Global Phosphoproteomic Analysis Reveals the Defense and Response Mechanisms of Japonica Rice under Low Nitrogen Stress
Abstract
1. Introduction
2. Results
2.1. Low Nitrogen Stress Affects the Morphology, Physiology, and Growth Characteristics of Rice Leaves
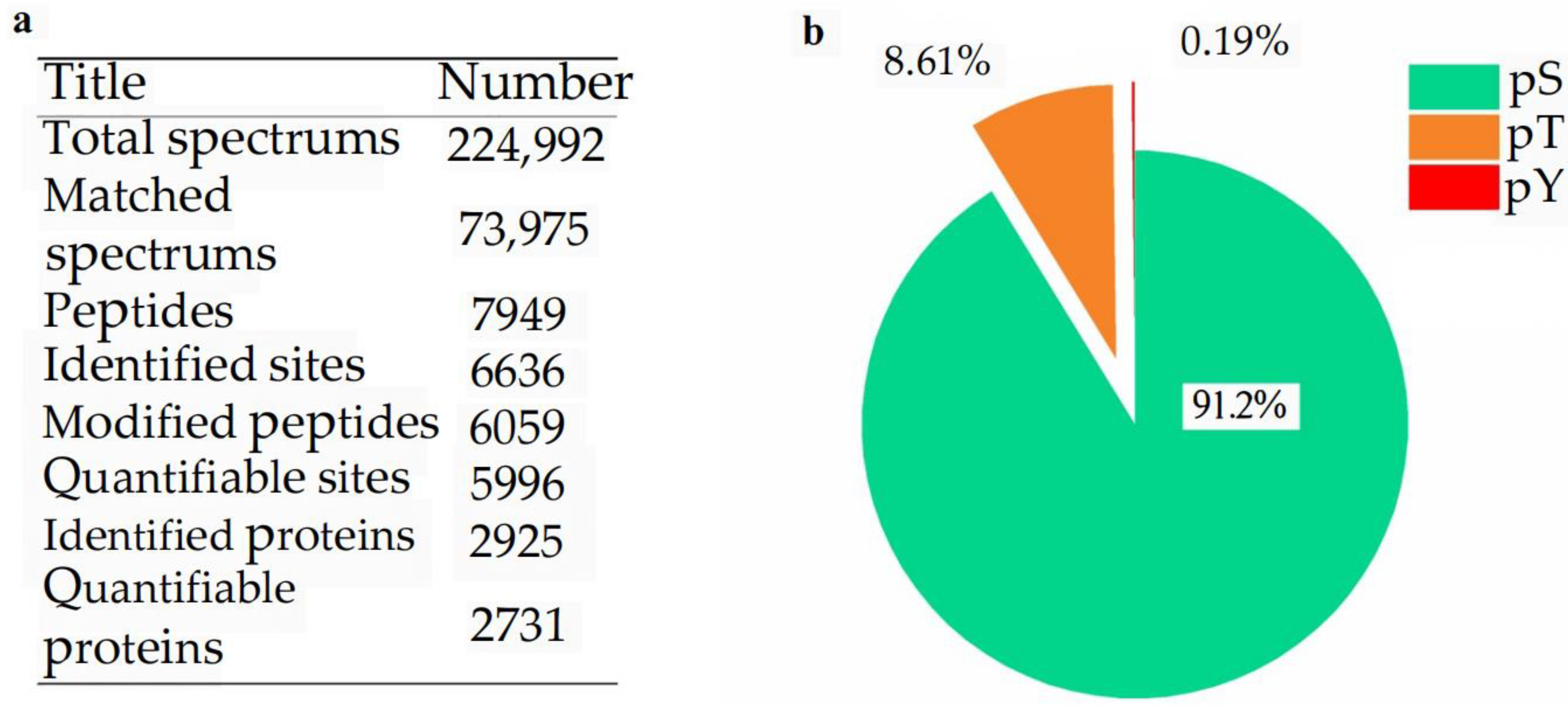
2.2. Phosphoprotein Identification and Phosphorylated Site Location
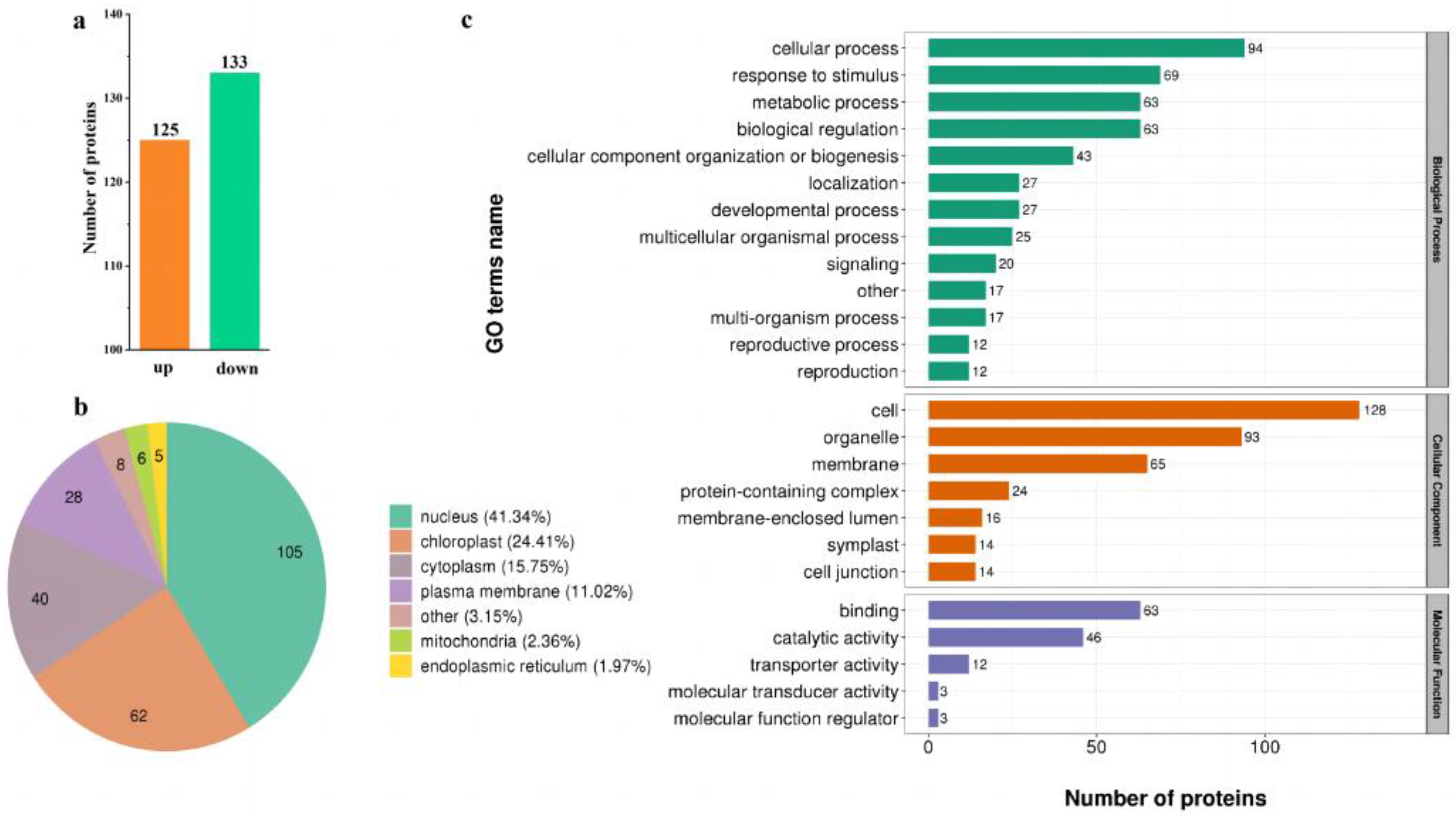
2.3. Features of Phosphorylated Proteins in Response to Low-N Stress
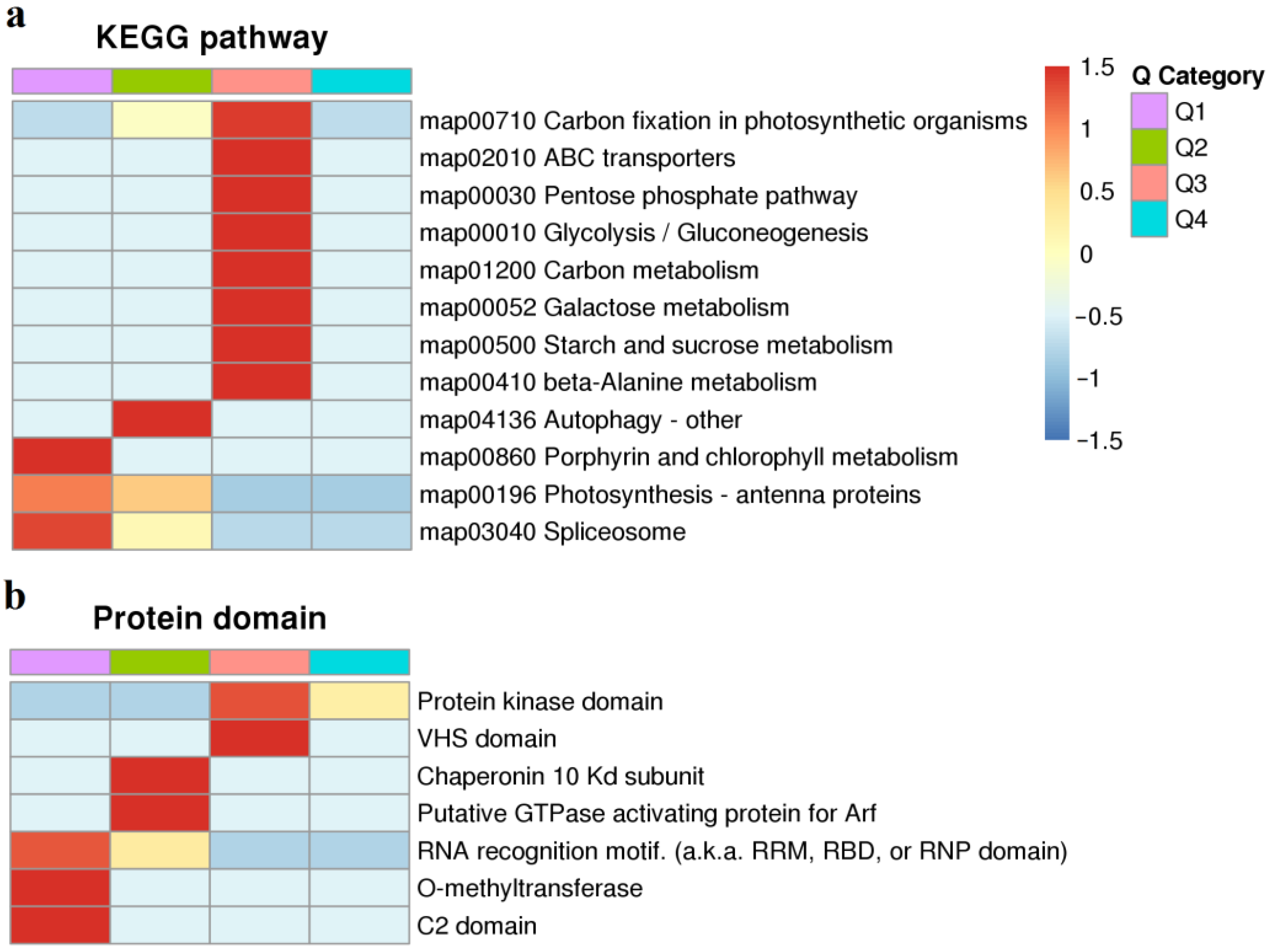
2.4. KEGG Pathway and Protein Domain Analysis
2.5. Protein Kinases in Response to N Deficiency
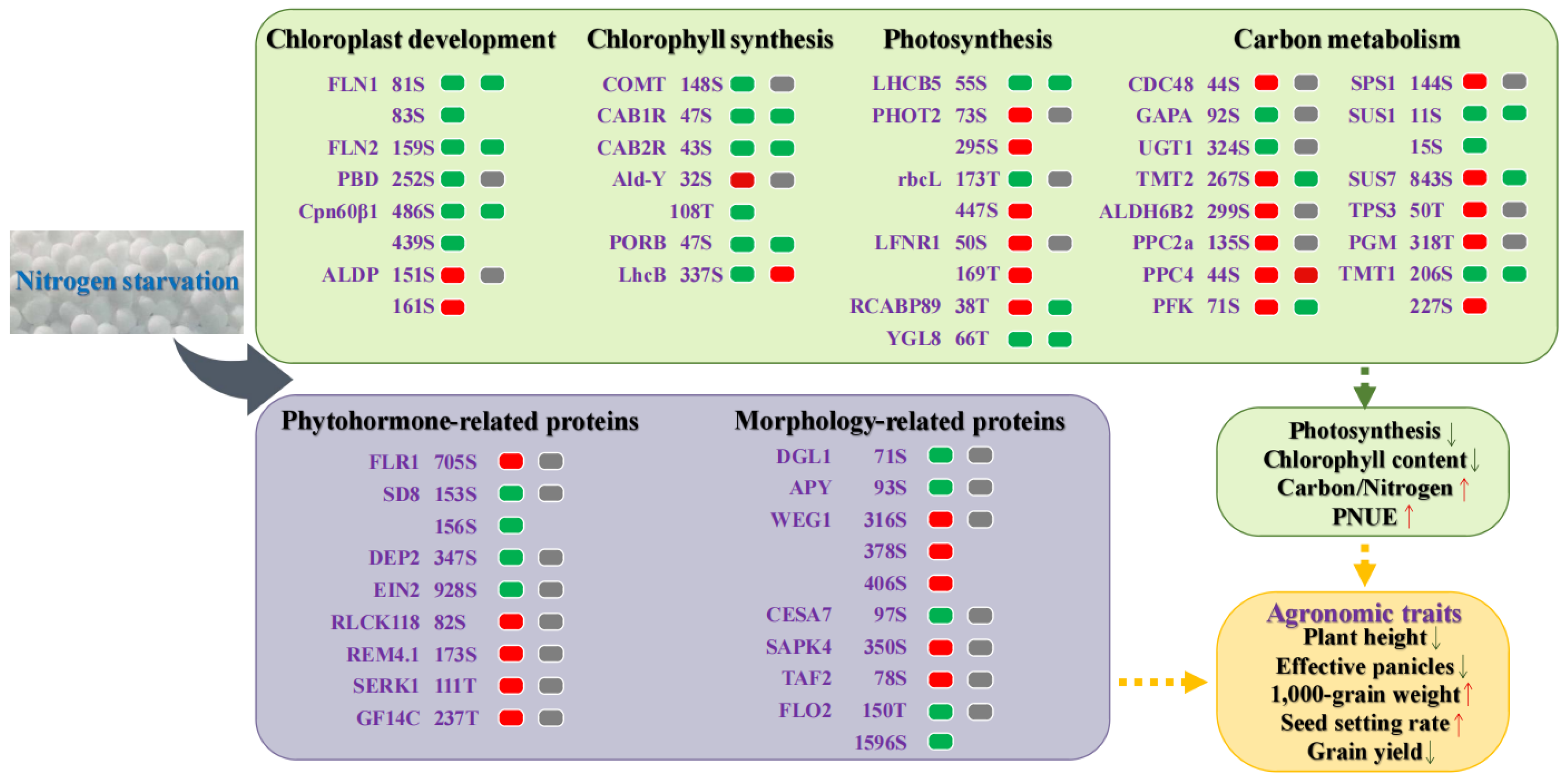
2.6. An Overview of Response and Adaptive Mechanism of Rice under Low-N Stress
3. Discussion
3.1. Effects of Low Nitrogen Stress on Carbon Metabolism in Rice
3.2. Effects of Low Nitrogen Stress on Agronomic Traits of Rice
4. Materials and Methods
4.1. Plant Material and Growth Conditions
4.2. Determination of Leaf Morphological and Physiological Characteristics
4.3. Determination of Yield and Plant Height at Maturity Stage
4.4. Protein Extraction and Digestion
4.5. Mass Spectrometry Analysis
4.6. Database Search
4.7. Bioinformatics Analysis
4.8. qRT-PCR Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Ladha, J.K.; Gjd, K.; Bennett, J.; Reddy, C.K.; Singh, U. Opportunities for increased nitrogen-use efficiency from improved lowland rice germplasm. Field Crops Res. 1998, 56, 41–71. [Google Scholar] [CrossRef]
- Hussain, F.; Bronson, K.F.; Yadvinder, S.; Bijay, S.; Peng, S. Use of chlorophyll meter sufficiency indices for nitrogen management of irrigated rice in asia. Agron. J. 2000, 92, 875–879. [Google Scholar] [CrossRef]
- Hussain, T.; Hussain, N.; Ahmed, M.; Nualsri, C.; Duangpan, S. Impact of nitrogen application rates on upland rice performance, planted under varying sowing times. Sustainability 2022, 14, 1997. [Google Scholar] [CrossRef]
- Ogawa, T.; Oikawa, S.; Hirose, T. Nitrogen-utilization efficiency in rice: An analysis at leaf, shoot, and whole-plant level. Plant Soil 2016, 404, 321–344. [Google Scholar] [CrossRef]
- Xin, W.; Zhang, L.; Zhang, W.; Gao, J.; Yi, J.; Zhen, X.; Li, Z.; Zhao, Y.; Peng, C.; Zhao, C. An Integrated Analysis of the Rice Transcriptome and Metabolome Reveals Differential Regulation of Carbon and Nitrogen Metabolism in Response to Nitrogen Availability. Int. J. Mol. Sci. 2019, 20, 2349. [Google Scholar] [CrossRef] [PubMed]
