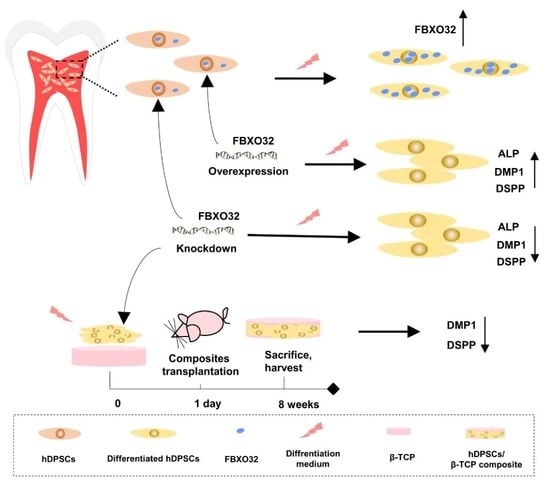
Promotive Effect of FBXO32 on the Odontoblastic Differentiation of Human Dental Pulp Stem Cells
Abstract
1. Introduction
2. Results
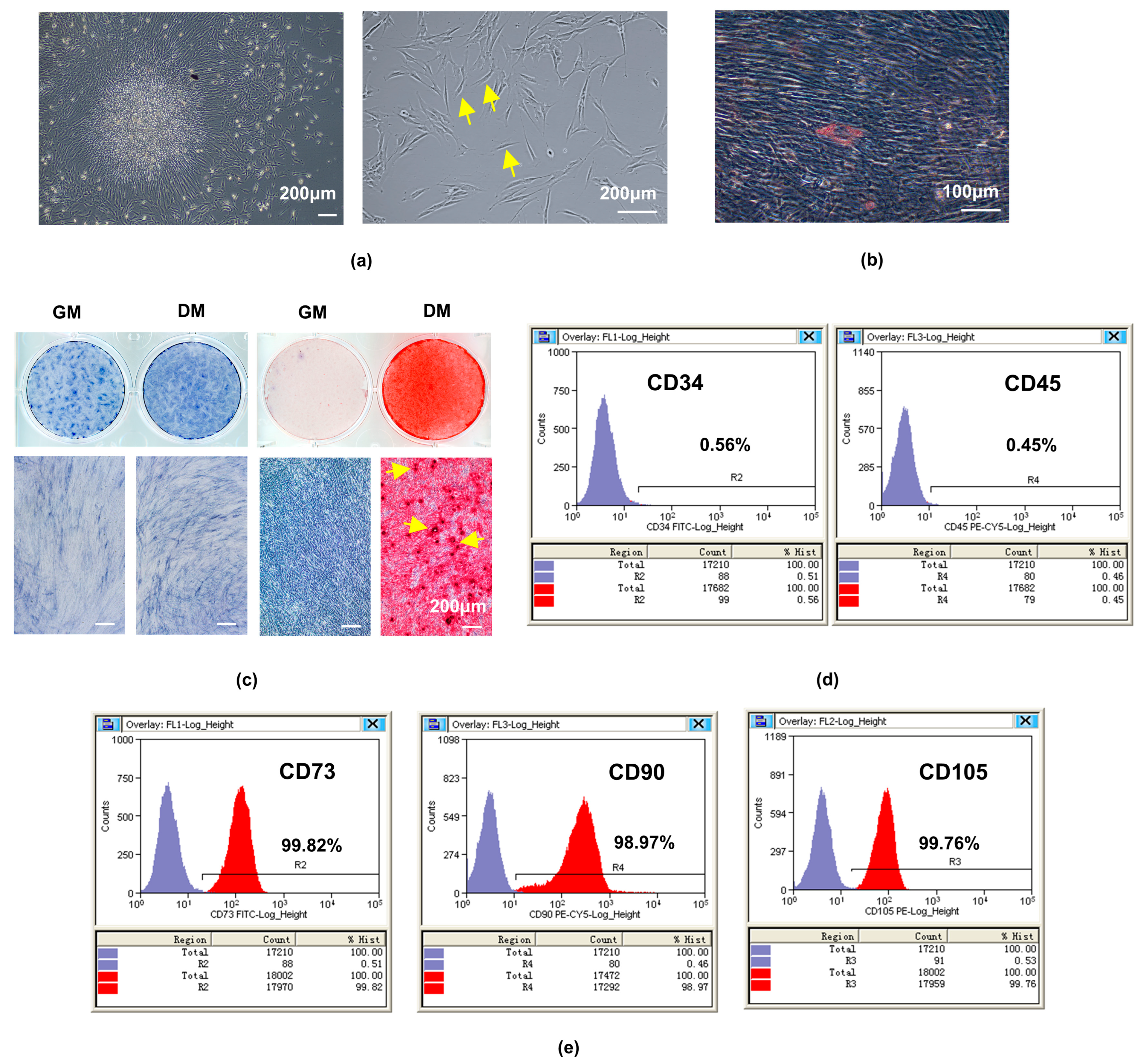
2.1. Isolation and Characterization of hDPSCs
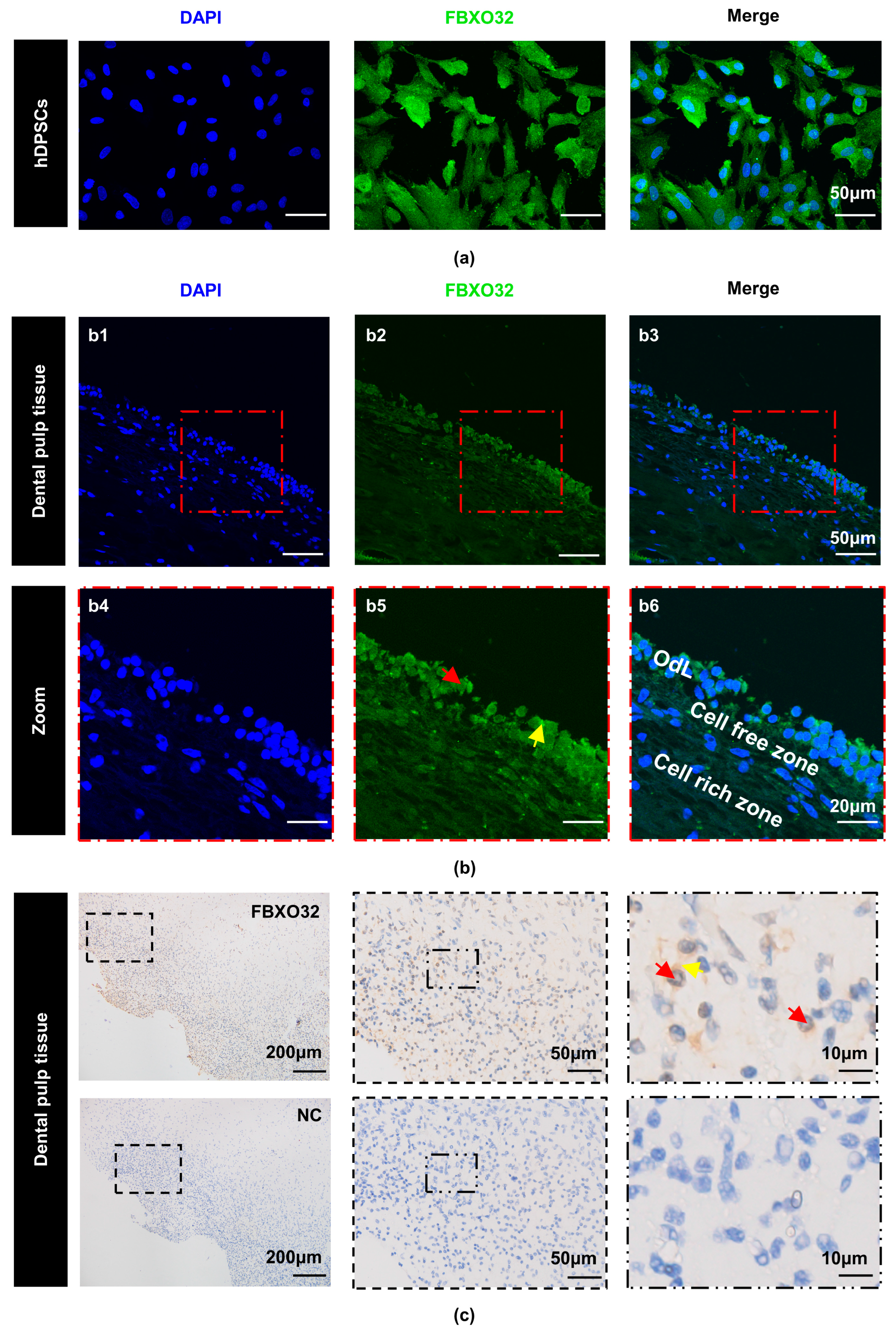
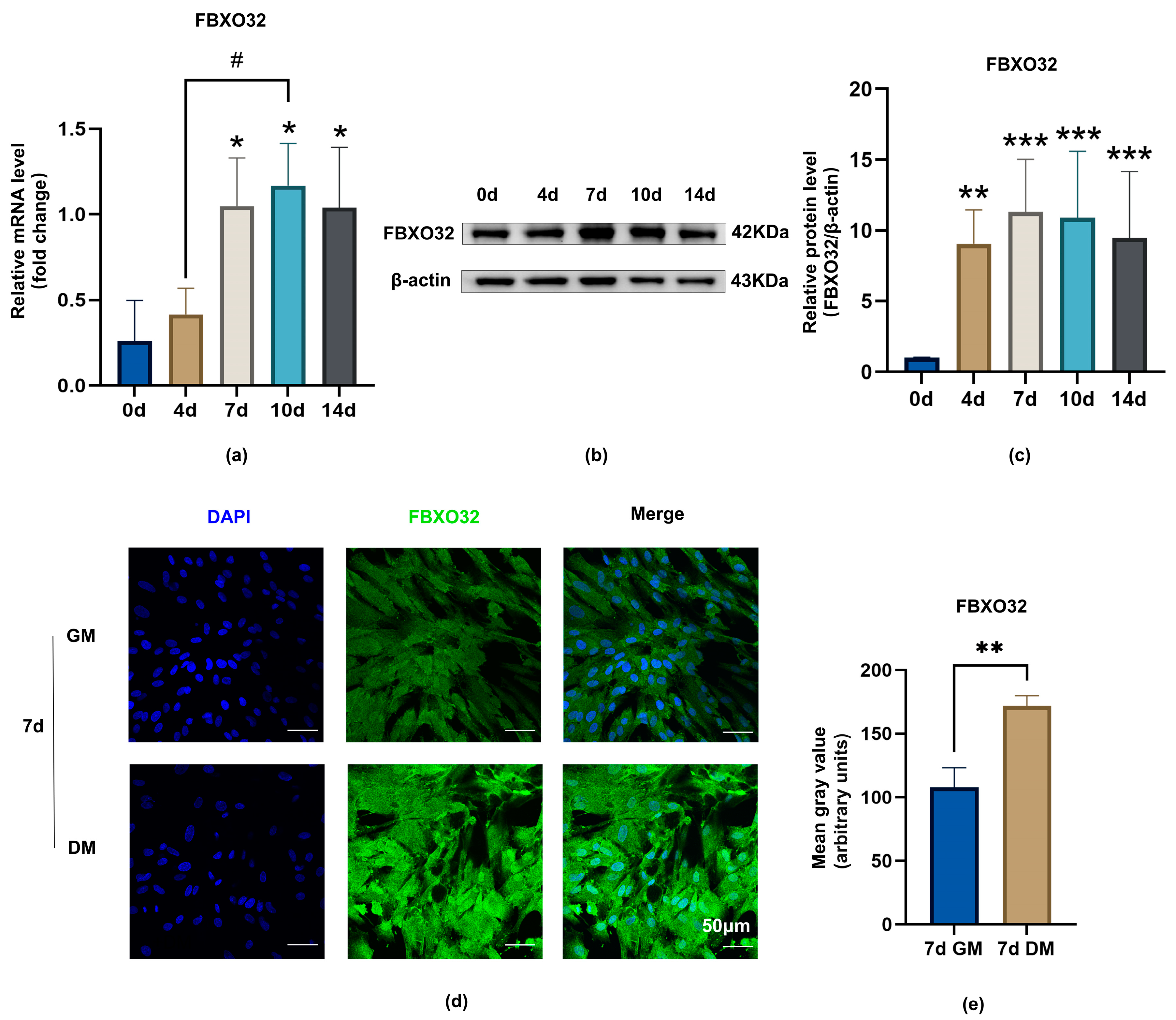
2.2. E3 Ubiquitination Ligase FBXO32 Was Expressed in Dental Pulp Cells/Tissues and Was Upregulated during Odontoblastic Differentiation of hDPSCs
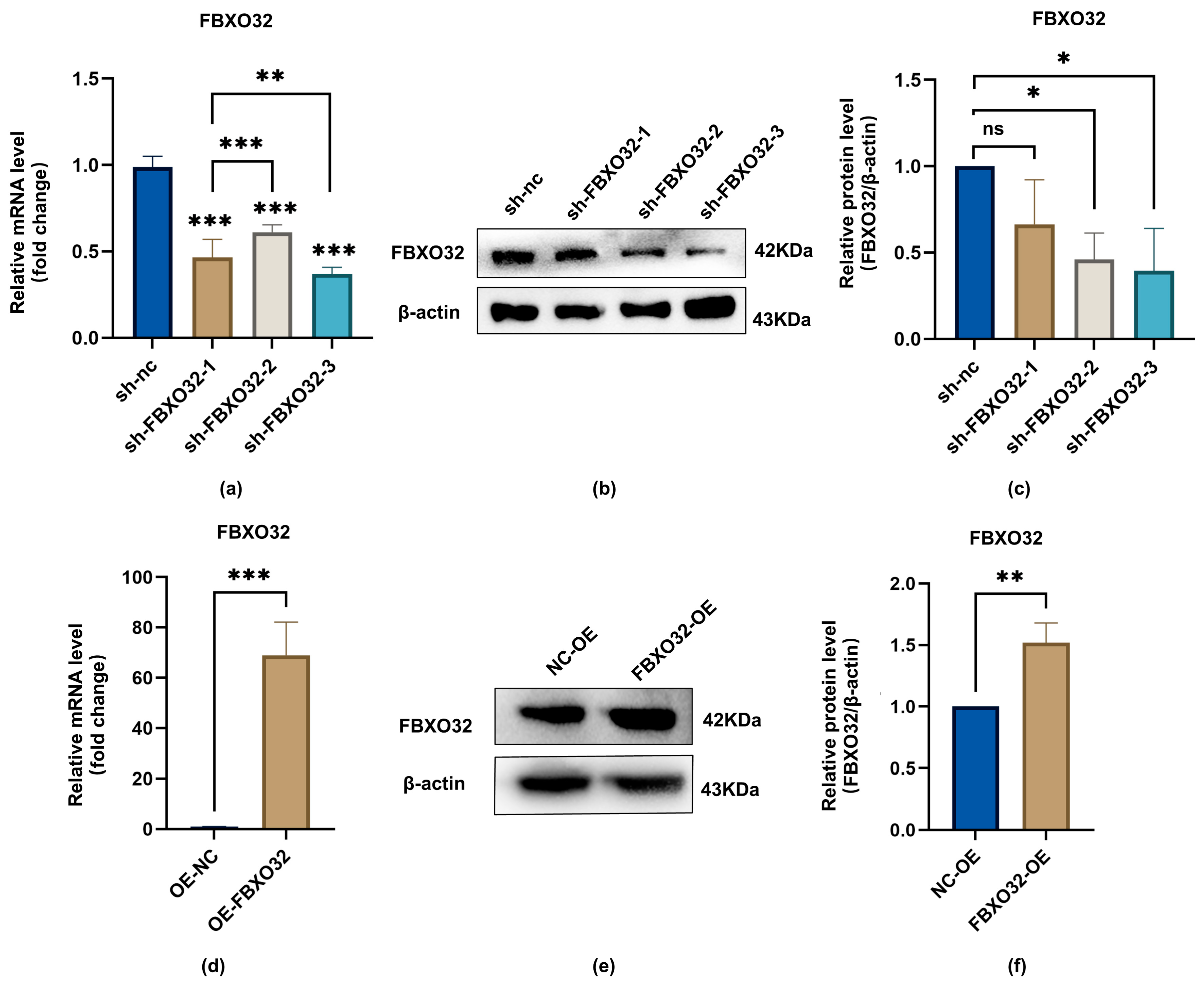
2.3. FBXO32 Positively Regulated Odontoblastic Differentiation of hDPSCs
2.3.1. FBXO32 Knockdown Impaired the Odontoblast Differentiation of hDPSCs In Vitro
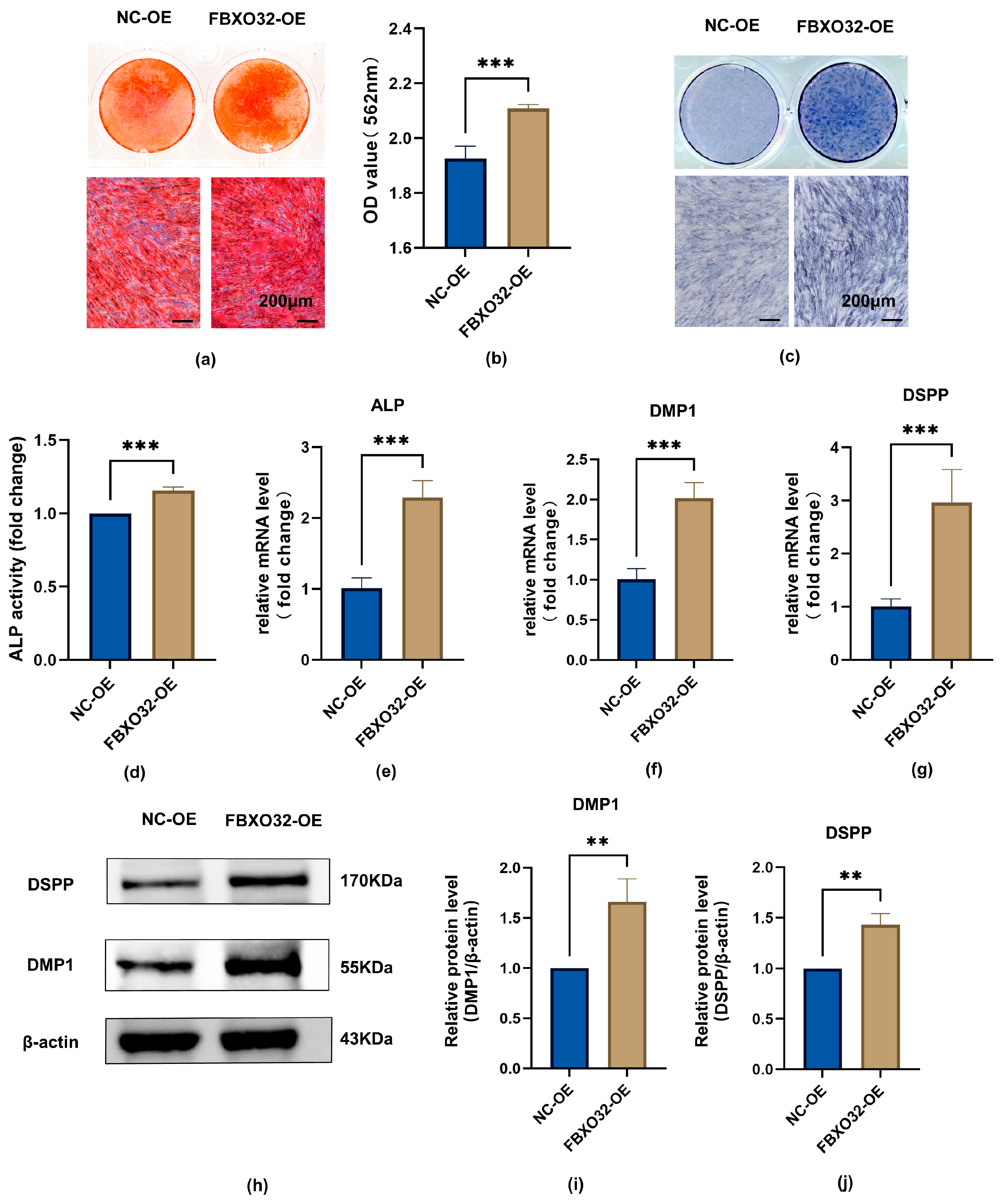
2.3.2. FBXO32 Overexpression Promoted the Odontoblast Differentiation of hDPSCs In Vitro
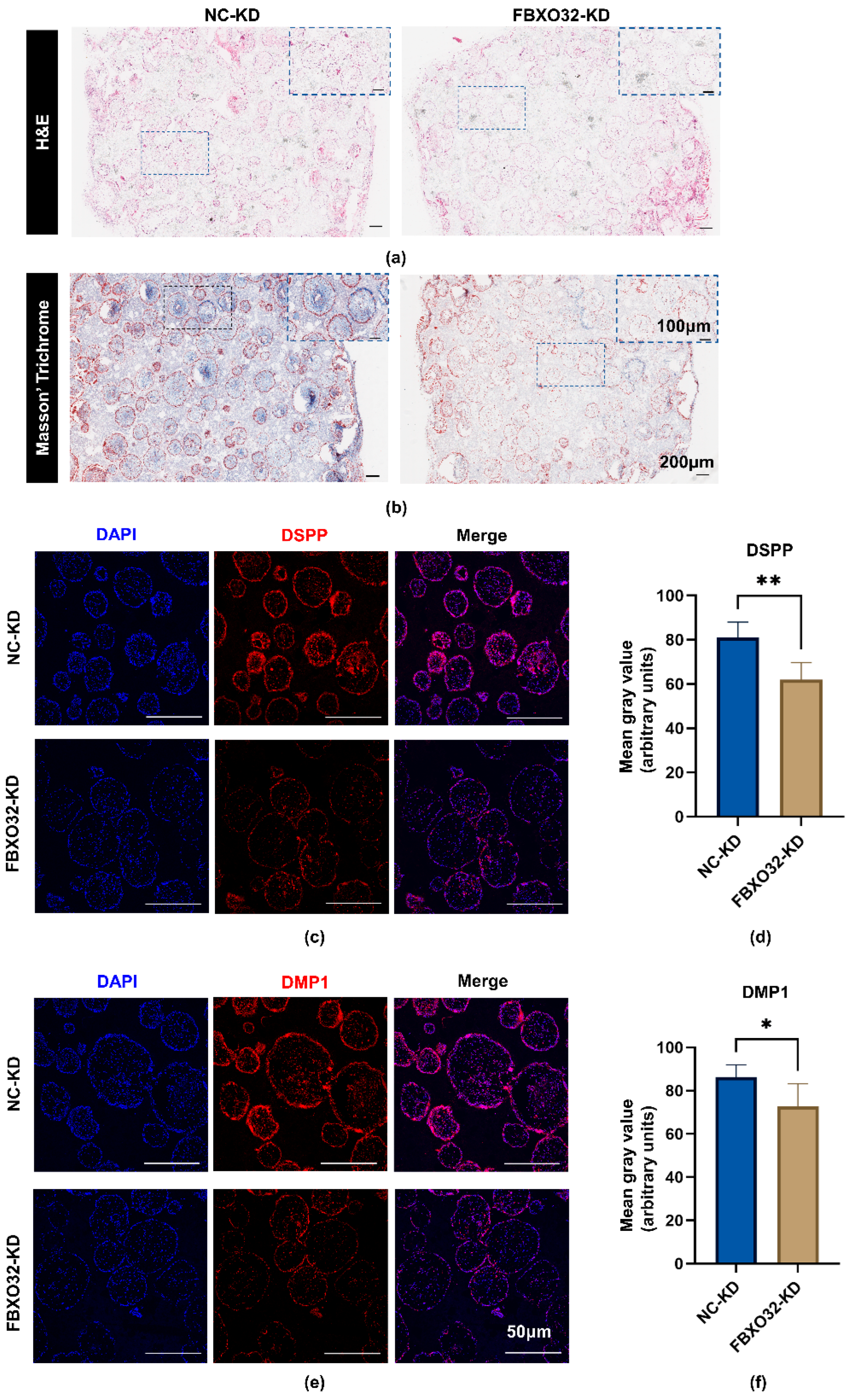
2.3.3. FBXO32 Knockdown Decreased the Mineralization Tissue Formation of hDPSCs after Subcutaneous Transplantation
2.3.4. FBXO32 Knockdown or Overexpression Does Not Influence the Proliferation and Migration of hDPSCs
3. Discussion
4. Materials and Methods
4.1. Isolation, Culture, and Identification of hDPSCs
4.2. RNA-Seq Data Analysis
4.3. Real-Time Quantitative PCR (RT-qPCR)
4.4. Western Blotting
4.5. Immunohistochemistry and Immunofluorescence Analysis
4.6. Lentivirus Packaging and Infection
4.7. ALP Staining and Activity Analysis
4.8. Alizarin Red Staining
4.9. Subcutaneous Transplantation in Nude Mice
4.10. Histological Analysis
4.11. Cell Proliferation Assay and Migration Assay
4.12. Statistical Analysis
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
References
- Ricucci, D.; Loghin, S.; Lin, L.M.; Spångberg, L.S.; Tay, F.R. Is hard tissue formation in the dental pulp after the death of the primary odontoblasts a regenerative or a reparative process? J. Dent. 2014, 42, 1156–1170. [Google Scholar] [CrossRef] [PubMed]
- Brizuela, C.; Huang, G.T.; Diogenes, A.; Botero, T.; Khoury, M. The Four Pillars for Successful Regenerative Therapy in Endodontics: Stem Cells, Biomaterials, Growth Factors, and Their Synergistic Interactions. Stem Cells Int. 2022, 2022, 1580842. [Google Scholar] [CrossRef] [PubMed]
