.; Rawling, T.; Ung, A.; Grau, G.E.; Bebawy, M. Cell-derived microparticles: New targets in the therapeutic management of disease. J. Pharm. Pharm. Sci. A Publ. Can. Soc. Pharm. Sci. Soc. Can. Des Sci. Pharm. 2013, 16, 238–253. [Google Scholar] [CrossRef]
- Atkin-Smith, G.K.; Poon, I.K.H. Disassembly of the Dying: Mechanisms and Functions. Trends Cell Biol. 2017, 27, 151–162. [Google Scholar] [CrossRef]
- Wang, J.; Zhuang, X.; Greene, K.S.; Si, H.; Antonyak, M.A.; Druso, J.E.; Wilson, K.F.; Cerione, R.A.; Feng, Q.; Wang, H. Cdc42 functions as a regulatory node for tumour-derived microvesicle biogenesis. J. Extracell. Vesicles 2021, 10, e12051. [Google Scholar] [CrossRef] [PubMed]
- Sedgwick, A.E.; Clancy, J.W.; Olivia Balmert, M.; D’Souza-Schorey, C. Extracellular microvesicles and invadopodia mediate non-overlapping modes of tumor cell invasion. Sci. Rep. 2015, 5, 14748. [Google Scholar] [CrossRef] [PubMed]
- Muralidharan-Chari, V.; Clancy, J.; Plou, C.; Romao, M.; Chavrier, P.; Raposo, G.; D’Souza-Schorey, C. ARF6-regulated shedding of tumor cell-derived plasma membrane microvesicles. Curr. Biol. CB 2009, 19, 1875–1885. [Google Scholar] [CrossRef]
- Clancy, J.W.; Tricarico, C.J.; Marous, D.R.; D’Souza-Schorey, C. Coordinated Regulation of Intracellular Fascin Distribution Governs Tumor Microvesicle Release and Invasive Cell Capacity. Mol. Cell. Biol. 2019, 39, e00264-18. [Google Scholar] [CrossRef]
- Clancy, J.W.; Schmidtmann, M.; D’Souza-Schorey, C. The ins and outs of microvesicles. FASEB BioAdv. 2021, 3, 399–406. [Google Scholar] [CrossRef]
- Clancy, J.W.; Sedgwick, A.; Rosse, C.; Muralidharan-Chari, V.; Raposo, G.; Method, M.; Chavrier, P.; D’Souza-Schorey, C. Regulated delivery of molecular cargo to invasive tumour-derived microvesicles. Nat. Commun. 2015, 6, 6919. [Google Scholar] [CrossRef]
- Clancy, J.W.; Zhang, Y.; Sheehan, C.; D’Souza-Schorey, C. An ARF6-Exportin-5 axis delivers pre-miRNA cargo to tumour microvesicles. Nat. Cell Biol. 2019, 21, 856–866. [Google Scholar] [CrossRef]
- Lee, H.; Li, C.; Zhang, Y.; Zhang, D.; Otterbein, L.E.; Jin, Y. Caveolin-1 selectively regulates microRNA sorting into microvesicles after noxious stimuli. J. Exp. Med. 2019, 216, 2202–2220. [Google Scholar] [CrossRef] [PubMed]
- Lee, H.; Carnino, J.M.; Jin, Y. Caveolin-1 regulates extracellular vesicle-miRNA packaging. Aging 2019, 11, 8733–8735. [Google Scholar] [CrossRef] [PubMed]
- Ni, K.; Wang, C.; Carnino, J.M.; Jin, Y. The Evolving Role of Caveolin-1: A Critical Regulator of Extracellular Vesicles. Med. Sci. 2020, 8, 46. [Google Scholar] [CrossRef] [PubMed]
- Wang, Q.; Yu, J.; Kadungure, T.; Beyene, J.; Zhang, H.; Lu, Q. ARMMs as a versatile platform for intracellular delivery of macromolecules. Nat. Commun. 2018, 9, 960. [Google Scholar] [CrossRef]
- Kuo, L.; Freed, E.O. ARRDC1 as a mediator of microvesicle budding. Proc. Natl. Acad. Sci. USA 2012, 109, 4025–4026. [Google Scholar] [CrossRef] [PubMed]
- Rauch, S.; Martin-Serrano, J. Multiple interactions between the ESCRT machinery and arrestin-related proteins: Implications for PPXY-dependent budding. J. Virol. 2011, 85, 3546–3556. [Google Scholar] [CrossRef]
- Nabhan, J.F.; Hu, R.; Oh, R.S.; Cohen, S.N.; Lu, Q. Formation and release of arrestin domain-containing protein 1-mediated microvesicles (ARMMs) at plasma membrane by recruitment of TSG101 protein. Proc. Natl. Acad. Sci. USA 2012, 109, 4146–4151. [Google Scholar] [CrossRef]
