Characterization of PcSTT3B as a Key Oligosaccharyltransferase Subunit Involved in N-glycosylation and Its Role in Development and Pathogenicity of Phytophthora capsici
Abstract
:1. Introduction
2. Results
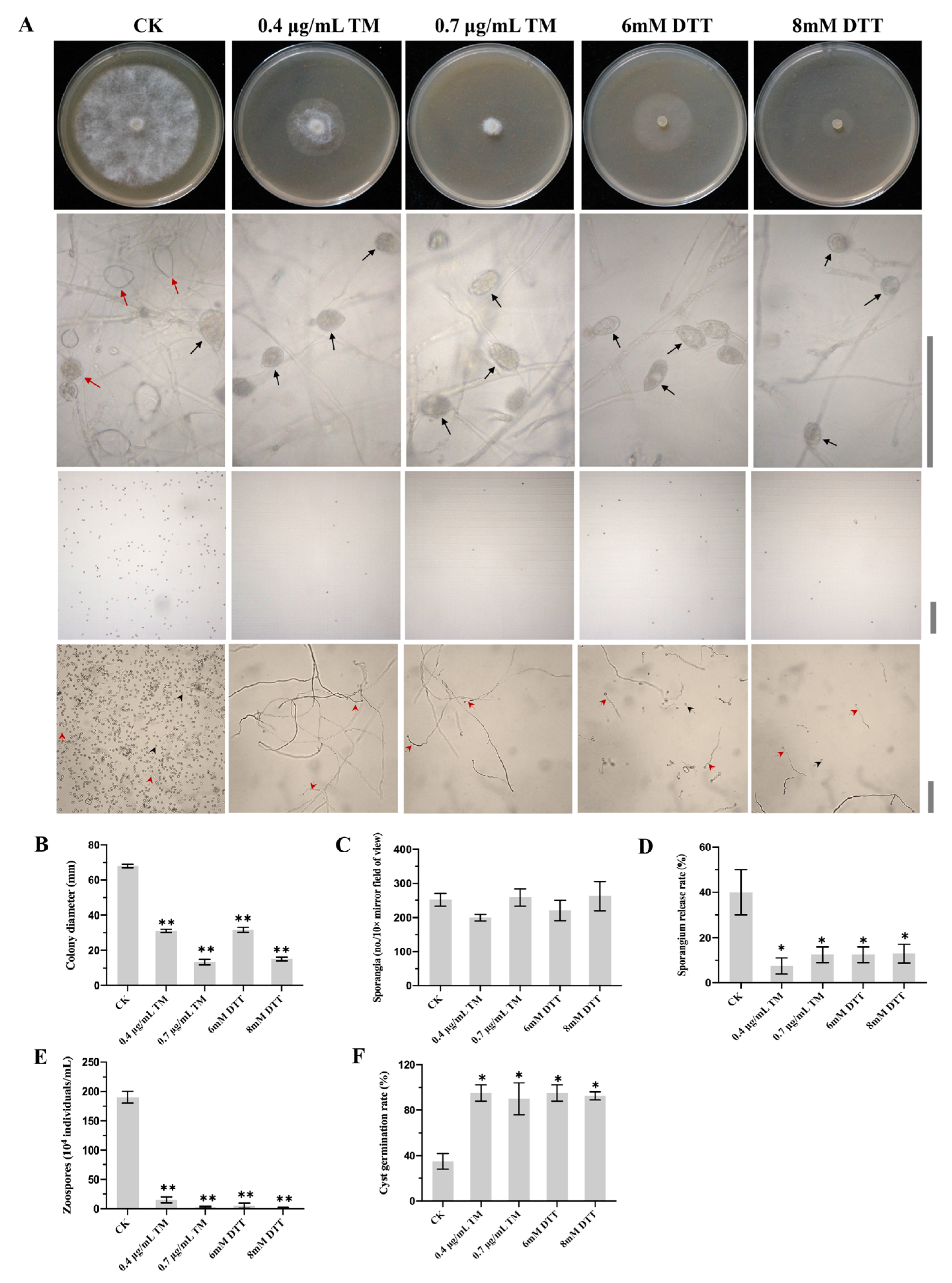
2.1. The Inhibition of TM and DTT in P. capsici
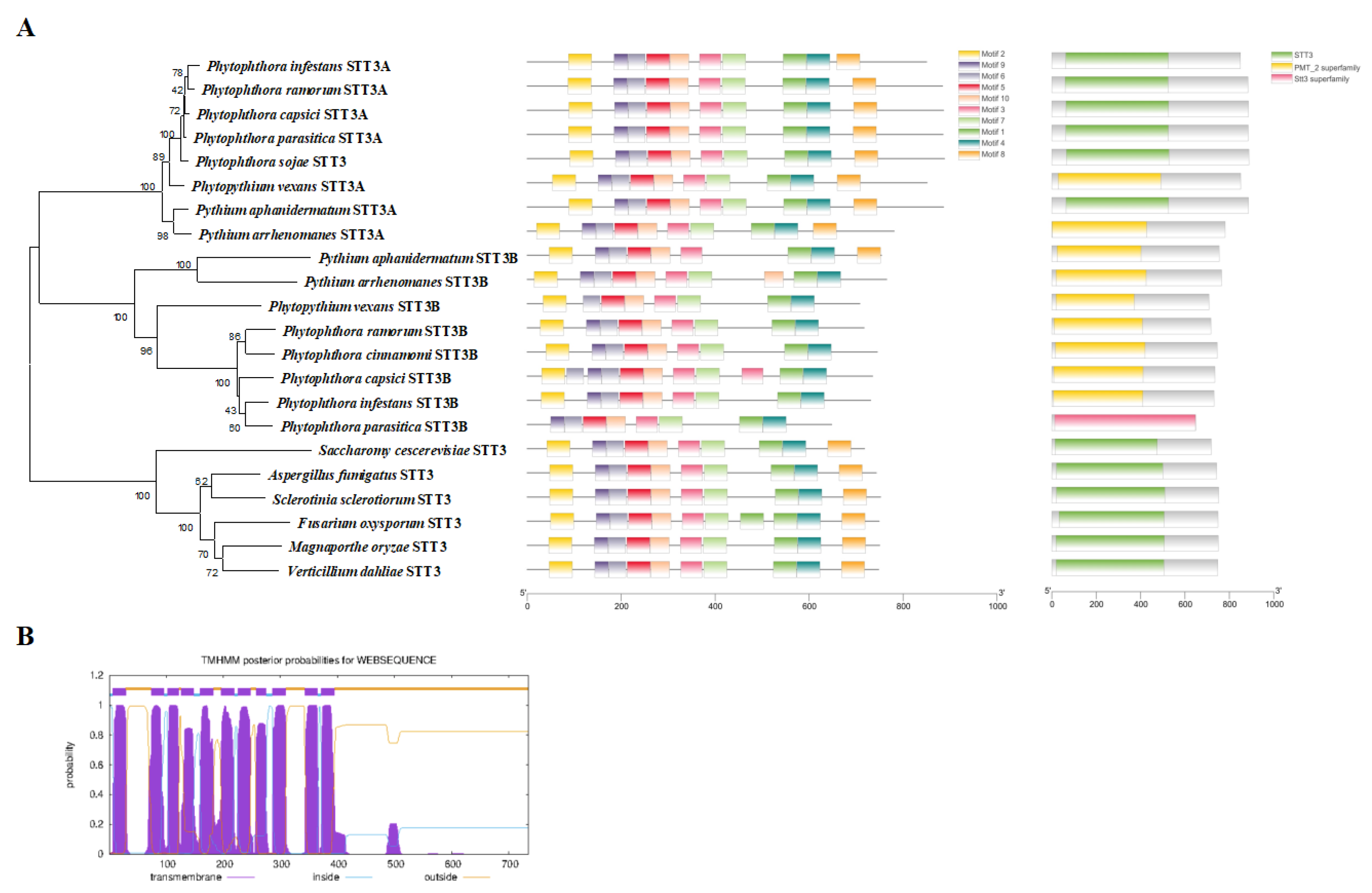
2.2. Sequence and Phylogenetic Analysis of PcSTT3B
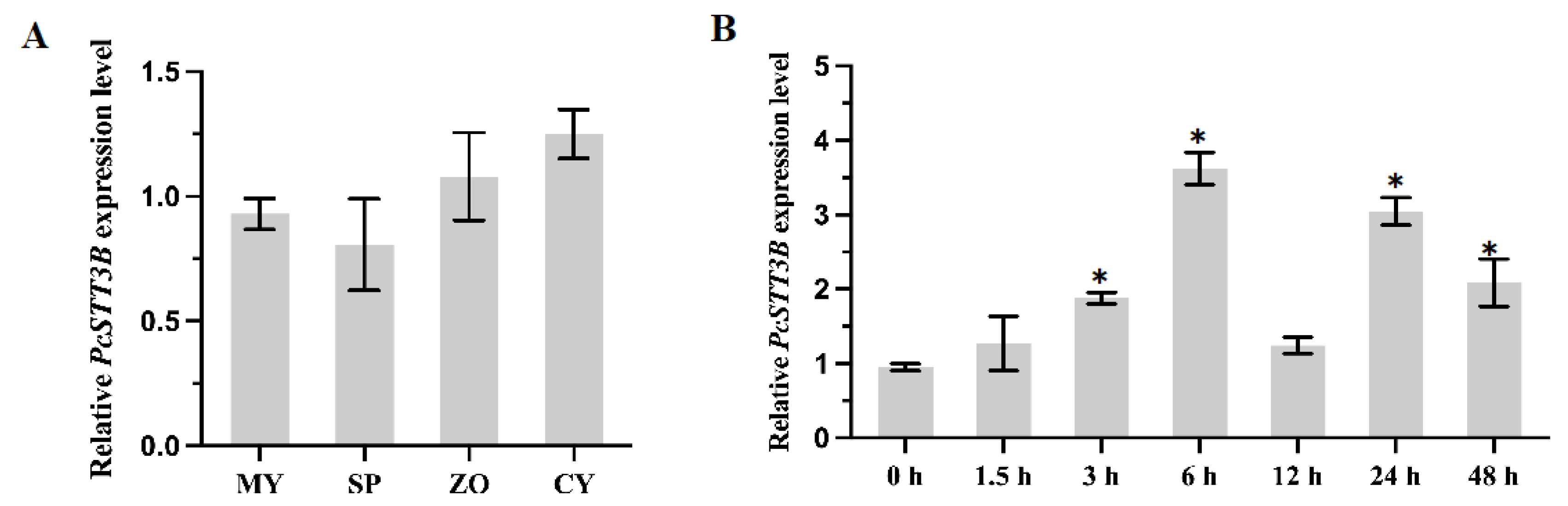
2.3. Transcription Profile of the PcSTT3B Gene
2.4. PcSTT3B Gene Disruption and Complementation
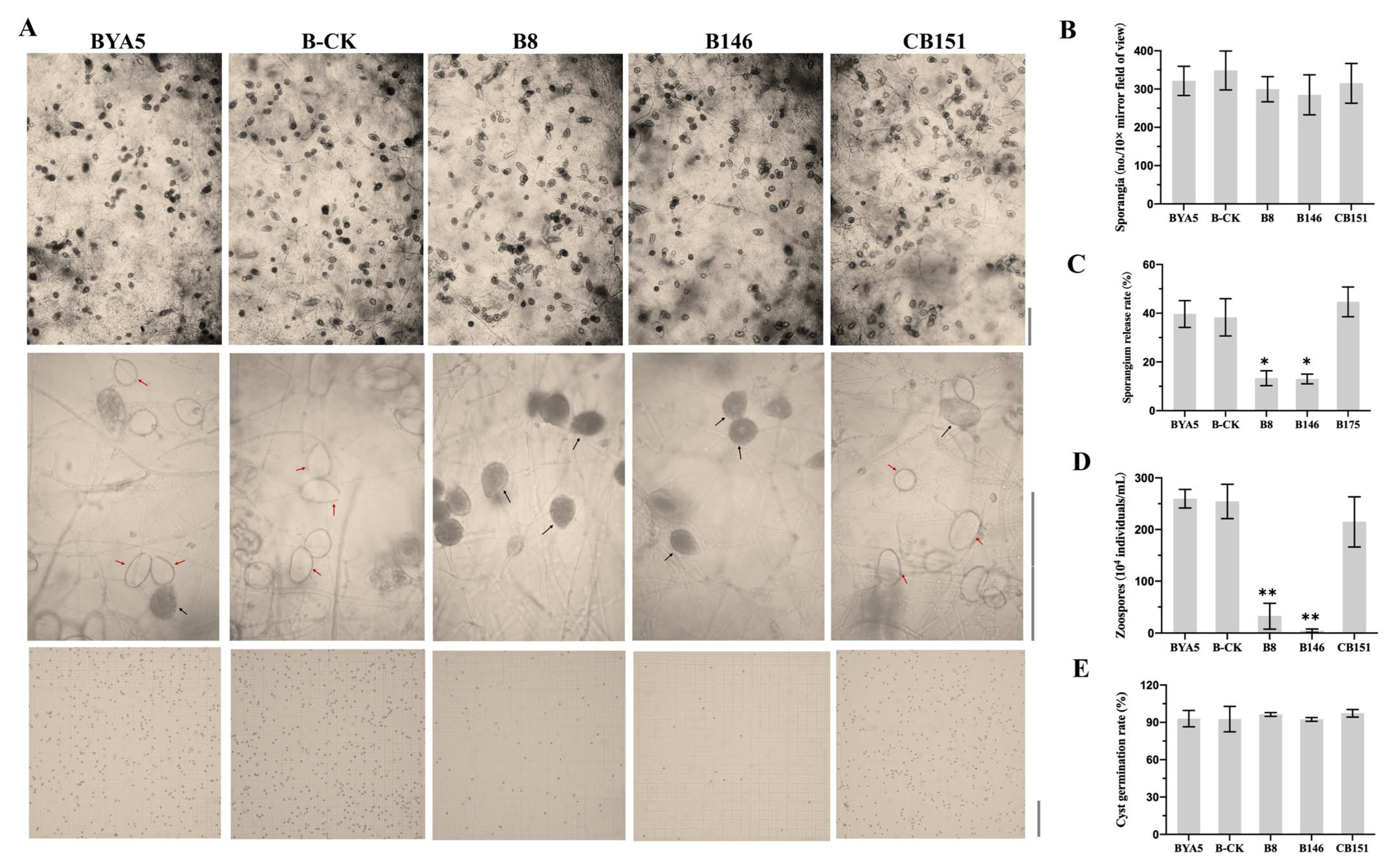
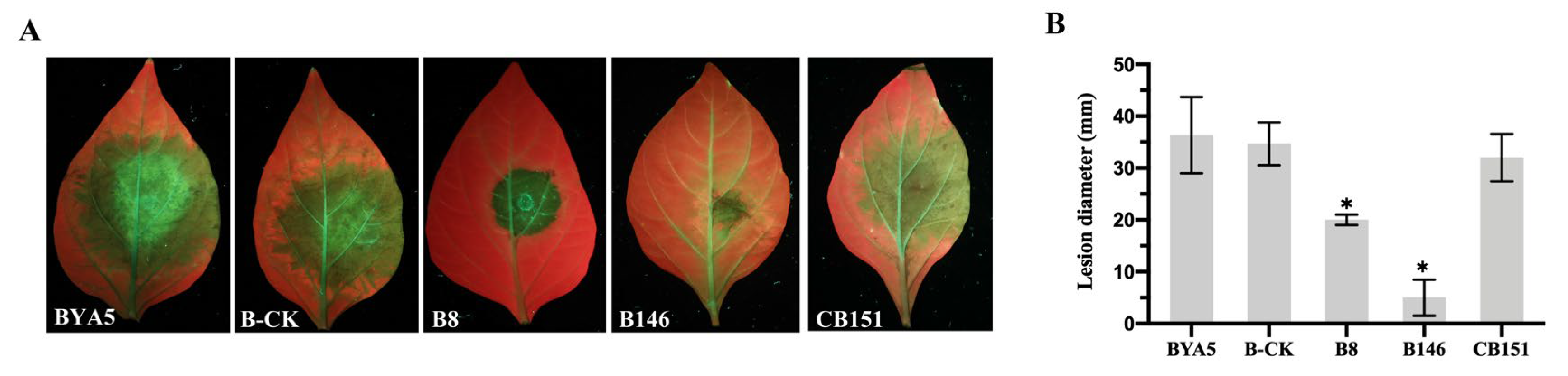
2.5. Biological Characteristics of the PcSTT3B-Deleted Transformants
2.6. Responses of PcSTT3B to ER Stress in P. capsici
2.7. Disruption of PcSTT3B Abolished the Attachment of Glycoproteins in Mycelium
3. Discussion
4. Materials and Methods
4.1. Pathogen and Plant
4.2. Analysis of the PcSTT3B Protein
4.3. Vector Construction
4.4. P. capsici Transformation
4.5. Transcription Profile of the PcSTT3B Gene
4.6. Phenotypic Characterization of the PsSTT3B-Deleted Transformants
4.7. Tolerance of the PcSTT3B-Deleted Transformants to ER Stress Inducers
4.8. Western Blotting
4.9. Data Statistics
5. Conclusions and Perspective
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Kamoun, S.; Furzer, O.; Jones, J.D.G.; Judelson, H.S.; Ali, G.S.; Dalio, R.J.D.; Roy, S.G.; Schena, L.; Zambounis, A.; Panabières, F.; et al. The Top 10 oomycete pathogens in molecular plant pathology. Mol. Plant Pathol. 2015, 16, 413–434. [Google Scholar] [CrossRef] [PubMed]
- Derevnina, L.; Petre, B.; Kellner, R.; Dagdas, Y.F.; Sarowar, M.N.; Giannakopoulou, A.; De la Concepcion, J.C.; Chaparro-Garcia, A.; Pennington, H.G.; Van West, P.; et al. Emerging oomycete threats to plants and animals. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2015, 371, 20150459. [Google Scholar] [CrossRef] [PubMed]
- Fry, W.E.; Birch, P.R.J.; Judelson, H.S.; Griinwald, N.J.; Danies, G.; Everts, K.L.; Gevens, A.J.; Gugino, B.K.; Johnson, D.A.; Johnson, S.B.; et al. Five reasons to consider Phytophthora infestans a reemerging pathogen. Phytopathology 2015, 105, 966–981. [Google Scholar] [CrossRef]