- Bjørndal, L.; Simon, S.; Tomson, P.L.; Duncan, H.F. Management of deep caries and the exposed pulp. Int. Endod. J. 2019, 52, 949–973. [Google Scholar] [CrossRef] [PubMed]
- Gronthos, S.; Brahim, J.; Li, W.; Fisher, L.W.; Cherman, N.; Boyde, A.; DenBesten, P.; Robey, P.G.; Shi, S. Stem Cell Properties of Human Dental Pulp Stem Cells. J. Dent. Res. 2002, 81, 531–535. [Google Scholar] [CrossRef]
- Nuti, N.; Corallo, C.; Chan, B.M.; Ferrari, M.; Gerami-Naini, B. Multipotent Differentiation of Human Dental Pulp Stem Cells: A Literature Review. Stem Cell Rev. Rep. 2016, 12, 511–523. [Google Scholar] [CrossRef]
- Fang, F.; Zhang, K.; Chen, Z.; Wu, B. Noncoding RNAs: New insights into the odontogenic differentiation of dental tissue-derived mesenchymal stem cells. Stem Cell Res. Ther. 2019, 10, 297. [Google Scholar] [CrossRef]
- Kearney, M.; Cooper, P.R.; Smith, A.J.; Duncan, H.F. Epigenetic Approaches to the Treatment of Dental Pulp Inflammation and Repair: Opportunities and Obstacles. Front. Genet. 2018, 9, 311. [Google Scholar] [CrossRef]
- Fu, T.; Liu, Y.; Huang, X.; Guo, Y.; Shen, J.; Shen, H. lncRNA SNHG1 regulates odontogenic differentiation of human dental pulp stem cells via miR-328-3p/Wnt/β-catenin pathway. Stem Cell Res. Ther. 2022, 13, 311. [Google Scholar] [CrossRef]
- Yamauchi, Y.; Cooper, P.R.; Shimizu, E.; Kobayashi, Y.; Smith, A.J.; Duncan, H.F. Histone Acetylation as a Regenerative Target in the Dentine-Pulp Complex. Front. Genet. 2020, 11, 1. [Google Scholar] [CrossRef]
- Wang, N.; Han, X.; Yang, H.; Xia, D.; Fan, Z. miR-6807-5p Inhibited the Odontogenic Differentiation of Human Dental Pulp Stem Cells through Directly Targeting METTL7A. Front. Cell Dev. Biol. 2021, 9, 759192. [Google Scholar] [CrossRef]
- Qi, X.; Xiao, Q.; Sheng, R.; Jiang, S.; Yuan, Q.; Liu, W. Endogenous GDF11 regulates odontogenic differentiation of dental pulp stem cells. J. Cell. Mol. Med. 2020, 24, 11457–11464. [Google Scholar] [CrossRef]