- Xin, W.; Zhang, L.; Gao, J.; Zhang, W.; Yi, J.; Zhen, X.; Bi, C.; He, D.; Liu, S.; Zhao, X. Adaptation Mechanism of Roots to Low and High Nitrogen Revealed by Proteomic Analysis. Rice 2021, 14, 5. [Google Scholar] [CrossRef] [PubMed]
- Liu, Q.; Wu, K.; Song, W.; Zhong, N.; Wu, Y.; Fu, X. Improving Crop Nitrogen Use Efficiency Toward Sustainable Green Revolution. Annu. Rev. Plant Biol. 2022, 73, 523–551. [Google Scholar] [CrossRef]
- Subudhi, P.K.; Garcia, R.S.; Coronejo, S.; Tapia, R. Comparative Transcriptomics of Rice Genotypes with Contrasting Responses to Nitrogen Stress Reveals Genes Influencing Nitrogen Uptake through the Regulation of Root Architecture. Int. J. Mol. Sci. 2020, 21, 5759. [Google Scholar] [CrossRef]
- Yang, W.; Yoon, J.; Choi, H.; Fan, Y.; Chen, R.; An, G. Transcriptome analysis of nitrogen-starvation-responsive genes in rice. BMC Plant Biol. 2015, 15, 31. [Google Scholar]
- Subodh, S.; Amitha, S.V.; Saurabh, C.; Punit, T.; Sureshkumar, V.; Manju, R.; Pranab, M. Transcriptome Analysis of Two Rice Varieties Contrasting for Nitrogen Use Efficiency under Chronic N Starvation Reveals Differences in Chloroplast and Starch Metabolism-Related Genes. Genes 2018, 9, 206. [Google Scholar] [CrossRef]
- Chitteti, B.R.; Peng, Z. Proteome and phosphoproteome differential expression under salinity stress in rice (Oryza sativa) roots. J. Proteome Res. 2007, 6, 1718–1727. [Google Scholar] [CrossRef] [PubMed]
- Gupta, R.; Min, C.W.; Kim, Y.J.; Kim, S.T. Identification of Msp1-Induced Signaling Components in Rice Leaves by Integrated Proteomic and Phosphoproteomic Analysis. Int. J. Mol. Sci. 2019, 20, 4135. [Google Scholar] [CrossRef]
- Nohzadeh Malakshah, S.; Habibi Rezaei, M.; Heidari, M.; Salekdeh, G.H. Proteomics reveals new salt responsive proteins associated with rice plasma membrane. Biosci. Biotechnol. Biochem. 2004, 71, 2144–2154. [Google Scholar] [CrossRef] [PubMed]
- Mateo Sánchez, S.; Freeman, S.D.; Delacroix, L.; Malgrange, B. The role of post-translational modifications in hearing and deafness. Cell. Mol. Life Sci. CMLS 2016, 73, 3521–3533. [Google Scholar] [CrossRef]
- Abolore, A.; Amara, C.; Shakeel, A.; Wang, Y.; Shu, Y.; Li, S.; Liu, X.; Babatunde, K.; Sani, M.; Tong, X.; et al. Protein Phosphorylation and Phosphoproteome: An Overview of Rice. Rice Sci. 2020, 27, 23–39. [Google Scholar]