- Mackenzie, K.; Foot, N.J.; Anand, S.; Dalton, H.E.; Chaudhary, N.; Collins, B.M.; Mathivanan, S.; Kumar, S. Regulation of the divalent metal ion transporter via membrane budding. Cell Discov. 2016, 2, 16011. [Google Scholar] [CrossRef]
- Farooq, A.U.; Gembus, K.; Sandow, J.J.; Webb, A.; Mathivanan, S.; Manning, J.A.; Shah, S.S.; Foot, N.J.; Kumar, S. K-29 linked ubiquitination of Arrdc4 regulates its function in extracellular vesicle biogenesis. J. Extracell. Vesicles 2022, 11, e12188. [Google Scholar] [CrossRef]
- Kanada, M.; Bachmann, M.H.; Hardy, J.W.; Frimannson, D.O.; Bronsart, L.; Wang, A.; Sylvester, M.D.; Schmidt, T.L.; Kaspar, R.L.; Butte, M.J.; et al. Differential fates of biomolecules delivered to target cells via extracellular vesicles. Proc. Natl. Acad. Sci. USA 2015, 112, E1433–E1442. [Google Scholar] [CrossRef]
- Kanada, M.; Kim, B.D.; Hardy, J.W.; Ronald, J.A.; Bachmann, M.H.; Bernard, M.P.; Perez, G.I.; Zarea, A.A.; Ge, T.J.; Withrow, A.; et al. Microvesicle-Mediated Delivery of Minicircle DNA Results in Effective Gene-Directed Enzyme Prodrug Cancer Therapy. Mol. Cancer Ther. 2019, 18, 2331–2342. [Google Scholar] [CrossRef] [PubMed]
- D’Arcy, M.S. Cell death: A review of the major forms of apoptosis, necrosis and autophagy. Cell Biol. Int. 2019, 43, 582–592. [Google Scholar] [CrossRef] [PubMed]
- Xu, X.; Lai, Y.; Hua, Z.C. Apoptosis and apoptotic body: Disease message and therapeutic target potentials. Biosci. Rep. 2019, 39, BSR20180992. [Google Scholar] [CrossRef]
- Galluzzi, L.; Vitale, I.; Aaronson, S.A.; Abrams, J.M.; Adam, D.; Agostinis, P.; Alnemri, E.S.; Altucci, L.; Amelio, I.; Andrews, D.W.; et al. Molecular mechanisms of cell death: Recommendations of the Nomenclature Committee on Cell Death 2018. Cell Death Differ. 2018, 25, 486–541. [Google Scholar] [CrossRef] [PubMed]
- Battistelli, M.; Falcieri, E. Apoptotic Bodies: Particular Extracellular Vesicles Involved in Intercellular Communication. Biology 2020, 9, 21. [Google Scholar] [CrossRef] [PubMed]
- Santavanond, J.P.; Rutter, S.F.; Atkin-Smith, G.K.; Poon, I.K.H. Apoptotic Bodies: Mechanism of Formation, Isolation and Functional Relevance. In New Frontiers: Extracellular Vesicles; Mathivanan, S., Fonseka, P., Nedeva, C., Atukorala, I., Eds.; Springer International Publishing: Cham, Switzerland, 2021; pp. 61–88. [Google Scholar]
- Moss, D.K.; Betin, V.M.; Malesinski, S.D.; Lane, J.D. A novel role for microtubules in apoptotic chromatin dynamics and cellular fragmentation. J. Cell Sci. 2006, 119, 2362–2374. [Google Scholar] [CrossRef]
- Tixeira, R.; Phan, T.K.; Caruso, S.; Shi, B.; Atkin-Smith, G.K.; Nedeva, C.; Chow, J.D.Y.; Puthalakath, H.; Hulett, M.D.; Herold, M.J.; et al. ROCK1 but not LIMK1 or PAK2 is a key regulator of apoptotic membrane blebbing and cell disassembly. Cell Death Differ. 2020, 27, 102–116. [Google Scholar] [CrossRef]
- Suzuki, J.; Denning, D.P.; Imanishi, E.; Horvitz, H.R.; Nagata, S. Xk-related protein 8 and CED-8 promote phosphatidylserine exposure in apoptotic cells. Science 2013, 341, 403–406. [Google Scholar] [CrossRef]
- Suzuki, J.; Imanishi, E.; Nagata, S. Xkr8 phospholipid scrambling complex in apoptotic phosphatidylserine exposure. Proc. Natl. Acad. Sci. USA 2016, 113, 9509–9514. [Google Scholar] [CrossRef]
- Sakuragi, T.; Kanai, R.; Tsutsumi, A.; Narita, H.; Onishi, E.; Nishino, K.; Miyazaki, T.; Baba, T.; Kosako, H.; Nakagawa, A.; et al. The tertiary structure of the human Xkr8-Basigin complex that scrambles phospholipids at plasma membranes. Nat. Struct. Mol. Biol. 2021, 28, 825–834. [Google Scholar] [CrossRef]
