Genome-Wide Identification and Expression Analysis of NCED Gene Family in Pear and Its Response to Exogenous Gibberellin and Paclobutrazol
Abstract
:1. Introduction
2. Results
2.1. Identification and Analysis of PbNCED Genes
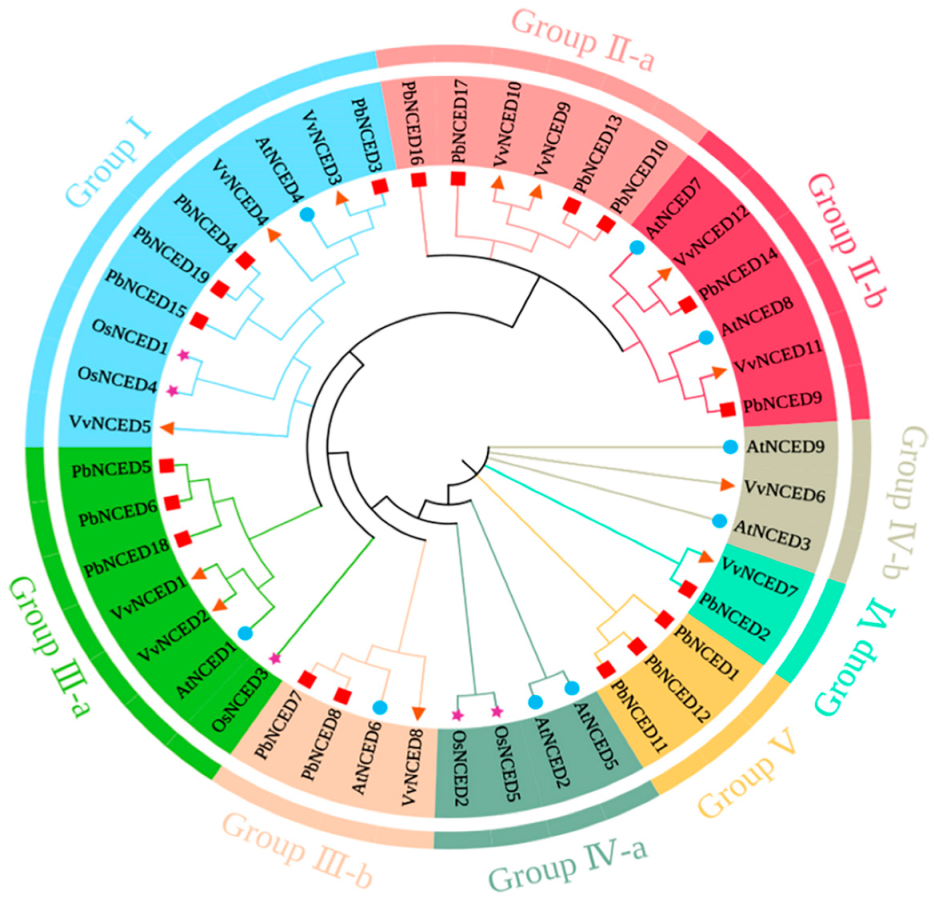
2.2. Multiple Sequence Alignment and Phylogenetic Analysis of PbNCED Genes
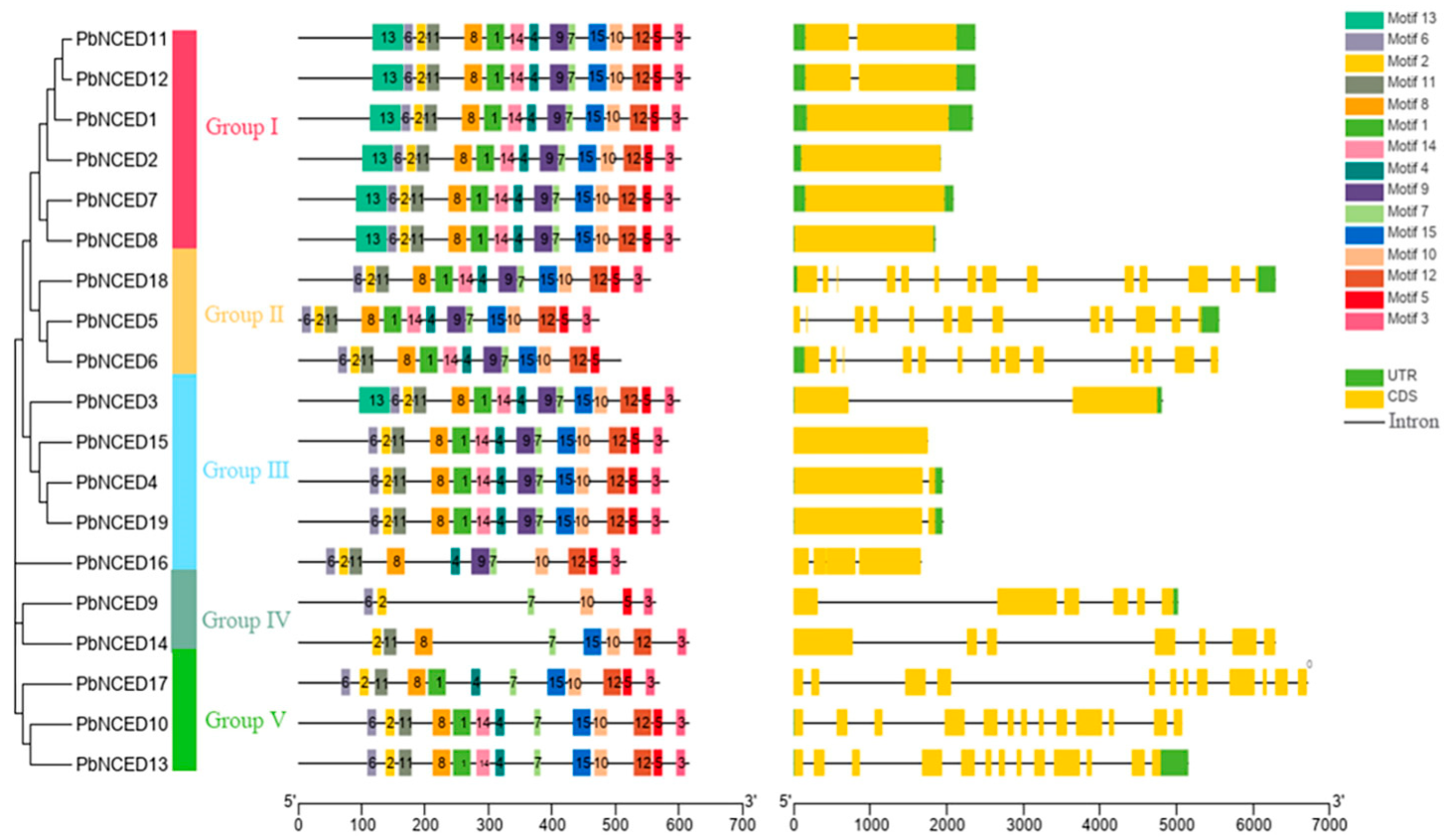
2.3. Gene Structure and Motif Composition
2.4. Chromosomal Location, Gene Duplication and Synteny Analysis
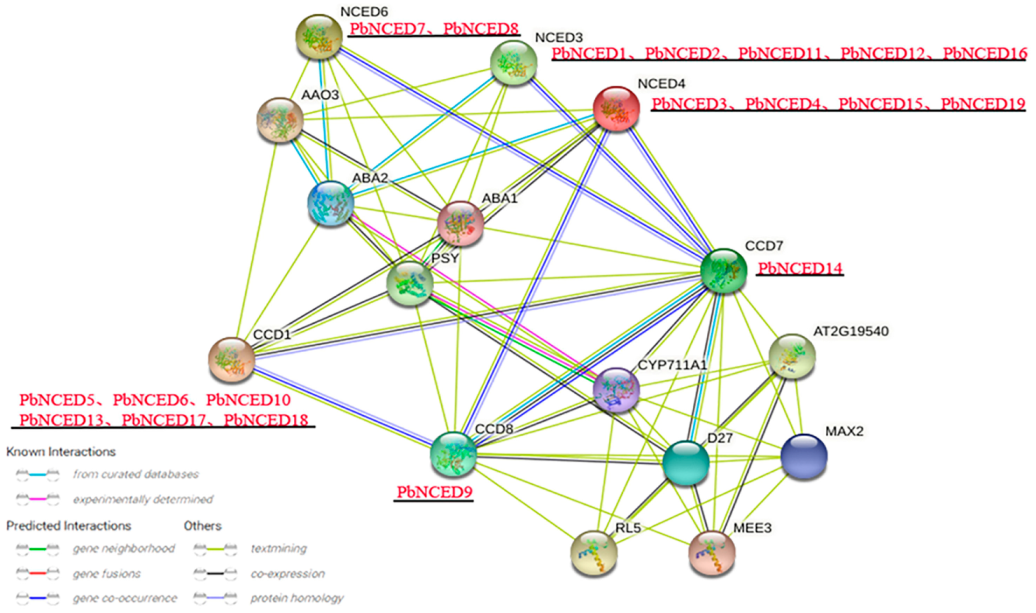
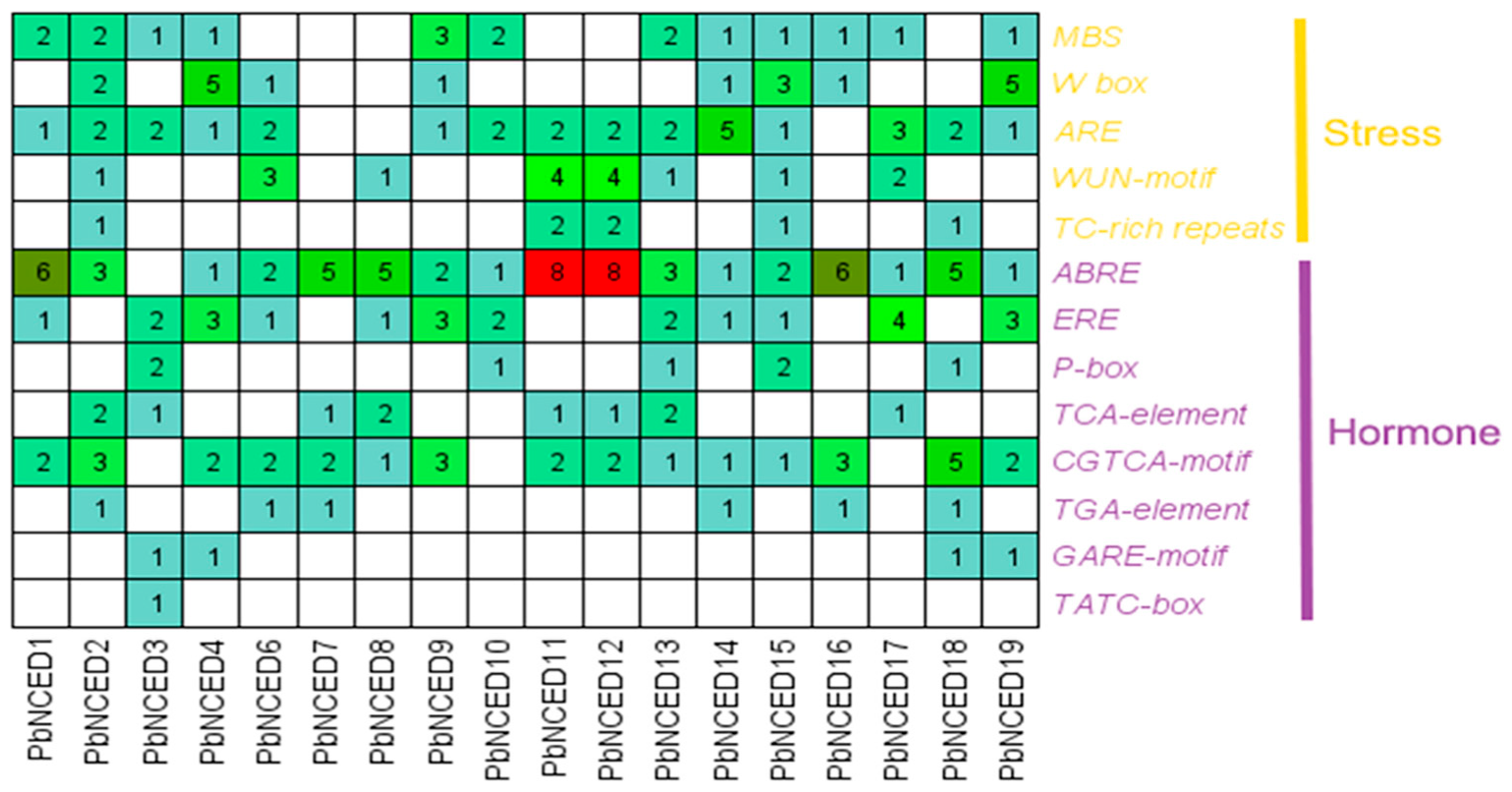
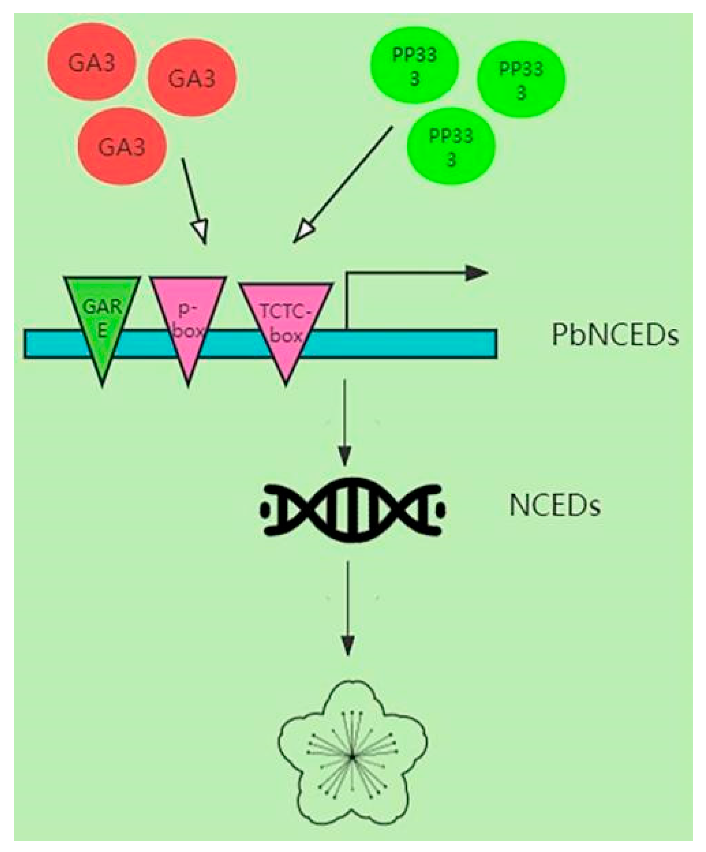
2.5. Interaction Network and Cis-Regulatory Element Analysis
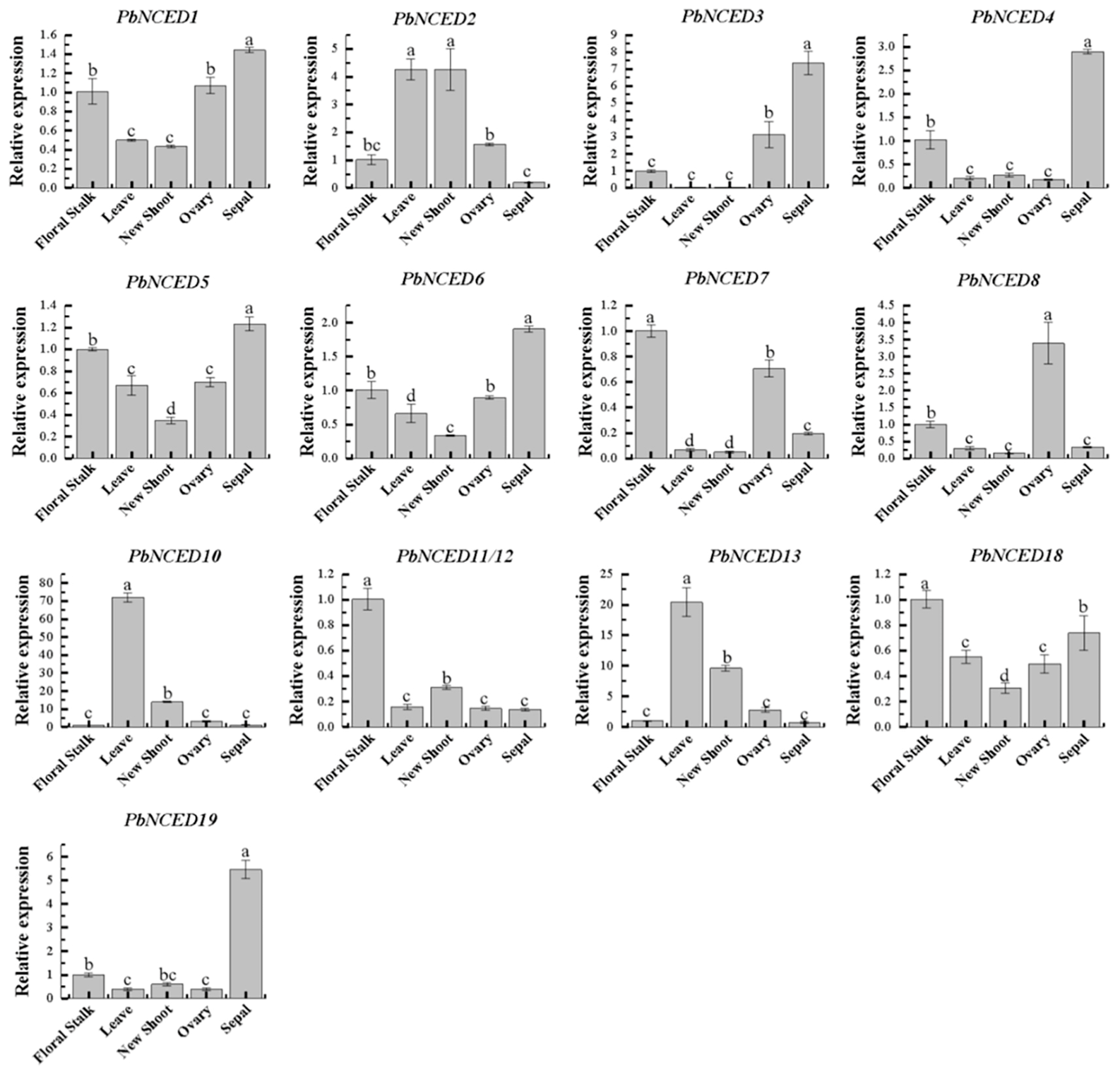
2.6. Tissue-Specific Expression of PbNCED Genes
2.7. Effect of Exogenous GA3 and PP333 on Content of GA3, IAA and ABA in ‘Kuerle Xiangli’
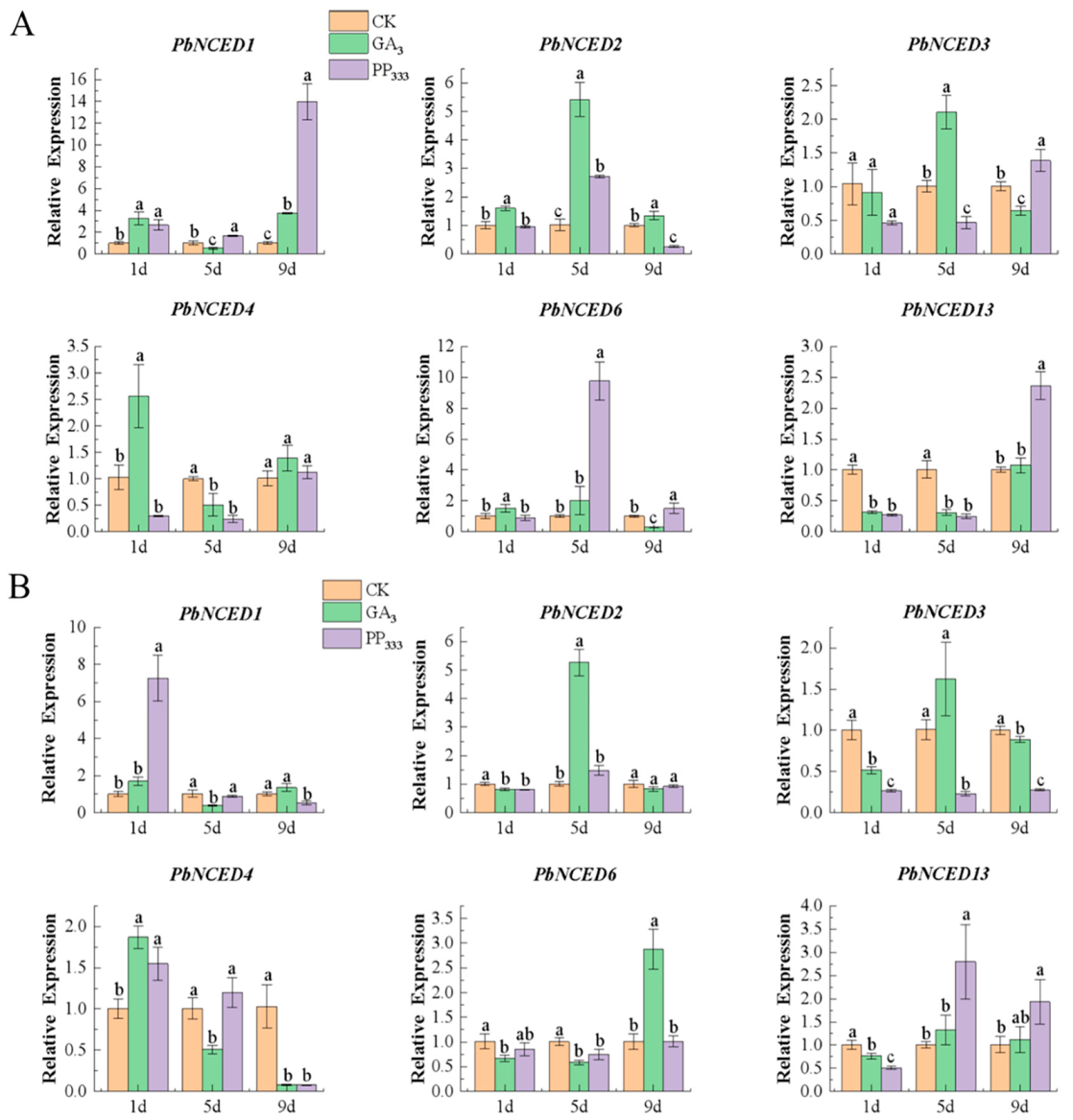
2.8. Expression Analysis of PbNCED Genes under Exogenous GA3 and PP333
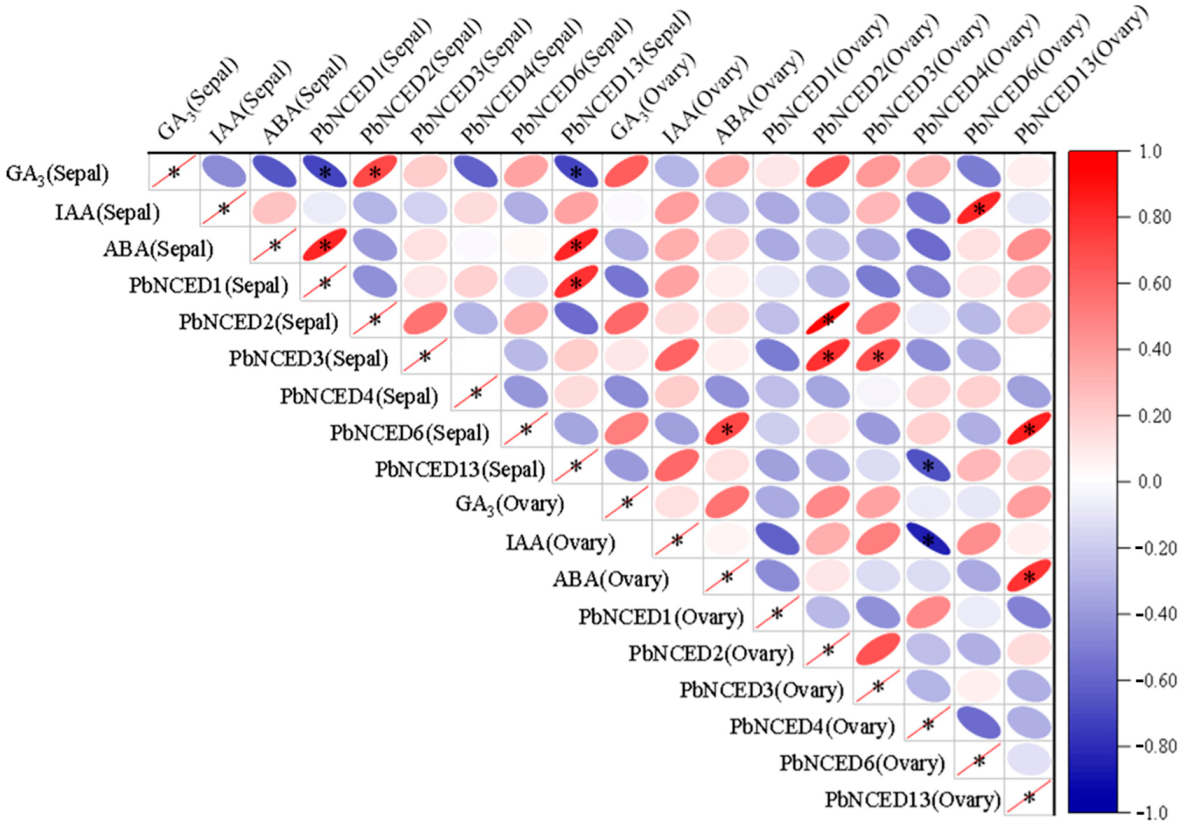
2.9. Correlation Analysis for PbNCED Gene Expresssion Level, Content of GA3, IAA, and ABA in ‘Kuerle Xiangli’ during Sepal and Ovary Development
3. Discussion
4. Materials and Methods
4.1. Plant Materials
4.2. Genome-Wide Identification and Protein Properties of NCED Family in Pear
4.3. Sequence Analyses and Phylogenetic Tree Construction
4.4. Gene Duplication and Synteny Analysis
4.5. Interaction Network and Cis-Regulatory Element Analyses
4.6. Determination of Phytohormones
4.7. RNA Extraction and Gene Expression Analysis
4.8. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Raghavendra, A.S.; Gonugunta, V.K.; Christmann, A.; Grill, E. ABA perception and signalling. Trends Plant Sci. 2010, 15, 395–401. [Google Scholar] [CrossRef] [PubMed]