- Li, X.; Wang, L.; Su, Q.; Ye, L.; Zhou, X.; Zhang, L.; Song, D.; Huang, D. Potential Roles of Bone Morphogenetic Protein 9 in the Odontogenic Differentiation of Dental Pulp Cells. J. Endod. 2021, 47, 436–443. [Google Scholar] [CrossRef]
- Li, J.; Wang, Z.; Wang, J.; Guo, Q.; Fu, Y.; Dai, Z.; Wang, M.; Bai, Y.; Liu, X.; Cooper, P.R.; et al. Amphiregulin regulates odontogenic differentiation of dental pulp stem cells by activation of mitogen-activated protein kinase and the phosphatidylinositol 3-kinase signaling pathways. Stem Cell Res. Ther. 2022, 13, 304. [Google Scholar] [CrossRef]
- He, P.; Zheng, L.; Zhou, X. IGFs in Dentin Formation and Regeneration: Progress and Remaining Challenges. Stem Cells Int. 2022, 2022, 3737346. [Google Scholar] [CrossRef]
- Lou, Y.; Liu, Y.; Zhao, J.; Tian, W.; Xu, N.; Zang, C.; Que, K. Activation of Transient Receptor Potential Ankyrin 1 and Vanilloid 1 Channels Promotes Odontogenic Differentiation of Human Dental Pulp Cells. J. Endod. 2021, 47, 1409–1416. [Google Scholar] [CrossRef]
- Chen, Y.; Koshy, R.; Guirado, E.; George, A. STIM1 a calcium sensor promotes the assembly of an ECM that contains Extracellular vesicles and factors that modulate mineralization. Acta Biomater. 2021, 120, 224–239. [Google Scholar] [CrossRef]
- Yang, X.; Song, Z.; Chen, L.; Wang, R.; Huang, S.; Qin, W.; Guo, J.; Lin, Z. Role of transient receptor potential channel 6 in the odontogenic differentiation of human dental pulp cells. Exp. Ther. Med. 2017, 14, 73–78. [Google Scholar] [CrossRef]
- Song, Z.; Chen, L.; Guo, J.; Qin, W.; Wang, R.; Huang, S.; Yang, X.; Tian, Y.; Lin, Z. The Role of Transient Receptor Potential Cation Channel, Subfamily C, Member 1 in the Odontoblast-like Differentiation of Human Dental Pulp Cells. J. Endod. 2017, 43, 315–320. [Google Scholar] [CrossRef]
- Ju, Y.; Ge, J.; Ren, X.; Zhu, X.; Xue, Z.; Feng, Y.; Zhao, S. Cav1.2 of L-type Calcium Channel Is a Key Factor for the Differentiation of Dental Pulp Stem Cells. J. Endod. 2015, 41, 1048–1055. [Google Scholar] [CrossRef]
- Arnold, A.; Dennison, E.; Kovacs, C.S.; Mannstadt, M.; Rizzoli, R.; Brandi, M.L.; Clarke, B.; Thakker, R.V. Hormonal regulation of biomineralization. Nat. Rev. Endocrinol. 2021, 17, 261–275. [Google Scholar] [CrossRef]