- Pontejo, S.M.; Murphy, P.M. Chemokines act as phosphatidylserine-bound "find-me" signals in apoptotic cell clearance. PLoS Biol. 2021, 19, e3001259. [Google Scholar] [CrossRef] [PubMed]
- Caruso, S.; Poon, I.K.H. Apoptotic Cell-Derived Extracellular Vesicles: More Than Just Debris. Front. Immunol. 2018, 9, 1486. [Google Scholar] [CrossRef] [PubMed]
- Jiang, L.; Paone, S.; Caruso, S.; Atkin-Smith, G.K.; Phan, T.K.; Hulett, M.D.; Poon, I.K.H. Determining the contents and cell origins of apoptotic bodies by flow cytometry. Sci. Rep. 2017, 7, 14444. [Google Scholar] [CrossRef] [PubMed]
- Phan, T.K.; Fonseka, P.; Tixeira, R.; Pathan, M.; Ang, C.S.; Ozkocak, D.C.; Mathivanan, S.; Poon, I.K.H. Pannexin-1 channel regulates nuclear content packaging into apoptotic bodies and their size. Proteomics 2021, 21, e2000097. [Google Scholar] [CrossRef]
- Phan, T.K.; Ozkocak, D.C.; Poon, I.K.H. Unleashing the therapeutic potential of apoptotic bodies. Biochem. Soc. Trans. 2020, 48, 2079–2088. [Google Scholar] [CrossRef]
- Kakarla, R.; Hur, J.; Kim, Y.J.; Kim, J.; Chwae, Y.J. Apoptotic cell-derived exosomes: Messages from dying cells. Exp. Mol. Med. 2020, 52, 1–6. [Google Scholar] [CrossRef] [PubMed]
- Beillevaire, D.; Migneault, F.; Turgeon, J.; Gingras, D.; Rimbaud, A.K.; Marcoux, G.; Spino, C.; Thibodeau, N.; Bonneil, E.; Thibault, P.; et al. Autolysosomes and caspase-3 control the biogenesis and release of immunogenic apoptotic exosomes. Cell Death Dis. 2022, 13, 145. [Google Scholar] [CrossRef] [PubMed]
- Baxter, A.A.; Phan, T.K.; Hanssen, E.; Liem, M.; Hulett, M.D.; Mathivanan, S.; Poon, I.K.H. Analysis of extracellular vesicles generated from monocytes under conditions of lytic cell death. Sci. Rep. 2019, 9, 7538. [Google Scholar] [CrossRef] [PubMed]
- Giri, K.R.; de Beaurepaire, L.; Jegou, D.; Lavy, M.; Mosser, M.; Dupont, A.; Fleurisson, R.; Dubreil, L.; Collot, M.; Van Endert, P.; et al. Molecular and Functional Diversity of Distinct Subpopulations of the Stressed Insulin-Secreting Cell’s Vesiculome. Front. Immunol. 2020, 11, 1814. [Google Scholar] [CrossRef]
- Liu, M.L.; Reilly, M.P.; Casasanto, P.; McKenzie, S.E.; Williams, K.J. Cholesterol enrichment of human monocyte/macrophages induces surface exposure of phosphatidylserine and the release of biologically-active tissue factor-positive microvesicles. Arter. Thromb Vasc. Biol. 2007, 27, 430–435. [Google Scholar] [CrossRef]
- Skovronova, R.; Grange, C.; Dimuccio, V.; Deregibus, M.C.; Camussi, G.; Bussolati, B. Surface Marker Expression in Small and Medium/Large Mesenchymal Stromal Cell-Derived Extracellular Vesicles in Naive or Apoptotic Condition Using Orthogonal Techniques. Cells 2021, 10, 2948. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Pang, J.; Wang, Q.; Yan, L.; Wang, L.; Xing, Z.; Wang, C.; Zhang, J.; Dong, L. Delivering Antisense Oligonucleotides across the Blood-Brain Barrier by Tumor Cell-Derived Small Apoptotic Bodies. Adv. Sci. 2021, 8, 2004929. [Google Scholar] [CrossRef] [PubMed]
- Tang, K.; Zhang, Y.; Zhang, H.; Xu, P.; Liu, J.; Ma, J.; Lv, M.; Li, D.; Katirai, F.; Shen, G.X.; et al. Delivery of chemotherapeutic drugs in tumour cell-derived microparticles. Nat. Commun. 2012, 3, 1282. [Google Scholar] [CrossRef] [PubMed]
- Ma, J.; Zhang, Y.; Tang, K.; Zhang, H.; Yin, X.; Li, Y.; Xu, P.; Sun, Y.; Ma, R.; Ji, T.; et al. Reversing drug resistance of soft tumor-repopulating cells by tumor cell-derived chemotherapeutic microparticles. Cell Res. 2016, 26, 713–727. [Google Scholar] [CrossRef]