- Nonogaki, M.; Sall, K.; Nambara, E.; Nonogaki, H. Amplification of ABA biosynthesis and signaling through a positive feedback mechanism in seeds. Plant J. 2014, 78, 527–539. [Google Scholar] [CrossRef] [PubMed]
- Manzi, M.; Lado, J.; Rodrigo, M.J.; Arbona, V.; Gómez-Cadenas, A. ABA accumulation in water-stressed Citrus roots does not rely on carotenoid content in this organ. Plant Sci. 2016, 252, 151–161. [Google Scholar] [CrossRef] [PubMed]
- McAdam, S.A.M.; Brodribb, T.J.; Banks, J.A.; Hedrich, R.; Atallah, N.M.; Cai, C.; Geringer, M.A.; Lind, C.; Nichols, D.S.; Stachowski, K.; et al. Abscisic acid controlled sex before transpiration in vascular plants. Proc. Natl. Acad. Sci. USA 2016, 113, 12862–12867. [Google Scholar] [CrossRef]
- Santosh Kumar, V.V.; Yadav, S.K.; Verma, R.K.; Shrivastava, S.; Ghimire, O.; Pushkar, S.; Rao, M.V.; Senthil Kumar, T.; Chinnusamy, V. The abscisic acid receptor OsPYL6 confers drought tolerance to indica rice through dehydration avoidance and tolerance mechanisms. J. Exp. Bot. 2021, 72, 1411–1431. [Google Scholar] [CrossRef]
- Iuchi, S.; Kobayashi, M.; Taji, T.; Naramoto, M.; Seki, M.; Kato, T.; Tabata, S.; Kakubari, Y.; Yamaguchi-Shinozaki, K.; Shinozaki, K. Regulation of drought tolerance by gene manipulation of 9-cis-epoxycarotenoid dioxygenase, a key enzyme in abscisic acid biosynthesis in Arabidopsis. Plant J. 2001, 27, 325–333. [Google Scholar] [CrossRef]
- Seiler, C.; Harshavardhan, V.T.; Rajesh, K.; Reddy, P.S.; Strickert, M.; Rolletschek, H.; Scholz, U.; Wobus, U.; Sreenivasulu, N. ABA biosynthesis and degradation contributing to ABA homeostasis during barley seed development under control and terminal drought-stress conditions. J. Exp. Bot. 2011, 62, 2615–2632. [Google Scholar] [CrossRef]
- Schwartz, S.H.; Tan, B.C.; Gage, D.A.; Zeevaart, J.A.D.; McCarty, D.R. Specific oxidative cleavage of carotenoids by VP14 of maize. Science 1997, 276, 1872–1874. [Google Scholar] [CrossRef]