- Kovacs, C.S.; Chaussain, C.; Osdoby, P.; Brandi, M.L.; Clarke, B.; Thakker, R.V. The role of biomineralization in disorders of skeletal development and tooth formation. Nat. Rev. Endocrinol. 2021, 17, 336–349. [Google Scholar] [CrossRef] [PubMed]
- Fisher, L.W.; Fedarko, N.S. Six genes expressed in bones and teeth encode the current members of the SIBLING family of proteins. Connect. Tissue Res. 2003, 44 (Suppl. S1), 33–40. [Google Scholar] [CrossRef] [PubMed]
- Butler, W.T.; Brunn, J.C.; Qin, C. Dentin extracellular matrix (ECM) proteins: Comparison to bone ECM and contribution to dynamics of dentinogenesis. Connect. Tissue Res. 2003, 44 (Suppl. S1), 171–178. [Google Scholar] [CrossRef] [PubMed]
- Vijaykumar, A.; Dyrkacz, P.; Vidovic-Zdrilic, I.; Maye, P.; Mina, M. Expression of BSP-GFPtpz Transgene during Osteogenesis and Reparative Dentinogenesis. J. Dent. Res. 2020, 99, 89–97. [Google Scholar] [CrossRef]
- Vital, S.O.; Gaucher, C.; Bardet, C.; Rowe, P.S.; George, A.; Linglart, A.; Chaussain, C. Tooth dentin defects reflect genetic disorders affecting bone mineralization. Bone 2012, 50, 989–997. [Google Scholar] [CrossRef]
- Severe, N.; Dieudonne, F.X.; Marie, P.J. E3 ubiquitin ligase-mediated regulation of bone formation and tumorigenesis. Cell Death Dis. 2013, 4, e463. [Google Scholar] [CrossRef]
- Shen, J.; Fu, B.; Li, Y.; Wu, Y.; Sang, H.; Zhang, H.; Lin, H.; Liu, H.; Huang, W. E3 Ubiquitin Ligase-Mediated Regulation of Osteoblast Differentiation and Bone Formation. Front. Cell Dev. Biol. 2021, 9, 706395. [Google Scholar] [CrossRef]
- Yang, X.; Hao, D.; He, B. The Regulation of E3 Ubiquitin Ligases Cbl and its Cross-talking in Bone Homeostasis. Curr. Stem Cell Res. Ther. 2021, 16, 683–687. [Google Scholar] [CrossRef]
- Xu, K.; Chu, Y.; Liu, Q.; Fan, W.; He, H.; Huang, F. NEDD4 E3 Ligases: Functions and Mechanisms in Bone and Tooth. Int. J. Mol. Sci. 2022, 23, 9937. [Google Scholar] [CrossRef]
- Rape, M. Ubiquitylation at the crossroads of development and disease. Nat. Rev. Mol. Cell Biol. 2018, 19, 59–70. [Google Scholar] [CrossRef]