- Judelson, H.S.; Blanco, F.A. The spores of Phytophthora: Weapons of the plant destroyer. Nat. Rev. Microbiol. 2005, 3, 47–58. [Google Scholar] [CrossRef] [PubMed]
- Granke, L.L.; Quesada-Ocampo, L.; Lamour, K.; Hausbeck, M.K. Advances in research on Phytophthora capsici on vegetable crops in the united states. Plant Dis. 2012, 96, 1588–1600. [Google Scholar] [CrossRef]
- Lamour, K.H.; Stam, R.; Jupe, J.; Huitema, E. The oomycete broad-host-range pathogen Phytophthora capsici. Mol. Plant Pathol. 2012, 13, 329–337. [Google Scholar] [CrossRef]
- Feng, B.; Li, P.; Wang, H.; Zhang, X. Functional analysis of pcpme6 from oomycete plant pathogen Phytophthora capsici. Microb. Pathog. 2010, 49, 23–31. [Google Scholar] [CrossRef]
- Li, P.; Feng, B.; Wang, H.; Tooley, P.W.; Zhang, X. Isolation of nine Phytophthora capsici pectin methylesterase genes which are differentially expressed in various plant species. J. Basic Microbiol. 2011, 51, 61–70. [Google Scholar] [CrossRef]
- Ristaino, J.B.; Johnston, S.A. Ecologically based approaches to management of Phytophthora blight on bell pepper. Plant Dis. 1999, 83, 1080–1089. [Google Scholar] [CrossRef]
- Helenius, A. Intracellular functions of N-linked glycans. Science 2001, 291, 2364–2369. [Google Scholar] [CrossRef]
- Helenius, A.; Aebi, M. Roles of N-linked glycans in the endoplasmic reticulum. Annu. Rev. Biochem. 2004, 73, 1019–1049. [Google Scholar] [CrossRef]
- Takatsuki, A.; Arima, K.; Tamura, G. Tunicamycin, a new antibiotic. I. Isolation and characterization of tunicamycin. J. Antibiot. 1971, 24, 215–223. [Google Scholar] [CrossRef]
- Elbein, A.D. Inhibitors of the biosynthesis and processing of N-linked oligosaccharide chains. Ann. Rev. Biochem. 1987, 56, 497–534. [Google Scholar] [CrossRef]
- Wild, R.; Kowal, J.; Eyring, J.; Ngwa, E.M.; Aebi, M.; Locher, K.P. Structure of the yeast oligosaccharyltransferase complex gives insight into eukaryotic N-glycosylation. Science 2018, 359, 545–550. [Google Scholar] [CrossRef]
- Bai, L.; Wang, T.; Zhao, G.P. The atomic structure of a eukaryotic oligosaccharyltransferase complex. Nature 2018, 555, 328–332. [Google Scholar] [CrossRef]