- Zhang, L.; Ma, G.; Kato, M.; Yamawaki, K.; Takagi, T.; Kiriiwa, Y.; Ikoma, Y.; Matsumoto, H.; Yoshioka, T.; Nesumi, H. Regulation of carotenoid accumulation and the expression of carotenoid metabolic genes in citrus juice sacs in vitro. J. Exp. Bot. 2012, 63, 871–886. [Google Scholar] [CrossRef]
- Kato, M. Mechanism of carotenoid accumulation in citrus fruit. J. Jpn. Soc. Hortic. Sci. 2012, 81, 843–851. [Google Scholar] [CrossRef]
- Jia, H.-F.; Chai, Y.-M.; Li, C.-L.; Lu, D.; Luo, J.-J.; Qin, L.; Shen, Y.-Y. Abscisic acid plays an important role in the regulation of strawberry fruit ripening. Plant Physiol. 2011, 157, 188–199. [Google Scholar] [CrossRef]
- Jia, H.-F.; Lu, D.; Sun, J.-H.; Li, C.-L.; Xing, Y.; Qin, L.; Shen, Y.-Y. Type 2C protein phosphatase ABI1 is a negative regulator of strawberry fruit ripening. J. Exp. Bot. 2013, 64, 1677–1687. [Google Scholar] [CrossRef]
- Zhang, M.; Leng, P.; Zhang, G.; Li, X. Cloning and functional analysis of 9-cis-epoxycarotenoid dioxygenase (NCED) genes encoding a key enzyme during abscisic acid biosynthesis from peach and grape fruits. J. Plant Physiol. 2009, 166, 1241–1252. [Google Scholar] [CrossRef]
- Huang, Y.; Jiao, Y.; Xie, N.; Guo, Y.; Zhang, F.; Xiang, Z.; Wang, R.; Wang, F.; Gao, Q.; Tian, L.; et al. OsNCED5, a 9-cis-epoxycarotenoid dioxygenase gene, regulates salt and water stress tolerance and leaf senescence in rice. Plant Sci. 2019, 287, 110–188. [Google Scholar] [CrossRef]
- Bao, G.; Zhuo, C.; Qian, C.; Xiao, T.; Guo, Z.; Lu, S. Co-expression of NCED and ALO improves vitamin C level and tolerance to drought and chilling in transgenic tobacco and stylo plants. Plant Biotechnol. J. 2016, 14, 206–214. [Google Scholar] [CrossRef]