- Zheng, N.; Shabek, N. Ubiquitin Ligases: Structure, Function, and Regulation. Annu. Rev. Biochem. 2017, 86, 129–157. [Google Scholar] [CrossRef]
- Komander, D.; Rape, M. The Ubiquitin Code. Annu. Rev. Biochem. 2012, 81, 203–229. [Google Scholar] [CrossRef]
- Zheng, H.; Fu, J.; Chen, Z.; Yang, G.; Yuan, G. Dlx3 Ubiquitination by Nuclear Mdm2 Is Essential for Dentinogenesis in Mice. J. Dent. Res. 2022, 101, 1064–1074. [Google Scholar] [CrossRef]
- Fu, J.; Zhang, X.; Zheng, H.; Yang, G.; Chen, Z.; Yuan, G. A WWP2–PTEN–KLF5 signaling axis regulates odontoblast differentiation and dentinogenesis in mice. J. Biol. Chem. 2022, 298, 102220. [Google Scholar] [CrossRef]
- Fu, J.; Zheng, H.; Xue, Y.; Jin, R.; Yang, G.; Chen, Z.; Yuan, G. WWP2 Promotes Odontoblastic Differentiation by Monoubiquitinating KLF5. J. Dent. Res. 2021, 100, 432–439. [Google Scholar] [CrossRef]
- Zheng, H.; Yang, G.; Fu, J.; Chen, Z.; Yuan, G. Mdm2 Promotes Odontoblast-like Differentiation by Ubiquitinating Dlx3 and p53. J. Dent. Res. 2020, 99, 320–328. [Google Scholar] [CrossRef]
- Zhao, L.D.; Xu, W.C.; Cui, J.; Liang, Y.C.; Cheng, W.Q.; Xin, B.C.; Song, J. Long non-coding RNA maternally expressed gene 3 inhibits osteogenic differentiation of human dental pulp stem cells via microRNA-543/smad ubiquitin regulatory factor 1/runt-related transcription factor 2 axis. Arch. Oral Biol. 2020, 118, 104838. [Google Scholar] [CrossRef]
- Yang, F.; Xu, N.; Li, D.; Guan, L.; He, Y.; Zhang, Y.; Lu, Q.; Zhang, X. A Feedback Loop between RUNX2 and the E3 Ligase SMURF1 in Regulation of Differentiation of Human Dental Pulp Stem Cells. J. Endod. 2014, 40, 1579–1586. [Google Scholar] [CrossRef]
- Lee, D.S.; Yoon, W.J.; Cho, E.S.; Kim, H.J.; Gronostajski, R.M.; Cho, M.I.; Park, J.C. Crosstalk between Nuclear Factor I-C and Transforming Growth Factor-β1 Signaling Regulates Odontoblast Differentiation and Homeostasis. PLoS ONE 2011, 6, e29160. [Google Scholar] [CrossRef]
- Cardozo, T.; Pagano, M. The SCF ubiquitin ligase: Insights into a molecular machine. Nat. Rev. Mol. Cell Biol. 2004, 5, 739–751. [Google Scholar] [CrossRef]