- Gao, Y.; Zhang, H.; Zhou, N.; Xu, P.; Wang, J.; Gao, Y.; Jin, X.; Liang, X.; Lv, J.; Zhang, Y.; et al. Methotrexate-loaded tumour-cell-derived microvesicles can relieve biliary obstruction in patients with extrahepatic cholangiocarcinoma. Nat. Biomed. Eng. 2020, 4, 743–753. [Google Scholar] [CrossRef]
- Zhou, M.; Li, Y.J.; Tang, Y.C.; Hao, X.Y.; Xu, W.J.; Xiang, D.X.; Wu, J.Y. Apoptotic bodies for advanced drug delivery and therapy. J. Control. Release Off. J. Control. Release Soc. 2022, 351, 394–406. [Google Scholar] [CrossRef]
- Liu, J.; Qiu, X.; Lv, Y.; Zheng, C.; Dong, Y.; Dou, G.; Zhu, B.; Liu, A.; Wang, W.; Zhou, J.; et al. Apoptotic bodies derived from mesenchymal stem cells promote cutaneous wound healing via regulating the functions of macrophages. Stem Cell Res. Ther. 2020, 11, 507. [Google Scholar] [CrossRef]
- Dou, G.; Tian, R.; Liu, X.; Yuan, P.; Ye, Q.; Liu, J.; Liu, S.; Zhou, J.; Deng, Z.; Chen, X.; et al. Chimeric apoptotic bodies functionalized with natural membrane and modular delivery system for inflammation modulation. Sci. Adv. 2020, 6, eaba2987. [Google Scholar] [CrossRef]
- Bao, L.; Dou, G.; Tian, R.; Lv, Y.; Ding, F.; Liu, S.; Zhao, R.; Zhao, L.; Zhou, J.; Weng, L.; et al. Engineered neutrophil apoptotic bodies ameliorate myocardial infarction by promoting macrophage efferocytosis and inflammation resolution. Bioact. Mater. 2022, 9, 183–197. [Google Scholar] [CrossRef]
- Oshchepkova, A.; Markov, O.; Evtushenko, E.; Chernonosov, A.; Kiseleva, E.; Morozova, K.; Matveeva, V.; Artemyeva, L.; Vlassov, V.; Zenkova, M. Tropism of Extracellular Vesicles and Cell-Derived Nanovesicles to Normal and Cancer Cells: New Perspectives in Tumor-Targeted Nucleic Acid Delivery. Pharmaceutics 2021, 13, 1911. [Google Scholar] [CrossRef]
- Termini, C.M.; Gillette, J.M. Tetraspanins Function as Regulators of Cellular Signaling. Front. Cell Dev. Biol. 2017, 5, 34. [Google Scholar] [CrossRef] [PubMed]
- Horodecka, K.; Düchler, M. CRISPR/Cas9: Principle, Applications, and Delivery through Extracellular Vesicles. Int. J. Mol. Sci. 2021, 22, 6072. [Google Scholar] [CrossRef] [PubMed]
- Ye, Y.; Zhang, X.; Xie, F.; Xu, B.; Xie, P.; Yang, T.; Shi, Q.; Zhang, C.Y.; Zhang, Y.; Chen, J.; et al. An engineered exosome for delivering sgRNA:Cas9 ribonucleoprotein complex and genome editing in recipient cells. Biomater. Sci. 2020, 8, 2966–2976. [Google Scholar] [CrossRef] [PubMed]
- Osteikoetxea, X.; Silva, A.; Lázaro-Ibáñez, E.; Salmond, N.; Shatnyeva, O.; Stein, J.; Schick, J.; Wren, S.; Lindgren, J.; Firth, M.; et al. Engineered Cas9 extracellular vesicles as a novel gene editing tool. J. Extracell. Vesicles 2022, 11, e12225. [Google Scholar] [CrossRef]
- Li, Z.; Zhou, X.; Wei, M.; Gao, X.; Zhao, L.; Shi, R.; Sun, W.; Duan, Y.; Yang, G.; Yuan, L. In Vitro and in Vivo RNA Inhibition by CD9-HuR Functionalized Exosomes Encapsulated with miRNA or CRISPR/dCas9. Nano Lett. 2019, 19, 19–28. [Google Scholar] [CrossRef]
- Es-Haghi, M.; Neustroeva, O.; Chowdhury, I.; Laitinen, P.; Väänänen, M.A.; Korvenlaita, N.; Malm, T.; Turunen, M.P.; Turunen, T.A. Construction of Fusion Protein for Enhanced Small RNA Loading to Extracellular Vesicles. Genes 2023, 14, 261. [Google Scholar] [CrossRef]
- Durán-Jara, E.; Vera-Tobar, T.; Lobos-González, L.L. Lactadherin: From a Well-Known Breast Tumor Marker to a Possible Player in Extracellular Vesicle-Mediated Cancer Progression. Int. J. Mol. Sci. 2022, 23, 3855. [Google Scholar] [CrossRef]