- Martín, M.L.; Busconi, L. Membrane localization of a rice calcium-dependent protein kinase (cdpk) is mediated by myristoylation and palmitoylation. Plant J. 2010, 24, 429–435. [Google Scholar]
- Miyata, K.; Hayafune, M.; Kobae, Y.; Kaku, H.; Nishizawa, Y.; Masuda, Y.; Shibuya, N.; Nakagawa, T. Evaluation of the Role of the LysM Receptor-Like Kinase, OsNFR5/OsRLK2 for AM Symbiosis in Rice. Plant Cell Physiol. 2016, 57, 2283–2290. [Google Scholar] [CrossRef] [PubMed]
- Liu, K.H.; Niu, Y.; Konishi, M.; Wu, Y.; Du, H.; Sun, C.H.; Li, L.; Boudsocq, M.; McCormack, M.; Maekawa, S.; et al. Discovery of nitrate-CPK-NLP signalling in central nutrient-growth networks. Nature 2017, 545, 311–316. [Google Scholar] [CrossRef]
- Kumar, K.; Rao, K.P.; Sharma, P.; Sinha, A.K. Differential regulation of rice mitogen activated protein kinase kinase (MKK) by abiotic stress. Plant Physiol Biochem. 2008, 46, 891–897. [Google Scholar] [CrossRef] [PubMed]
- Harkenrider, M.; Sharma, R.; De Vleesschauwer, D.; Tsao, L.; Zhang, X.; Chern, M.; Canlas, P.; Zuo, S.; Ronald, P.C. Overexpression of Rice Wall-Associated Kinase 25 (OsWAK25) Alters Resistance to Bacterial and Fungal Pathogens. PLoS ONE 2016, 11, e0147310. [Google Scholar] [CrossRef] [PubMed]
- Banerjee, A.; Roychoudhury, A. The gymnastics of epigenomics in rice. Plant Cell Rep. 2018, 37, 25–49. [Google Scholar] [PubMed]
- Higo, A.; Saihara, N.; Miura, F.; Higashi, Y.; Yamada, M.; Tamaki, S.; Ito, T.; Tarutani, Y.; Sakamoto, T.; Fujiwara, M.; et al. DNA methylation is reconfigured at the onset of reproduction in rice shoot apical meristem. Nat. Commun. 2020, 11, 4079. [Google Scholar] [CrossRef] [PubMed]
- Aburajab, R.; Pospiech, M.; Alachkar, H. Profiling the epigenetic landscape of the antigen receptor repertoire: The missing epi-immunogenomics data. Nat. Methods 2023, 20, 477–481. [Google Scholar] [CrossRef]
- Das, A.B.; Seddon, A.R.; O’Connor, K.M.; Hampton, M.B. Regulation of the epigenetic landscape by immune cell oxidants. Free Radic. Biol. Med. 2021, 170, 131–149. [Google Scholar] [CrossRef]
- Kusumi, K.; Hirotsuka, S.; Shimada, H.; Chono, Y.; Matsuda, O.; Iba, K. Contribution of chloroplast biogenesis to carbon-nitrogen balance during early leaf development in rice. J. Plant Res. 2010, 123, 617–622. [Google Scholar] [CrossRef] [PubMed]