- Hebert, D.N.; Garman, S.C.; Molinari, M. The glycan code of the endoplasmic reticulum: Asparagine-linked carbohydrates as protein maturation and quality-control tags. Trends Cell Biol. 2005, 15, 364–370. [Google Scholar] [CrossRef]
- Schirawski, J.; Bohnert, H.U.; Steinberg, G.; Snetselaar, K.; Adamikowa, L.; Kahmann, R. Endoplasmic reticulum glucosidase II is required for pathogenicity of Ustilago maydis. Plant Cell 2005, 17, 3532–3543. [Google Scholar] [CrossRef]
- Braunger, K.; Pfeffer, S.; Shrimal, S.; Gilmore, R.; Berninghausen, O.; Mandon, E.C.; Becker, T.; Förster, F.; Beckmann, R. Structural basis for coupling protein transport and N-glycosylation at the mammalian endoplasmic reticulum. Science 2018, 360, 215–219. [Google Scholar] [CrossRef]
- Lu, H.; Fermaintt, C.S.; Cherepanova, N.A.; Gilmore, R.; Yan, N.; Lehrman, M.A. Mammalian STT3A/B oligosaccharyltransferases segregate N-glycosylation at the translocon from lipid-linked oligosaccharide hydrolysis. Proc. Natl. Acad. Sci. USA 2018, 115, 9557–9562. [Google Scholar] [CrossRef]
- Shrimal, S.; Cherepanova, N.A.; Gilmore, R. Cotranslational and posttranslocational N-glycosylation of proteins in the endoplasmic reticulum. Semin. Cell Dev. Biol. 2015, 41, 71–78. [Google Scholar] [CrossRef]
- Cherepanova, N.A.; Venev, S.V.; Leszyk, J.D.; Shaffer, S.A.; Gilmore, R. Quantitative glycoproteomics reveals new classes of STT3A- and STT3B-dependent N-glycosylation sites. J. Cell Biol. 2019, 218, 2782–2796. [Google Scholar] [CrossRef] [PubMed]
- Koiwa, H.; Li, F.; McCully, M.G.; Mendoza, I.; Koizumi, N.; Manabe, Y.; Nakagawa, Y.; Zhu, J.H.; Rus, A.; Pardo, J.M.; et al. The STT3a subunit isoform of the Arabidopsis thaliana oligosaccharyltransferase controls adaptive responses to salt/osmotic stress. Plant Cell 2003, 15, 2273–2284. [Google Scholar] [CrossRef] [PubMed]
- Li, K.; Ouyang, H.; Lü, Y.; Liang, J.; Wilson, I.B.H.; Jin, C. Repression of N-glycosylation triggers the unfolded protein response (UPR) and overexpression of cell wall protein and chitin in Aspergillus fumigatus. Microbiology 2011, 157, 1968–1979. [Google Scholar] [CrossRef] [PubMed]