- Xia, H.; Wu, S.; Ma, F. Cloning and expression of two 9-cis-epoxycarotenoid dioxygenase genes during fruit development and under stress conditions from Malus. Mol. Biol. Rep. 2014, 41, 6795–6802. [Google Scholar] [CrossRef]
- Zhang, T.; Gao, Y.; Han, M.; Yang, L. Changes in the physiological characteristics of Panax ginseng embryogenic calli and molecular mechanism of ginsenoside biosynthesis under cold stress. Planta 2021, 253, 79. [Google Scholar] [CrossRef]
- Tan, B.-C.; Joseph, L.M.; Deng, W.-T.; Liu, L.; Li, Q.-B.; Cline, K.; McCarty, D.R. Molecular characterization of the Arabidopsis 9-cis epoxycarotenoid dioxygenase gene family. Plant J. 2003, 35, 44–56. [Google Scholar] [CrossRef]
- Li, Q.; Yu, X.; Chen, L.; Zhao, G.; Li, S.; Zhou, H.; Dai, Y.; Sun, N.; Xie, Y.; Gao, J.; et al. Genome-wide identification and expression analysis of the NCED family in cotton (Gossypium hirsutum L.). PLoS ONE 2021, 16, e0246021. [Google Scholar] [CrossRef]
- Chernys, J.T.; Zeevaart, J.A. Characterization of the 9-cis-epoxycarotenoid dioxygenase gene family and the regulation of abscisic acid biosynthesis in avocado. Plant Physiol. 2000, 124, 343–353. [Google Scholar] [CrossRef]
- Iuchi, S.; Kobayashi, M.; Yamaguchi-Shinozaki, K.; Shinozaki, K. A stress-inducible gene for 9-cis-epoxycarotenoid dioxygenase involved in abscisic acid biosynthesis under water stress in drought-tolerant cowpea. Plant Physiol. 2000, 123, 553–562. [Google Scholar] [CrossRef]
- Gan, Z.; Shan, N.; Fei, L.; Wan, C.; Chen, J. Isolation of the 9-cis-epoxycarotenoid dioxygenase (NCED) gene from kiwifruit and its effects on postharvest softening and ripening. Sci. Hortic. 2019, 261, 109020. [Google Scholar] [CrossRef]
- Wang, X.; Liu, F.; Shi, X.; Wang, X.; Ji, X.; Wang, Z. Evolution and expression of NCED family genes in Vitis vinifera. Chin. Bull. Bot. 2019, 54, 474–485. [Google Scholar] [CrossRef]