- Vincentz, M.; Moureaux, T.; Leydecker, M.T.; Vaucheret, H.; Caboche, M. Regulation of nitrate and nitrite reductase expression in Nicotiana plumbaginifolia leaves by nitrogen and carbon metabolites. Plant J. 2010, 3, 315–324. [Google Scholar] [CrossRef]
- Foyer, C.H.; Noctor, G.; Hodges, M. Respiration and nitrogen assimilation: Targeting mitochondria-associated metabolism as a means to enhance nitrogen use efficiency. J. Exp. Bot. 2011, 62, 1467–1482. [Google Scholar] [PubMed]
- Cong, F.; Diehl, B.G.; Hill, J.L.; Brown, N.R.; Tien, M. Covalent bond formation between amino acids and lignin: Cross-coupling between proteins and lignin. Phytochemistry 2013, 96, 449–456. [Google Scholar] [CrossRef]
- Ray, D.; Sheshshayee, M.S.; Mukhopadhyay, K.; Bindumadhava, H.; Prasad, T.G.; Kumar, M.U. High Nitrogen Use Efficiency in Rice Genotypes is Associated with Higher Net Photosynthetic Rate at Lower Rubisco Content. Biol. Plant. 2003, 46, 251–256. [Google Scholar] [CrossRef]
- Zhang, X.; Misra, A.; Nargund, S.; Coleman, G.D.; Sriram, G. Concurrent isotope-assisted metabolic flux analysis and transcriptome profiling reveal responses of poplar cells to altered nitrogen and carbon supply. Plant J. 2018, 93, 472–488. [Google Scholar] [CrossRef]
- He, L.; Zhang, S.; Qiu, Z.; Zhao, J.; Nie, W.; Lin, H.; Zhu, Z.; Zeng, D.; Qian, Q.; Zhu, L. FRUCTOKINASE-LIKE PROTEIN 1 interacts with TRXz to regulate chloroplast development in rice. J. Integr. Plant Biol. 2018, 60, 94–111. [Google Scholar] [CrossRef]
- Lv, Y.; Shao, G.; Qiu, J.; Jiao, G.; Sheng, Z.; Xie, L.; Wu, Y.; Tang, S.; Wei, X.; Hu, P. White Leaf and Panicle 2, encoding a PEP-associated protein, is required for chloroplast biogenesis under heat stress in rice. J. Exp. Bot. 2017, 68, 5147–5160. [Google Scholar] [CrossRef] [PubMed]
- Huangfu, L.; Chen, R.; Lu, Y.; Zhang, E.; Miao, J.; Zuo, Z.; Zhao, Y.; Zhu, M.; Zhang, Z.; Li, P.; et al. OsCOMT, encoding a caffeic acid O-methyltransferase in melatonin biosynthesis, increases rice grain yield through dual regulation of leaf senescence and vascular development. Plant Biotechnol. J. 2022, 20, 1122–1139. [Google Scholar] [CrossRef] [PubMed]
- Wei, S.; Li, X.; Lu, Z.; Zhang, H.; Ye, X.; Zhou, Y.; Li, J.; Yan, Y.; Pei, H.; Duan, F.; et al. A transcriptional regulator that boosts grain yields and shortens the growth duration of rice. Science 2022, 377, eabi8455. [Google Scholar] [CrossRef] [PubMed]
- Ermakova, M.; Danila, F.R.; Furbank, R.T.; von Caemmerer, S. On the road to C4 rice: Advances and perspectives. Plant J. 2020, 101, 940–950. [Google Scholar] [CrossRef]