- Li, Q.; Hu, W.; Huang, Q.; Yang, J.; Li, B.; Ma, K.; Wei, Q.; Wang, Y.; Su, J.; Sun, M.; et al. MiR146a-loaded engineered exosomes released from silk fibroin patch promote diabetic wound healing by targeting IRAK1. Signal Transduct. Target. Ther. 2023, 8, 62. [Google Scholar] [CrossRef]
- Si, K.; Dai, Z.; Li, Z.; Ye, Z.; Ding, B.; Feng, S.; Sun, B.; Shen, Y.; Xiao, Z. Engineered exosome-mediated messenger RNA and single-chain variable fragment delivery for human chimeric antigen receptor T-cell engineering. Cytotherapy 2023, in press. [Google Scholar] [CrossRef]
- Elsharkasy, O.M.; Nordin, J.Z.; Hagey, D.W.; de Jong, O.G.; Schiffelers, R.M.; Andaloussi, S.E.; Vader, P. Extracellular vesicles as drug delivery systems: Why and how? Adv. Drug Deliv. Rev. 2020, 159, 332–343. [Google Scholar] [CrossRef]
- Lamichhane, T.N.; Jeyaram, A.; Patel, D.B.; Parajuli, B.; Livingston, N.K.; Arumugasaamy, N.; Schardt, J.S.; Jay, S.M. Oncogene Knockdown via Active Loading of Small RNAs into Extracellular Vesicles by Sonication. Cell Mol. Bioeng. 2016, 9, 315–324. [Google Scholar] [CrossRef]
- Biscans, A.; Haraszti, R.A.; Echeverria, D.; Miller, R.; Didiot, M.C.; Nikan, M.; Roux, L.; Aronin, N.; Khvorova, A. Hydrophobicity of Lipid-Conjugated siRNAs Predicts Productive Loading to Small Extracellular Vesicles. Mol. Ther. 2018, 26, 1520–1528. [Google Scholar] [CrossRef]
- Xu, H.; Liao, C.; Liang, S.; Ye, B.C. A Novel Peptide-Equipped Exosomes Platform for Delivery of Antisense Oligonucleotides. ACS Appl. Mater. Interfaces 2021, 13, 10760–10767. [Google Scholar] [CrossRef] [PubMed]
- Didiot, M.-C.; Hall, L.M.; Coles, A.H.; Haraszti, R.A.; Godinho, B.M.; Chase, K.; Sapp, E.; Ly, S.; Alterman, J.F.; Hassler, M.R.; et al. Exosome-mediated Delivery of Hydrophobically Modified siRNA for Huntingtin mRNA Silencing. Mol. Ther. 2016, 24, 1836–1847. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Wu, J.; Wu, J.; Fan, Q.; Zhou, J.; Wu, J.; Liu, S.; Zang, J.; Ye, J.; Xiao, M.; et al. Exosome-mediated targeted delivery of miR-210 for angiogenic therapy after cerebral ischemia in mice. J. Nanobiotechnol. 2019, 17, 29. [Google Scholar] [CrossRef] [PubMed]
- Chernikov, I.V.; Vlassov, V.V.; Chernolovskaya, E.L. Current Development of siRNA Bioconjugates: From Research to the Clinic. Front. Pharmacol. 2019, 10, 444. [Google Scholar] [CrossRef] [PubMed]
- Osborn, M.F.; Khvorova, A. Improving siRNA Delivery In Vivo Through Lipid Conjugation. Nucleic Acid Ther. 2018, 28, 128–136. [Google Scholar] [CrossRef]
- Biscans, A.; Coles, A.; Haraszti, R.; Echeverria, D.; Hassler, M.; Osborn, M.; Khvorova, A. Diverse lipid conjugates for functional extra-hepatic siRNA delivery in vivo. Nucleic Acids Res. 2019, 47, 1082–1096. [Google Scholar] [CrossRef]
- Chernikov, I.V.; Gladkikh, D.V.; Meschaninova, M.I.; Ven’yaminova, A.G.; Zenkova, M.A.; Vlassov, V.V.; Chernolovskaya, E.L. Cholesterol-Containing Nuclease-Resistant siRNA Accumulates in Tumors in a Carrier-free Mode and Silences MDR1 Gene. Mol. Nucleic Acids 2017, 6, 209–220. [Google Scholar] [CrossRef]
- Biscans, A.; Caiazzi, J.; Davis, S.; McHugh, N.; Sousa, J.; Khvorova, A. The chemical structure and phosphorothioate content of hydrophobically modified siRNAs impact extrahepatic distribution and efficacy. Nucleic Acids Res. 2020, 48, 7665–7680. [Google Scholar] [CrossRef]