- Chevreau, E.; Skirvin, R.M.; Abu-Qaoud, H.A.; Korban, S.S.; Sullivan, J.G. Adventitious shoot regeneration from leaf tissue of three pear (Pyrus sp.) cultivars in vitro. Plant Cell Rep. 1989, 7, 688–691. [Google Scholar] [CrossRef]
- Ma, L.; Zhou, L.; Quan, S.; Xu, H.; Yang, J.; Niu, J. Integrated analysis of mRNA-seq and miRNA-seq in calyx abscission zone of Korla fragrant pear involved in calyx persistence. BMC Plant Biol. 2019, 19, 192. [Google Scholar] [CrossRef] [PubMed]
- Wu, J.; Wang, Z.; Shi, Z.; Zhang, S.; Ming, R.; Zhu, S.; Khan, M.A.; Tao, S.; Korban, S.S.; Wang, H.; et al. The genome of the pear (Pyrus bretschneideri Rehd.). Genome Res. 2013, 23, 396–408. [Google Scholar] [CrossRef]
- Klepikova, A.V.; Kasianov, A.S.; Gerasimov, E.S.; Logacheva, M.D.; Penin, A.A. A high resolution map of the Arabidopsis thaliana developmental transcriptome based on RNA-seq profiling. Plant J. 2016, 88, 1058–1070. [Google Scholar] [CrossRef]
- Auldridge, M.E.; Block, A.; Vogel, J.T.; Dabney-Smith, C.; Mila, I.; Bouzayen, M.; Magallanes-Lundback, M.; DellaPenna, D.; McCarty, D.R.; Klee, H.J. Characterization of three members of the Arabidopsis carotenoid cleavage dioxygenase family demonstrates the divergent roles of this multifunctional enzyme family. Plant J. 2006, 45, 982–993. [Google Scholar] [CrossRef]
- Pei, X.; Wang, X.; Fu, G.; Chen, B.; Nazir, M.F.; Pan, Z.; He, S.; Du, X. Identification and functional analysis of 9-cis-epoxy carotenoid dioxygenase (NCED) homologs in G. hirsutum. Int. J. Biol. Macromol. 2021, 182, 298–310. [Google Scholar] [CrossRef]
- Priya, R.; Siva, R. Analysis of phylogenetic and functional diverge in plant nine-cis epoxycarotenoid dioxygenase gene family. J. Plant Res. 2015, 128, 519–534. [Google Scholar] [CrossRef]