- Nayak, L.; Panda, D.; Dash, G.K.; Lal, M.K.; Swain, P.; Baig, M.J.; Kumar, A. A chloroplast Glycolate catabolic pathway bypassing the endogenous photorespiratory cycle enhances photosynthesis, biomass and yield in rice (Oryza sativa L.). Plant Sci. 2022, 314, 111103. [Google Scholar] [CrossRef]
- Bown, H.E.; Watt, M.S.; Mason, E.G.; Clinton, P.E.; Whitehead, D. The influence of nitrogen and phosphorus supply and genotype on mesophyll conductance limitations to photosynthesis in Pinus radiata. Tree Physiol. 2009, 29, 1143–1151. [Google Scholar] [CrossRef]
- Suganami, M.; Suzuki, Y.; Tazoe, Y.; Yamori, W.; Makino, A. Co-overproducing Rubisco and Rubisco activase enhances photosynthesis in the optimal temperature range in rice. Plant Physiol. 2021, 185, 108–119. [Google Scholar] [CrossRef]
- Yoon, D.K.; Ishiyama, K.; Suganami, M.; Tazoe, Y.; Watanabe, M.; Imaruoka, S.; Ogura, M.; Ishida, H.; Suzuki, Y.; Obara, M.; et al. Transgenic rice overproducing Rubisco exhibits increased yields with improved nitrogen-use efficiency in an experimental paddy field. Nat. Food 2020, 1, 134–139. [Google Scholar] [CrossRef]
- Li, Y.; Yang, X.; Ren, B.; Shen, Q.; Guo, S. Why nitrogen use efficiency decreases under high nitrogen supply in rice (Oryza sativa L.) seedlings. J. Plant Growth Regul. 2012, 31, 47–52. [Google Scholar] [CrossRef]
- Suzuki, Y.; Makino, A. Availability of Rubisco small subunit up-regulates the transcript levels of large subunit for stoichiometric assembly of its holoenzyme in rice. Plant Physiol. 2012, 160, 533–540. [Google Scholar] [CrossRef] [PubMed]
- Yang, C.; Hu, H.; Ren, H.; Kong, Y.; Lin, H.; Guo, J.; Wang, L.; He, Y.; Ding, X.; Grabsztunowicz, M.; et al. LIGHT-INDUCED RICE1 Regulates Light-Dependent Attachment of Leaf-Type Ferredoxin-NADP+ Oxidoreductase to the Thylakoid Membrane in Rice and Arabidopsis. Plant Cell 2016, 28, 712–728. [Google Scholar] [CrossRef] [PubMed]
- Sasaki, A.; Ashikari, M.; Ueguchi-Tanaka, M.; Itoh, H.; Nishimura, A.; Swapan, D.; Ishiyama, K.; Saito, T.; Kobayashi, M.; Khush, G.S.; et al. Green revolution: A mutant gibberellin-synthesis gene in rice. Nature 2002, 416, 701–702. [Google Scholar] [CrossRef] [PubMed]
- Uga, Y.; Sugimoto, K.; Ogawa, S.; Rane, J.; Ishitani, M.; Hara, N.; Kitomi, Y.; Inukai, Y.; Ono, K.; Kanno, N.; et al. Control of root system architecture by DEEPER ROOTING 1 increases rice yield under drought conditions. Nat. Genet. 2013, 45, 1097–1102. [Google Scholar] [CrossRef]