- Gomes, M.D.; Lecker, S.H.; Jagoe, R.T.; Navon, A.; Goldberg, A.L. Atrogin-1, a muscle-specific F-box protein highly expressed during muscle atrophy. Proc. Natl. Acad. Sci. USA 2001, 98, 14440–14445. [Google Scholar] [CrossRef] [PubMed]
- Tintignac, L.A.; Lagirand, J.; Batonnet, S.; Sirri, V.; Leibovitch, M.P.; Leibovitch, S.A. Degradation of MyoD Mediated by the SCF (MAFbx) Ubiquitin Ligase. J. Biol. Chem. 2005, 280, 2847–2856. [Google Scholar] [CrossRef] [PubMed]
- Csibi, A.; Leibovitch, M.P.; Cornille, K.; Tintignac, L.A.; Leibovitch, S.A. MAFbx/Atrogin-1 Controls the Activity of the Initiation Factor eIF3-f in Skeletal Muscle Atrophy by Targeting Multiple C-terminal Lysines. J. Biol. Chem. 2009, 284, 4413–4421. [Google Scholar] [CrossRef]
- Li, H.H.; Kedar, V.; Zhang, C.; McDonough, H.; Arya, R.; Wang, D.Z.; Patterson, C. Atrogin-1/muscle atrophy F-box inhibits calcineurin-dependent cardiac hypertrophy by participating in an SCF ubiquitin ligase complex. J. Clin. Investig. 2004, 114, 1058–1071. [Google Scholar] [CrossRef] [PubMed]
- Li, H.H.; Willis, M.S.; Lockyer, P.; Miller, N.; McDonough, H.; Glass, D.J.; Patterson, C. Atrogin-1 inhibits Akt-dependent cardiac hypertrophy in mice via ubiquitin-dependent coactivation of Forkhead proteins. J. Clin. Investig. 2007, 117, 3211–3223. [Google Scholar] [CrossRef]
- Al-Yacoub, N.; Shaheen, R.; Awad, S.M.; Kunhi, M.; Dzimiri, N.; Nguyen, H.C.; Xiong, Y.; Al-Buraiki, J.; Al-Habeeb, W.; Alkuraya, F.S.; et al. FBXO32, encoding a member of the SCF complex, is mutated in dilated cardiomyopathy. Genome Biol. 2016, 17, 2. [Google Scholar] [CrossRef]
- Zhou, H.; Liu, Y.; Zhu, R.; Ding, F.; Wan, Y.; Li, Y.; Liu, Z. FBXO32 suppresses breast cancer tumorigenesis through targeting KLF4 to proteasomal degradation. Oncogene 2017, 36, 3312–3321. [Google Scholar] [CrossRef]
- Zhang, N.; Liao, Y.; Lv, W.; Zhu, S.; Qiu, Y.; Chen, N.; Xiao, M.; Zhang, H. FBXO32 targets PHPT1 for ubiquitination to regulate the growth of EGFR mutant lung cancer. Cell. Oncol. 2022, 45, 293–307. [Google Scholar] [CrossRef]