- Xu, P.; Cai, W. Functional characterization of the BnNCED3 gene in Brassica napus. Plant Sci. 2017, 256, 16–24. [Google Scholar] [CrossRef]
- Xian, L.; Sun, P.; Hu, S.; Wu, J.; Liu, J.-H. Molecular cloning and characterization of CrNCED1, a gene encoding 9-cis-epoxycarotenoid dioxygenase in Citrus reshni, with functions in tolerance to multiple abiotic stresses. Planta 2014, 239, 61–77. [Google Scholar] [CrossRef]
- Wan, X.-R.; Li, L. Regulation of ABA level and water-stress tolerance of Arabidopsis by ectopic expression of a peanut 9-cis-epoxycarotenoid dioxygenase gene. Biochem. Biophys. Res. Commun. 2006, 347, 1030–1038. [Google Scholar] [CrossRef]
- Wittkopp, P.J.; Kalay, G. Cis-regulatory elements: Molecular mechanisms and evolutionary processes underlying divergence. Nat. Rev. Genet. 2011, 13, 59–69. [Google Scholar] [CrossRef]
- Wang, P.; Lu, S.; Zhang, X.; Hyden, B.; Qin, L.; Liu, L.; Bai, Y.; Han, Y.; Wen, Z.; Xu, J.; et al. Double NCED isozymes control ABA biosynthesis for ripening and senescent regulation in peach fruits. Plant Sci. 2021, 304, 110739. [Google Scholar] [CrossRef]
- Ju, Y.-L.; Min, Z.; Yue, X.-F.; Zhang, Y.-L.; Zhang, J.-X.; Zhang, Z.-Q.; Fang, Y.-L. Overexpression of grapevine VvNAC08 enhances drought tolerance in transgenic Arabidopsis. Plant Physiol. Biochem. 2014, 151, 214–222. [Google Scholar] [CrossRef]
- Wang, L.; Hua, D.; He, J.; Duan, Y.; Chen, Z.; Hong, X.; Gong, Z. Auxin Response Factor2 (ARF2) and its regulated homeodomain gene HB33 mediate abscisic acid response in Arabidopsis. PLoS Genet. 2011, 7, e1002172. [Google Scholar] [CrossRef]
- Zhao, Y.; Xing, L.; Wang, X.; Hou, Y.-J.; Gao, J.; Wang, P.; Duan, C.-G.; Zhu, X.; Zhu, J.-K. The ABA receptor PYL8 promotes lateral root growth by enhancing MYB77-dependent transcription of auxin-responsive genes. Sci. Signal. 2014, 7, ra53. [Google Scholar] [CrossRef]