- Fageria, N.K.; Santos, A.D.; Cutrim, V. Nitrogen uptake and its association with grain yield in lowland rice genotypes. J. Plant Nutr. 2009, 32, 1965–1974. [Google Scholar] [CrossRef]
- Cho, L.H.; Yoon, J.; Tun, W.; Baek, G.; Peng, X.; Hong, W.J.; Mori, I.C.; Hojo, Y.; Matsuura, T.; Kim, S.R.; et al. Cytokinin increases vegetative growth period by suppressing florigen expression in rice and maize. Plant J. 2022, 110, 1619–1635. [Google Scholar] [CrossRef]
- Li, C.; Wang, L.; Cui, Y.; He, L.; Qi, Y.; Zhang, J.; Lin, J.; Liao, H.; Lin, Q.; Yang, T.; et al. Two FERONIA-like receptor (FLR) genes are required to maintain architecture, fertility, and seed yield in rice. Mol. Breed. 2016, 36, 151. [Google Scholar] [CrossRef]
- Qu, R.; Zhang, P.; Liu, Q.; Wang, Y.; Guo, W.; Du, Z.; Li, X.; Yang, L.; Yan, S.; Gu, X. Genome-edited ATP binding cassette B1 transporter SD8 knockouts show optimized rice architecture without yield penalty. Plant Commun. 2022, 3, 100347. [Google Scholar] [CrossRef]
- Zhou, X.; Wang, J.; Peng, C.; Zhu, X.; Yin, J.; Li, W.; He, M.; Wang, J.; Chern, M.; Yuan, C.; et al. Four receptor-like cytoplasmic kinases regulate development and immunity in rice. Plant Cell Environ. 2016, 39, 1381–1392. [Google Scholar] [CrossRef]
- Ma, B.; He, S.J.; Duan, K.X.; Yin, C.C.; Chen, H.; Yang, C.; Xiong, Q.; Song, Q.X.; Lu, X.; Chen, H.W.; et al. Identification of rice ethylene-response mutants and characterization of MHZ7/OsEIN2 in distinct ethylene response and yield trait regulation. Mol. Plant 2013, 6, 1830–1848. [Google Scholar] [CrossRef]
- Wang, D.; Qin, Y.; Fang, J.; Yuan, S.; Peng, L.; Zhao, J.; Li, X. A Missense Mutation in the Zinc Finger Domain of OsCESA7 Deleteriously Affects Cellulose Biosynthesis and Plant Growth in Rice. PLoS ONE 2016, 11, e0153993. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Wang, R.; Xing, Y.; Xu, Y.; Xiong, D.; Wang, Y.; Yao, S. Separable regulation of POW1 in grain size and leaf angle development in rice. Plant Biotechnol. J. 2021, 19, 2517–2531. [Google Scholar] [CrossRef] [PubMed]
- She, K.C.; Kusano, H.; Koizumi, K.; Yamakawa, H.; Hakata, M.; Imamura, T.; Fukuda, M.; Naito, N.; Tsurumaki, Y.; Yaeshima, M.; et al. A novel factor FLOURY ENDOSPERM2 is involved in regulation of rice grain size and starch quality. Plant Cell 2010, 22, 3280–3294. [Google Scholar] [CrossRef] [PubMed]