- Haraszti, R.A.; Miller, R.; Didiot, M.-C.; Biscans, A.; Alterman, J.F.; Hassler, M.R.; Roux, L.; Echeverria, D.; Sapp, E.; DiFiglia, M.; et al. Optimized Cholesterol-siRNA Chemistry Improves Productive Loading onto Extracellular Vesicles. Mol. Ther. 2018, 26, 1973–1982. [Google Scholar] [CrossRef] [PubMed]
- Lamichhane, T.N.; Raiker, R.S.; Jay, S.M. Exogenous DNA Loading into Extracellular Vesicles via Electroporation is Size-Dependent and Enables Limited Gene Delivery. Mol. Pharm. 2015, 12, 3650–3657. [Google Scholar] [CrossRef] [PubMed]
- Izco, M.; Blesa, J.; Schleef, M.; Schmeer, M.; Porcari, R.; Al-Shawi, R.; Ellmerich, S.; de Toro, M.; Gardiner, C.; Seow, Y.; et al. Systemic Exosomal Delivery of shRNA Minicircles Prevents Parkinsonian Pathology. Mol. Ther. 2019, 27, 2111–2122. [Google Scholar] [CrossRef] [PubMed]
- Usman, W.M.; Pham, T.C.; Kwok, Y.Y.; Vu, L.T.; Ma, V.; Peng, B.; Chan, Y.S.; Wei, L.; Chin, S.M.; Azad, A.; et al. Efficient RNA drug delivery using red blood cell extracellular vesicles. Nat. Commun. 2018, 9, 2359. [Google Scholar] [CrossRef] [PubMed]
- Aqil, F.; Munagala, R.; Jeyabalan, J.; Agrawal, A.K.; Kyakulaga, A.H.; Wilcher, S.A.; Gupta, R.C. Milk exosomes—Natural nanoparticles for siRNA delivery. Cancer Lett. 2019, 449, 186–195. [Google Scholar] [CrossRef]
- Shandilya, S.; Rani, P.; Onteru, S.K.; Singh, D. Natural ligand-receptor mediated loading of siRNA in milk derived exosomes. J. Biotechnol. 2020, 318, 1–9. [Google Scholar] [CrossRef]
- Ramanathan, S.; Shenoda, B.B.; Lin, Z.; Alexander, G.M.; Huppert, A.; Sacan, A.; Ajit, S.K. Inflammation potentiates miR-939 expression and packaging into small extracellular vesicles. J. Extracell. Vesicles 2019, 8, 1650595. [Google Scholar] [CrossRef]
- Pomatto, M.A.C.; Bussolati, B.; D’Antico, S.; Ghiotto, S.; Tetta, C.; Brizzi, M.F.; Camussi, G. Improved Loading of Plasma-Derived Extracellular Vesicles to Encapsulate Antitumor miRNAs. Molecular therapy. Methods Clin. Dev. 2019, 13, 133–144. [Google Scholar] [CrossRef]
- Kooijmans, S.A.A.; Stremersch, S.; Braeckmans, K.; de Smedt, S.C.; Hendrix, A.; Wood, M.J.A.; Schiffelers, R.M.; Raemdonck, K.; Vader, P. Electroporation-induced siRNA precipitation obscures the efficiency of siRNA loading into extracellular vesicles. J. Control. Release Off. J. Control. Release Soc. 2013, 172, 229–238. [Google Scholar] [CrossRef]
- Reshke, R.; Taylor, J.A.; Savard, A.; Guo, H.; Rhym, L.H.; Kowalski, P.S.; Trung, M.T.; Campbell, C.; Little, W.; Anderson, D.G.; et al. Reduction of the therapeutic dose of silencing RNA by packaging it in extracellular vesicles via a pre-microRNA backbone. Nat. Biomed. Eng. 2020, 4, 52–68. [Google Scholar] [CrossRef]
- Johnsen, K.B.; Gudbergsson, J.M.; Skov, M.N.; Christiansen, G.; Gurevich, L.; Moos, T.; Duroux, M. Evaluation of electroporation-induced adverse effects on adipose-derived stem cell exosomes. Cytotechnology 2016, 68, 2125–2138. [Google Scholar] [CrossRef] [PubMed]
- Zhang, D.; Lee, H.; Zhu, Z.; Minhas, J.K.; Jin, Y. Enrichment of selective miRNAs in exosomes and delivery of exosomal miRNAs in vitro and in vivo. Am. J. Physiol. Lung Cell Mol. Physiol. 2017, 312, L110–L121. [Google Scholar] [CrossRef] [PubMed]
- Zhang, D.; Lee, H.; Wang, X.; Rai, A.; Groot, M.; Jin, Y. Exosome-Mediated Small RNA Delivery: A Novel Therapeutic Approach for Inflammatory Lung Responses. Mol. Ther. 2018, 26, 2119–2130. [Google Scholar] [CrossRef] [PubMed]
- Asif, A.; Mohsin, H.; Tanvir, R.; Rehman, Y. Revisiting the Mechanisms Involved in Calcium Chloride Induced Bacterial Transformation. Front. Microbiol. 2017, 8, 2169. [Google Scholar] [CrossRef]