- Su, X.; Rehman, L.; Guo, H.; Li, X.; Cheng, H. The oligosaccharyl transferase subunit STT3 mediates fungal development and is required for virulence in Verticillium dahliae. Curr. Genet. 2018, 64, 235–246. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.L.; Liu, C.; Tang, B.; Ren, Z.; Wang, G.L.; Liu, W. Quantitative proteomics analysis reveals important roles of N-glycosylation on ER quality control system for development and pathogenesis in Magnaporthe oryzae. PLoS Pathog. 2020, 16, e1008355. [Google Scholar] [CrossRef]
- Cleland, W.W. Dithiothreitol, a New Protective Reagent for SH Groups. Biochemistry 1964, 3, 480–482. [Google Scholar] [CrossRef]
- Huntemann, M.; Ivanova, N.N.; Mavromatis, K.; Tripp, H.J.; Paez-Espino, D.; Palaniappan, K.; Szeto, E.; Pillay, M.; Chen, I.M.; Pati, A.; et al. The standard operating procedure of the DOE-JGI Microbial Genome Annotation Pipeline (MGAP v.4). Stand. Genomic Sci. 2015, 10, 86. [Google Scholar] [CrossRef]
- Lu, S.; Wang, J.Y.; Chitsaz, F.; Derbyshire, K.M.; Geer, C.R.; Gonzales, R.N.; Gwadz, M.; Hurwitz, I.D.; Marchler, H.G.; Song, S.J.; et al. CDD/SPARCLE: The conserved domain database in 2020. Nucleic Acids Res. 2020, 48, D265–D268. [Google Scholar] [CrossRef]
- Lizak, C.; Gerber, S.; Numao, S.; Aebi, M.; Locher, K.P. X-ray structure of a bacterial oligosaccharyltransferase. Nature 2011, 474, 350–355. [Google Scholar] [CrossRef]
- Fang, Y.; Cui, L.; Gu, B.; Arredondo, F.; Tyler, B.M. Efficient genome editing in the oomycete Phytophthora sojae using CRISPR/Cas9. Curr. Protoc. Microbiol. 2017, 44, 21A.1.1–21A.1.26. [Google Scholar] [CrossRef]
- Fang, Y.; Tyler, B.M. Efficient disruption and replacement of an effector gene in the oomycete Phytophthora sojae using CRISPR/Cas9. Mol. Plant Pathol. 2016, 17, 127–139. [Google Scholar] [CrossRef]
- Wang, W.Z.; Xue, Z.L.; Miao, J.Q.; Cai, M.; Zhang, C.; Li, T.J.; Zhang, B.R.; Tyler, B.M.; Liu, X.L. PcMuORP1, an OxathiapiprolinResistance Gene, Functions as a Novel Selection Marker for Phytophthora Transformation and CRISPR/Cas9 Mediated Genome Editing. Front. Microbiol. 2019, 10, 2042. [Google Scholar]
- Doyle, R.J.; Birdsell, D.C. Interaction of concanavalin a with the cell wall of Bacillus subtilis. J. Bacteriol. 1972, 109, 652–658. [Google Scholar] [CrossRef]