- Lorrai, R.; Boccaccini, A.; Ruta, V.; Possenti, M.; Costantino, P.; Vittorioso, P. Abscisic acid inhibits hypocotyl elongation acting on gibberellins, DELLA proteins and auxin. AoB Plants 2018, 10, ply061. [Google Scholar] [CrossRef]
- Kim, J.; Lee, J.G.; Hong, Y.; Lee, E.J. Analysis of eight phytohormone concentrations, expression levels of ABA biosynthesis genes, and ripening-related transcription factors during fruit development in strawberry. J. Plant Physiol. 2019, 239, 52–60. [Google Scholar] [CrossRef]
- Yan, B.; Hou, J.; Cui, J.; He, C.; Li, W.; Chen, X.; Li, M.; Wang, W. The Effects of Endogenous Hormones on the Flowering and Fruiting of Glycyrrhiza uralensis. Plants 2019, 8, 519. [Google Scholar] [CrossRef] [PubMed]
- Cheng, M.-Z.; Gong, C.; Zhang, B.; Qu, W.; Qi, H.-N.; Chen, X.-L.; Wang, X.-Y.; Zhang, Y.; Liu, J.-Y.; Ding, X.-D.; et al. Morphological and anatomical characteristics of exserted stigma sterility and the location and function of SlLst (Solanum lycopersicum Long styles) gene in tomato. Theor. Appl. Genet. 2021, 134, 505–518. [Google Scholar] [CrossRef] [PubMed]
- Ahmad, S.; Kamran, M.; Zhou, X.; Ahmad, I.; Meng, X.; Javed, T.; Iqbal, A.; Wang, G.; Su, W.; Wu, X.; et al. Melatonin improves the seed filling rate and endogenous hormonal mechanism in grains of summer maize. Physiol. Plant. 2021, 172, 1059–1072. [Google Scholar] [CrossRef] [PubMed]
- Zhang, W.; Wang, J.; Huang, Z.; Mi, L.; Xu, K.; Wu, J.; Fan, Y.; Ma, S.; Jiang, D. Effects of Low Temperature at Booting Stage on Sucrose Metabolism and Endogenous Hormone Contents in Winter Wheat Spikelet. Front. Plant Sci. 2019, 10, 498. [Google Scholar] [CrossRef]
- Procter, J.B.; Carstairs, G.M.; Soares, B.; Mourão, K.; Ofoegbu, T.C.; Barton, D.; Lui, L.; Menard, A.; Sherstnev, N.; Roldan-Martinez, D.; et al. Alignment of Biological Sequences with Jalview. Mult. Seq. Alignment 2021, 2231, 203–224. [Google Scholar] [CrossRef]