- Rezaei, R.; Baghaei, K.; Amani, D.; Piccin, A.; Hashemi, S.M.; Asadzadeh Aghdaei, H.; Zali, M.R. Exosome-mediated delivery of functionally active miRNA-375-3p mimic regulate epithelial mesenchymal transition (EMT) of colon cancer cells. Life Sci. 2021, 269, 119035. [Google Scholar] [CrossRef] [PubMed]
- Rezaei, R.; Baghaei, K.; Hashemi, S.M.; Zali, M.R.; Ghanbarian, H.; Amani, D. Tumor-Derived Exosomes Enriched by miRNA-124 Promote Anti-tumor Immune Response in CT-26 Tumor-Bearing Mice. Front. Med. 2021, 8, 619939. [Google Scholar] [CrossRef]
- Jamur, M.C.; Oliver, C. Permeabilization of cell membranes. Methods Mol. Biol. 2010, 588, 63–66. [Google Scholar] [CrossRef]
- Sutaria, D.S.; Badawi, M.; Phelps, M.A.; Schmittgen, T.D. Achieving the Promise of Therapeutic Extracellular Vesicles: The Devil is in Details of Therapeutic Loading. Pharm. Res. 2017, 34, 1053–1066. [Google Scholar] [CrossRef]
- Chen, H.; Wang, L.; Zeng, X.; Schwarz, H.; Nanda, H.S.; Peng, X.; Zhou, Y. Exosomes, a New Star for Targeted Delivery. Front. Cell Dev. Biol. 2021, 9, 751079. [Google Scholar] [CrossRef]
- Oshchepkova, A.; Neumestova, A.; Matveeva, V.; Artemyeva, L.; Morozova, K.; Kiseleva, E.; Zenkova, M.; Vlassov, V. Cytochalasin-B-Inducible Nanovesicle Mimics of Natural Extracellular Vesicles That Are Capable of Nucleic Acid Transfer. Micromachines 2019, 10, 750. [Google Scholar] [CrossRef]
- Qiao, H.; Hu, N.; Bai, J.; Ren, L.; Liu, Q.; Fang, L.; Wang, Z. Encapsulation of Nucleic Acids into Giant Unilamellar Vesicles by Freeze-Thaw: A Way Protocells May Form. Orig. Life Evol. Biosph. J. Int. Soc. Study Orig. Life 2017, 47, 499–510. [Google Scholar] [CrossRef]
- Zhuang, J.; Tan, J.; Wu, C.; Zhang, J.; Liu, T.; Fan, C.; Li, J.; Zhang, Y. Extracellular vesicles engineered with valency-controlled DNA nanostructures deliver CRISPR/Cas9 system for gene therapy. Nucleic Acids Res. 2020, 48, 8870–8882. [Google Scholar] [CrossRef] [PubMed]
- Gelibter, S.; Marostica, G.; Mandelli, A.; Siciliani, S.; Podini, P.; Finardi, A.; Furlan, R. The impact of storage on extracellular vesicles: A systematic study. J. Extracell. Vesicles 2022, 11, e12162. [Google Scholar] [CrossRef] [PubMed]
- Wu, J.Y.; Li, Y.J.; Hu, X.B.; Huang, S.; Xiang, D.X. Preservation of small extracellular vesicles for functional analysis and therapeutic applications: A comparative evaluation of storage conditions. Drug Deliv. 2021, 28, 162–170. [Google Scholar] [CrossRef] [PubMed]
- Bosch, S.; de Beaurepaire, L.; Allard, M.; Mosser, M.; Heichette, C.; Chrétien, D.; Jegou, D.; Bach, J.M. Trehalose prevents aggregation of exosomes and cryodamage. Sci. Rep. 2016, 6, 36162. [Google Scholar] [CrossRef] [PubMed]
- Görgens, A.; Corso, G.; Hagey, D.W.; Jawad Wiklander, R.; Gustafsson, M.O.; Felldin, U.; Lee, Y.; Bostancioglu, R.B.; Sork, H.; Liang, X.; et al. Identification of storage conditions stabilizing extracellular vesicles preparations. J. Extracell. Vesicles 2022, 11, e12238. [Google Scholar] [CrossRef]
- Claridge, B.; Lozano, J.; Poh, Q.H.; Greening, D.W. Development of Extracellular Vesicle Therapeutics: Challenges, Considerations, and Opportunities. Front. Cell Dev. Biol. 2021, 9, 734720. [Google Scholar] [CrossRef]
- Bertolino, G.M.; Maumus, M.; Jorgensen, C.; Noël, D. Recent Advances in Extracellular Vesicle-Based Therapies Using Induced Pluripotent Stem Cell-Derived Mesenchymal Stromal Cells. Biomedicines 2022, 10, 2281. [Google Scholar] [CrossRef]