- Zhang, C.; Cai, M.; Chen, S.S.; Zhang, F.; Cui, T.S.; Xue, Z.L.; Wang, W.Z.; Zhang, B.R.; Liu, X.L. The consensus Nglyco-X-S/T motif and a previously unknown Nglyco-N-linked glycosylation are necessary for growth and pathogenicity of Phytophthora. Environ. Microbiol. 2021, 23, 5147–5163. [Google Scholar] [CrossRef]
- Mohanty, S.; Chaudhary, B.P.; Zoetewey, D. Structural Insight into the Mechanism of N-Linked Glycosylation by Oligosaccharyltransferase. Biomolecules 2020, 10, 624. [Google Scholar] [CrossRef]
- Samuelson, J.; Banerjee, S.; Magnelli, P.; Cui, J.; Kelleher, D.J.; Gilmore, R.; Robbins, P.W. The diversity of dolichol-linked precursors to Asn-linked glycans likely results from secondary loss of sets of glycosyltransferases. Proc. Natl. Acad. Sci. USA 2005, 102, 1548–1553. [Google Scholar] [CrossRef]
- Castilho, A.; Beihammer, G.; Pfeiffer, C.; Göritzer, K.; Montero-Morales, L.; Vavra, U.; Maresch, D.; Grünwald-Gruber, C.; Altmann, F.; Steinkellner, H.; et al. An oligosaccharyltransferase from Leishmania major increases the N-glycan occupancy on recombinant glycoproteins produced in Nicotiana benthamiana. Plant Biotechnol. J. 2018, 16, 1700–1709. [Google Scholar] [CrossRef]
- Izquierdo, L.; Atrih, A.; Rodrigues, J.; Jones, D.; Ferguson, M. Trypanosoma brucei UDP-glucose: Glycoprotein glucosyltransferase has unusual substrate specificity and protects the parasite from stress. Eukaryot. Cell 2009, 8, 230–240. [Google Scholar] [CrossRef]
- Nasab, F.P.; Schulz, B.L.; Gamarro, F.; Parodi, A.J.; Aebi, M. All in one: Leishmania major STT3 proteins substitute for the whole oligosaccharyltransferase complex in Saccharomyces cerevisiae. Mol. Biol. Cell 2018, 19, 3758–3768. [Google Scholar] [CrossRef]
- Häweker, H.; Rips, S.; Koiwa, H.; Salomon, S.; Saijo, Y.; Chinchilla, D.; Robatzek, S.; von Schaewen, A. Pattern recognition receptors require N-glycosylation to mediate plant immunity. J. Biol. Chem. 2010, 285, 4629–4636. [Google Scholar] [CrossRef]
- In, S.J.; Sangmin, L.; Florian, B.; Jordan, T.; Akihito, F.; Yukihiro, N.; Kimberly, M.; Stephan, R.; Sang, Y.L.; Patrick, G.; et al. Purification and characterization of Arabidopsis thaliana oligosaccharyltransferase complexes from the native host: A protein super-expression system for structural studies. Plant J. 2018, 94, 131–145. [Google Scholar]