- Chen, C.; Chen, H.; Zhang, Y.; Thomas, H.R.; Frank, M.H.; He, Y.; Xia, R. TBtools: An Integrative Toolkit Developed for In-teractive Analyses of Big Biological Data. Mol. Plant 2020, 13, 1194–1202. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular Evolutionary Genetics Analysis across Computing Platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]


Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Liu, J.; Yuan, X.; Quan, S.; Zhang, M.; Kang, C.; Guo, C.; Zhang, Z.; Niu, J. Genome-Wide Identification and Expression Analysis of NCED Gene Family in Pear and Its Response to Exogenous Gibberellin and Paclobutrazol. Int. J. Mol. Sci. 2023, 24, 7566. https://doi.org/10.3390/ijms24087566
Liu J, Yuan X, Quan S, Zhang M, Kang C, Guo C, Zhang Z, Niu J. Genome-Wide Identification and Expression Analysis of NCED Gene Family in Pear and Its Response to Exogenous Gibberellin and Paclobutrazol. International Journal of Molecular Sciences. 2023; 24(8):7566. https://doi.org/10.3390/ijms24087566
Chicago/Turabian StyleLiu, Jinming, Xing Yuan, Shaowen Quan, Meng Zhang, Chao Kang, Caihua Guo, Zhongrong Zhang, and Jianxin Niu. 2023. "Genome-Wide Identification and Expression Analysis of NCED Gene Family in Pear and Its Response to Exogenous Gibberellin and Paclobutrazol" International Journal of Molecular Sciences 24, no. 8: 7566. https://doi.org/10.3390/ijms24087566
APA StyleLiu, J., Yuan, X., Quan, S., Zhang, M., Kang, C., Guo, C., Zhang, Z., & Niu, J. (2023). Genome-Wide Identification and Expression Analysis of NCED Gene Family in Pear and Its Response to Exogenous Gibberellin and Paclobutrazol. International Journal of Molecular Sciences, 24(8), 7566. https://doi.org/10.3390/ijms24087566