- Buzas, E.I. The roles of extracellular vesicles in the immune system. Nature reviews. Immunology 2022, 23, 236–250. [Google Scholar] [CrossRef]
- Kou, M.; Huang, L.; Yang, J.; Chiang, Z.; Chen, S.; Liu, J.; Guo, L.; Zhang, X.; Zhou, X.; Xu, X.; et al. Mesenchymal stem cell-derived extracellular vesicles for immunomodulation and regeneration: A next generation therapeutic tool? Cell Death Dis. 2022, 13, 580. [Google Scholar] [CrossRef]
- Dehghani, L.; Khojasteh, A.; Soleimani, M.; Oraee-Yazdani, S.; Keshel, S.H.; Saadatnia, M.; Saboori, M.; Zali, A.; Hashemi, S.M.; Soleimani, R. Safety of Intraparenchymal Injection of Allogenic Placenta Mesenchymal Stem Cells Derived Exosome in Patients Undergoing Decompressive Craniectomy Following Malignant Middle Cerebral Artery Infarct, A Pilot Randomized Clinical Trial. Int. J. Prev. Med. 2022, 13, 7. [Google Scholar] [CrossRef] [PubMed]
- Hade, M.D.; Suire, C.N.; Suo, Z. An Effective Peptide-Based Platform for Efficient Exosomal Loading and Cellular Delivery of a microRNA. ACS Appl Mater Interfaces 2023, 15, 3851–3866. [Google Scholar] [CrossRef] [PubMed]
- Ducrot, C.; Loiseau, S.; Wong, C.; Madec, E.; Volatron, J.; Piffoux, M. Hybrid extracellular vesicles for drug delivery. Cancer Lett. 2023, 558, 216107. [Google Scholar] [CrossRef] [PubMed]
- Shtam, T.A.; Kovalev, R.A.; Varfolomeeva, E.Y.; Makarov, E.M.; Kil, Y.V.; Filatov, M.V. Exosomes are natural carriers of exogenous siRNA to human cells in vitro. Cell Commun. Signal 2013, 11, 88. [Google Scholar] [CrossRef] [PubMed]
- Wahlgren, J.; De L Karlson, T.; Brisslert, M.; Vaziri Sani, F.; Telemo, E.; Sunnerhagen, P.; Valadi, H. Plasma exosomes can deliver exogenous short interfering RNA to monocytes and lymphocytes. Nucleic Acids Res. 2012, 40, e130. [Google Scholar] [CrossRef] [PubMed]
- Lin, Y.; Wu, J.; Gu, W.; Huang, Y.; Tong, Z.; Huang, L.; Tan, J. Exosome-Liposome Hybrid Nanoparticles Deliver CRISPR/Cas9 System in MSCs. Adv. Sci. 2018, 5, 1700611. [Google Scholar] [CrossRef] [PubMed]
| NCT Number | Trial Phase | Conditions | Cargo | Status | Location |
|---|---|---|---|---|---|
| NCT03608631 | I | Pancreatic cancer with KrasG12D mutation | KrasG12D siRNA | Recruiting | United States |
| NCT03384433 | I, II | Cerebrovascular disorders | miRNA-124 | Unknown | Iran |
| NCT05043181 | I | Homozygous familial hypercholesterolemia (HoFH) | Low-density lipoprotein receptor (ldlr)-encoding mRNA | Not yet recruiting | China |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Oshchepkova, A.; Zenkova, M.; Vlassov, V. Extracellular Vesicles for Therapeutic Nucleic Acid Delivery: Loading Strategies and Challenges. Int. J. Mol. Sci. 2023, 24, 7287. https://doi.org/10.3390/ijms24087287
Oshchepkova A, Zenkova M, Vlassov V. Extracellular Vesicles for Therapeutic Nucleic Acid Delivery: Loading Strategies and Challenges. International Journal of Molecular Sciences. 2023; 24(8):7287. https://doi.org/10.3390/ijms24087287
Chicago/Turabian StyleOshchepkova, Anastasiya, Marina Zenkova, and Valentin Vlassov. 2023. "Extracellular Vesicles for Therapeutic Nucleic Acid Delivery: Loading Strategies and Challenges" International Journal of Molecular Sciences 24, no. 8: 7287. https://doi.org/10.3390/ijms24087287
APA StyleOshchepkova, A., Zenkova, M., & Vlassov, V. (2023). Extracellular Vesicles for Therapeutic Nucleic Acid Delivery: Loading Strategies and Challenges. International Journal of Molecular Sciences, 24(8), 7287. https://doi.org/10.3390/ijms24087287