- Yan, A.X.; Lennarz, W.J. Two oligosaccharyltransferase complexes exist in yeast and associate with two different translocons. Glycobiology 2005, 15, 1407–1415. [Google Scholar] [CrossRef]
- Shrimal, S.; Trueman, S.F.; Gilmore, R. Extreme C-terminal sites are posttranslocationally glycosylated by the STT3B isoform of the OST. J. Cell Biol. 2013, 201, 81–95. [Google Scholar] [CrossRef] [PubMed]
- Kornfeld, R.; Kornfeld, S. Assembly of asparagine-linked oligosaccharides. Annu. Rev. Biochem. 1985, 54, 631–664. [Google Scholar] [CrossRef] [PubMed]
- Chavan, M.; Lennarz, W. The molecular basis of coupling of translocation and N-glycosylation. Trends Biochem. Sci. 2006, 31, 17–20. [Google Scholar] [CrossRef] [PubMed]
- Kelleher, D.J.; Gilmore, R. An evolving view of the eukaryotic oligosaccharyltransferase. Glycobiology 2006, 16, 47–62. [Google Scholar] [CrossRef]
- Mueller, S.; Wahlander, A.; Selevsek, N.; Otto, C.; Ngwa, E.M.; Poljak, K.; Frey, A.D.; Aebi, M.; Gauss, R. Protein degradation corrects for imbalanced subunit stoichiometry in OST complex assembly. Mol. Biol. Cell 2015, 26, 2596–2608. [Google Scholar] [CrossRef]
- Jiao, Q.; Chen, T.; Niu, G.; Zhang, H.; Zhou, C.F.; Hong, Z. N-glycosylation is involved in stomatal development by modulating the release of active abscisic acid and auxin in arabidopsis. J. Exp. Bot. 2020, 71, 5865–5879. [Google Scholar] [CrossRef]
- Xie, Q.; Chen, A.; Zheng, W.; Xu, H.; Shang, W.; Zheng, H.; Zhang, D.; Zhou, J.; Lu, G.; Li, G.; et al. Endosomal sorting complexes required for transport is essential for fungal development and pathogenicity in Fusarium graminearum. Environ. Microbiol. 2016, 18, 3742–3757. [Google Scholar] [CrossRef]
- Liu, Z.; Wu, S.; Chen, Y.; Han, X.; Gu, Q.; Yin, Y.; Ma, Z. The microtubule end-binding protein FgEB1 regulates polar growth and fungicide sensitivity via different interactors in Fusarium graminearum. Environ. Microbiol. 2017, 19, 1791–1807. [Google Scholar] [CrossRef]