- Jiang, Y.; Singh, P.; Yin, H.; Zhou, Y.X.; Gui, Y.; Wang, D.Z.; Zheng, X.L.; Smooth Muscle Research Group; Libin Cardiovascular Institute of Alberta. Opposite roles of myocardin and atrogin-1 in L6 myoblast differentiation. J. Cell. Physiol. 2013, 228, 1989–1995. [Google Scholar] [CrossRef]
- Suryadevara, V.; Krehbial, C.J.; Halsey, D.; Willis, M.S. The Role of Muscle Ring Finger-1 (MuRF1), MuRF2, MuRF3, and Atrogin-1 on Bone Microarchitecture In Vivo. Cell Biochem. Biophys. 2022, 80, 415–426. [Google Scholar] [CrossRef]
- Chen, Z.; Zhang, K.; Qiu, W.; Luo, Y.; Pan, Y.; Li, J.; Yang, Y.; Wu, B.; Fang, F. Genome-wide identification of long noncoding RNAs and their competing endogenous RNA networks involved in the odontogenic differentiation of human dental pulp stem cells. Stem Cell Res. Ther. 2020, 11, 114. [Google Scholar] [CrossRef]
- Wu, Y.; Sun, H.; Song, F.; Fu, D.; Wang, J. DDIT3 overexpression increases odontoblastic potential of human dental pulp cells. Cell Prolif. 2014, 47, 249–257. [Google Scholar] [CrossRef]
- Bodine, S.C.; Baehr, L.M. Skeletal muscle atrophy and the E3 ubiquitin ligases MuRF1 and MAFbx/atrogin-1. Am. J. Physiol.-Endocrinol. Metab. 2014, 307, E469–E484. [Google Scholar] [CrossRef]
- Galasso, G.; De Rosa, R.; Piscione, F.; Iaccarino, G.; Vosa, C.; Sorriento, D.; Piccolo, R.; Rapacciuolo, A.; Walsh, K.; Chiariello, M. Myocardial expression of FOXO3a–Atrogin-1 pathway in human heart failure. Eur. J. Heart Fail. 2010, 12, 1290–1296. [Google Scholar] [CrossRef]
- Zhang, D.M.; He, T.; Katusic, Z.S.; Lee, H.C.; Lu, T. Muscle-Specific F-Box Only Proteins Facilitate BK Channel β(1) Subunit Downregulation in Vascular Smooth Muscle Cells of Diabetes Mellitus. Circ. Res. 2010, 107, 1454–1459. [Google Scholar] [CrossRef]
- Cushing, L.; Costinean, S.; Xu, W.; Jiang, Z.; Madden, L.; Kuang, P.; Huang, J.; Weisman, A.; Hata, A.; Croce, C.M.; et al. Disruption of miR-29 Leads to Aberrant Differentiation of Smooth Muscle Cells Selectively Associated with Distal Lung Vasculature. PLoS Genet. 2015, 11, e1005238. [Google Scholar] [CrossRef]