- Young, M.D.; Wakefield, M.J.; Smyth, G.K.; Oshlack, A. Gene ontology analysis for RNA-seq: Accounting for selection bias. Genome Biol. 2010, 11, R14. [Google Scholar] [CrossRef] [PubMed]
| Treatments | Low Nitrogen | Control Nitrogen |
|---|---|---|
| Leaf biomass (g) | 1.58 ± 0.20 b | 2.35 ± 0.17 a |
| Leaf area (cm2) | 276.53 ± 18.38 b | 407.18 ± 11.65 a |
| Chlorophyll a (Chl a, mg g−1) | 0.93 ± 0.05 b | 1.76 ± 0.12 a |
| Chlorophyll b (Chl b, mg g−1) | 0.42 ± 0.02 b | 0.82 ± 0.07 a |
| Intercellular CO2 concentration (Ci, μmol mol−1) | 268.55 ± 8.13 a | 274.56 ± 16.95 a |
| Photosynthetic rate (Pn, μmol m−2 s−1) | 19.27 ± 0.72 b | 22.51 ± 1.21 a |
| Stomatal conductance (gs, mol m−2 s−1) | 639.23 ± 12.37 b | 723.56 ± 24.13 a |
| Electron transport rate (ETR, μmol m−2 s−1) | 11.83 ± 0.19 b | 12.29 ± 0.18 a |
| N content | 3.53 ± 0.21 c | 4.72 ± 0.32 b |
| C content | 36.11 ± 0.21 b | 42.16 ± 0.43 a |
| Carbon/Nitrogen (C/N) | 10.23 ± 0.14 a | 8.93 ± 0.12 b |
| Nitrogen use efficiency (NUE, g g−1) | 48.98 ± 3.14 a | 31.21 ± 3.25 b |
| Photosynthetic nitrogen use efficiency (PUNE, μmol g−1 s−1) | 9.55 ± 0.58 a | 8.26 ± 0.18 b |
| Grain yield (g plant−1) | 19.34 ± 1.78 b | 27.59 ± 0.91 a |
| Effective panicles | 8.18 ± 0.35 b | 12.33 ± 0.35 a |
| Grain per panicle | 121.32 ± 2.54 a | 119.92 ± 1.29 a |
| Seed setting rate (%) | 93.33 ± 1.78 a | 85.51 ± 2.78 b |
| 1000-grain weight (g) | 24.12 ± 0.43 a | 22.65 ± 0.43 b |
| Plant height at the mature stage (cm) | 86.16 ± 2.18 b | 97.34 ± 3.34 a |
| ID | Modified Sequence | LN/HN Ratio | Gene Name |
|---|---|---|---|
| LOC_Os03g57450 | VS(0.001)S(0.999)AGLLVGSVLK | 0.56 | OsCPK10 |
| LOC_Os07g38120 | DGS(1)LQLTTTQ | 1.513 | OsCPK20 |
| FT(0.002)S(0.993)LS(0.005)LK | 1.536 | ||
| FTS(0.001)LS(0.999)LK | 1.54 | ||
| LOC_Os04g38480 | LMDYKDT(0.999)HVT(0.86)T(0.141)AVR | 2.368 | OsSERK2 |
| LOC_Os03g24930 | LS(0.004)S(0.996)MTNSPASSVAGAAEGGK | 2.375 | OsRLCK109 |
| LOC_Os03g60710 | NFRPDS(1)VLGEGGFGSVYK | 2.634 | OsRLCK118 |
| LOC_Os06g46330 | AT(0.023)S(0.891)S(0.079)S(0.006)S(0.001)LLTSIMAR | 1.566 | OsRLCK213 |
| LOC_Os09g36320 | SIS(1)SLYEER | 1.668 | OsRLCK278 |
| LOC_Os11g10100 | LSETS(0.001)VS(0.999)PR | 0.58 | OsMAPKKKα |
| LOC_Os04g56530 | LDHHHS(0.917)S(0.083)GSLQSLQADADR | 0.575 | OsMAPKKKε |