- Li, T.J.; Cai, M.; Wang, W.Z.; Dai, T.; Zhang, C.; Zhang, B.R.; Shen, J.H.; Wang, Y.K.; Liu, X.L. PcCesA1 is involved in the polar growth, cellulose synthesis, and glycosidic linkage crosslinking in the cell wall of Phytophthora capsici. Int. J. Biol. Macromol. 2022, 208, 720–730. [Google Scholar] [CrossRef] [PubMed]
- Soussillane, P.; D’Alessio, C.; Paccalet, T.; Fitchette, A.C.; Parodi, A.J.; Williamson, R.; Plasson, C.; Faye, L.; Gomord, V. N-glycan trimming by glucosidase II is essential for Arabidopsis development. Glycoconj. J. 2009, 26, 597–607. [Google Scholar] [CrossRef]
- Martin, S.; Chang, J.S.; Kaufman, R.J. The unfolded protein response represses nitrogen-starvation induced developmental differentiation in yeast. Genes Dev. 2001, 14, 2962–2975. [Google Scholar]
- Stolz, A.; Wolf, D.H. Endoplasmic reticulum associated protein degradation: A chaperone assisted journey to hell. Biochim. Biophys. Acta 2010, 1803, 694–705. [Google Scholar] [CrossRef] [PubMed]
- Kong, Z.W.; Zhang, X.; Zhou, F.; Tang, L.; Chen, Y.T.; Li, S.J.; Zhang, X.K.; Kuai, L.T.; Su, W.J.; Cui, W.R.; et al. Structure-Aided Identification of an Inhibitor Targets Mps1 for the Management of Plant-Pathogenic Fungi. Mbio 2023, e0288322. [Google Scholar] [CrossRef]
- Miller, P.M. V8 juice agar as a general-purpose medium for fungi and bacteria. Phytopathology 1955, 45, 461–462. [Google Scholar]
- Stajich, J.E.; Harris, T.; Brunk, B.P.; Brestelli, J.; Fischer, S.; Harb, O.S.; Kissinger, J.C.; Li, W.; Nayak, V.; Pinney, D.F.; et al. FungiDB: An integrated functional genomics database for fungi. Nucleic Acids Res. 2012, 40, 675–681. [Google Scholar] [CrossRef]
- Tamura, K.; Stecher, G.; Peterson, D.; Filipski, A.; Kumar, S. Mega6: Molecular evolutionary genetics analysis version 6.0. Mol. Biol. Evol. 2013, 30, 2725–2729. [Google Scholar] [CrossRef]
- Bailey, T.L.; Boden, M.; Buske, F.A.; Frith, M.; Grant, C.E.; Clementi, L.; Ren, J.; Li, W.W.; Noble, W.S. MEME SUITE: Tools for motif discovery and searching. Nucleic Acids Res. 2009, 37, 202–208. [Google Scholar] [CrossRef]
- Bailey, T.L.; Johnson, J.; Grant, C.E.; Noble, W.S. The MEME Suite. Nucleic Acids Res. 2015, 43, 39–40. [Google Scholar] [CrossRef]
- Ye, W.; Wang, X.; Tao, K.; Lu, Y.; Dai, T.; Dong, S.; Dou, D.; Gijzen, M.; Wang, Y. Digital gene expression profiling of the Phytophthora sojae transcriptome. Mol. Plant Microbe Interact. 2011, 24, 1530–1539. [Google Scholar] [CrossRef]
- Yan, H.Z.; Liou, R.F. Selection of internal control genes for Real-Time quantitative RT-PCR assays in the oomycete plant pathogen Phytophthora parasitica. Fungal Genet. Biol. 2006, 43, 430–438. [Google Scholar] [CrossRef]
- Pang, Z.; Shao, J.; Chen, L.; Lu, X.; Hu, J.; Qin, Z.; Liu, X. Resistance to the novel fungicide pyrimorph in Phytophthora capsici: Risk assessment and detection of point mutations in CesA3 that confer resistance. PLoS ONE 2013, 8, e56513. [Google Scholar] [CrossRef]
- Bradford, M.M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 1976, 72, 248–254. [Google Scholar] [CrossRef]


Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Cui, T.; Ma, Q.; Zhang, F.; Chen, S.; Zhang, C.; Hao, J.; Liu, X. Characterization of PcSTT3B as a Key Oligosaccharyltransferase Subunit Involved in N-glycosylation and Its Role in Development and Pathogenicity of Phytophthora capsici. Int. J. Mol. Sci. 2023, 24, 7500. https://doi.org/10.3390/ijms24087500
Cui T, Ma Q, Zhang F, Chen S, Zhang C, Hao J, Liu X. Characterization of PcSTT3B as a Key Oligosaccharyltransferase Subunit Involved in N-glycosylation and Its Role in Development and Pathogenicity of Phytophthora capsici. International Journal of Molecular Sciences. 2023; 24(8):7500. https://doi.org/10.3390/ijms24087500
Chicago/Turabian StyleCui, Tongshan, Quanhe Ma, Fan Zhang, Shanshan Chen, Can Zhang, Jianjun Hao, and Xili Liu. 2023. "Characterization of PcSTT3B as a Key Oligosaccharyltransferase Subunit Involved in N-glycosylation and Its Role in Development and Pathogenicity of Phytophthora capsici" International Journal of Molecular Sciences 24, no. 8: 7500. https://doi.org/10.3390/ijms24087500
APA StyleCui, T., Ma, Q., Zhang, F., Chen, S., Zhang, C., Hao, J., & Liu, X. (2023). Characterization of PcSTT3B as a Key Oligosaccharyltransferase Subunit Involved in N-glycosylation and Its Role in Development and Pathogenicity of Phytophthora capsici. International Journal of Molecular Sciences, 24(8), 7500. https://doi.org/10.3390/ijms24087500