| LOC_Os04g35700 | VQS(1)PY(0.001)GS(0.999)PK | 0.282 | OsMAP3K.16 |
| LOC_Os06g05520 | FLTAS(0.001)GT(0.999)FKDGELR | 1.967 | OsMKK1 |
| LOC_Os01g64970 | EVHAS(1)GELR | 1.563 | OsSAPK4 |
| LOC_Os01g42294 | TTTEESEEGVRGT(0.003)S(0.997)EEER | 1.65 | OsRPK1 |
| LOC_Os01g28730 | S(0.933)FT(0.067)HINEDAALESPKEE | 1.696 | OsRKF3 |
| LOC_Os11g11490 | S(0.116)GT(0.884)DQFDLTDTD | 1.685 | OsCRR4 |
| LOC_Os05g47560 | TINES(1)MDELSSQSK | 1.674 | OsSTN7 |
| TINESMDELS(0.028)S(0.965)QS(0.007)K | 1.582 | ||
| VVRT(1)INES(1)MDELSSQSK | 2.062 | ||
| LOC_Os09g23570 | NADVDDFDS(0.002)VS(0.998)Q | 1.978 | |
| LOC_Os06g43840 | VAS(1)RENISPK | 0.53 | |
| LOC_Os07g43560 | HSTAMS(1)LNDVTVTEPEPR | 2.051 | |
| LSLSYS(0.867)S(0.133)R | 1.775 | ||
| SDS(0.979)S(0.021)SLDEILR | 1.812 | ||
| LOC_Os03g03570 | KPVES(1)PGVATAVVLR | 1.675 | |
| LOC_Os07g43570 | NRS(0.999)YT(0.001)ETMDVPLPSGPHSSITELEPR | 1.514 | |
| RLS(0.998)NCS(0.002)NQGLGQLK | 1.548 | ||
| LOC_Os03g27990 | NEPLTLRPIAS(1)GK | 0.533 | |
| LOC_Os12g01200 | ELPSSIHHLMS(1)K | 1.796 | |
| LGS(1)FFSEVATESAHR | 2.033 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Xie, S.; Liu, H.; Ma, T.; Shen, S.; Zheng, H.; Yang, L.; Liu, L.; Wei, Z.; Xin, W.; Zou, D.; et al. Global Phosphoproteomic Analysis Reveals the Defense and Response Mechanisms of Japonica Rice under Low Nitrogen Stress. Int. J. Mol. Sci. 2023, 24, 7699. https://doi.org/10.3390/ijms24097699
Xie S, Liu H, Ma T, Shen S, Zheng H, Yang L, Liu L, Wei Z, Xin W, Zou D, et al. Global Phosphoproteomic Analysis Reveals the Defense and Response Mechanisms of Japonica Rice under Low Nitrogen Stress. International Journal of Molecular Sciences. 2023; 24(9):7699. https://doi.org/10.3390/ijms24097699
Chicago/Turabian StyleXie, Shupeng, Hualong Liu, Tianze Ma, Shen Shen, Hongliang Zheng, Luomiao Yang, Lichao Liu, Zhonghua Wei, Wei Xin, Detang Zou, and et al. 2023. "Global Phosphoproteomic Analysis Reveals the Defense and Response Mechanisms of Japonica Rice under Low Nitrogen Stress" International Journal of Molecular Sciences 24, no. 9: 7699. https://doi.org/10.3390/ijms24097699
APA StyleXie, S., Liu, H., Ma, T., Shen, S., Zheng, H., Yang, L., Liu, L., Wei, Z., Xin, W., Zou, D., & Wang, J. (2023). Global Phosphoproteomic Analysis Reveals the Defense and Response Mechanisms of Japonica Rice under Low Nitrogen Stress. International Journal of Molecular Sciences, 24(9), 7699. https://doi.org/10.3390/ijms24097699