- Bdolah, Y.; Segal, A.; Tanksale, P.; Karumanchi, S.A.; Lecker, S.H. Atrophy-related ubiquitin ligases atrogin-1 and MuRF-1 are associated with uterine smooth muscle involution in the postpartum period. Am. J. Physiol. Regul. Integr. Comp. Physiol. 2007, 292, R971–R976. [Google Scholar] [CrossRef]
- Zhang, Y.; Liu, Q.; Cui, M.; Wang, M.; Hua, S.; Gao, J.; Liao, Q. Comprehensive Analysis of Expression, Prognostic Value, and Immune Infiltration for Ubiquitination-Related FBXOs in Pancreatic Ductal Adenocarcinoma. Front. Immunol. 2021, 12, 774435. [Google Scholar] [CrossRef]
- García-Valverde, A.; Rosell, J.; Sayols, S.; Gómez-Peregrina, D.; Pilco-Janeta, D.F.; Olivares-Rivas, I.; de Álava, E.; Maurel, J.; Rubió-Casadevall, J.; Esteve, A.; et al. E3 ubiquitin ligase Atrogin-1 mediates adaptive resistance to KIT-targeted inhibition in gastrointestinal stromal tumor. Oncogene 2021, 40, 6614–6626. [Google Scholar] [CrossRef]
- Sun, H.L.; Wu, Y.R.; Huang, C.; Wang, J.W.; Fu, D.J.; Liu, Y.C. The Effect of SIRT6 on the Odontoblastic Potential of Human Dental Pulp Cells. J. Endod. 2014, 40, 393–398. [Google Scholar] [CrossRef]
- Petroski, M.D.; Deshaies, R.J. Function and regulation of cullin–RING ubiquitin ligases. Nat. Rev. Mol. Cell Biol. 2005, 6, 9–20. [Google Scholar] [CrossRef] [PubMed]
- Blondelle, J.; Shapiro, P.; Domenighetti, A.A.; Lange, S. Cullin E3 Ligase Activity Is Required for Myoblast Differentiation. J. Mol. Biol. 2017, 429, 1045–1066. [Google Scholar] [CrossRef] [PubMed]
- Rodrigues, T.L.; Foster, B.L.; Silverio, K.G.; Martins, L.; Casati, M.Z.; Sallum, E.A.; Somerman, M.J.; Nociti, F.H., Jr. Hypophosphatasia-associated Deficiencies in Mineralization and Gene Expression in Cultured Dental Pulp Cells Obtained from Human Teeth. J. Endod. 2012, 38, 907–912. [Google Scholar] [CrossRef] [PubMed]
- Foster, B.L.; Nagatomo, K.J.; Tso, H.W.; Tran, A.B.; Nociti, F.H., Jr.; Narisawa, S.; Yadav, M.C.; McKee, M.D.; Millán, J.I.; Somerman, M.J. Tooth root dentin mineralization defects in a mouse model of hypophosphatasia. J. Bone Miner. Res. 2013, 28, 271–282. [Google Scholar] [CrossRef] [PubMed]
- Guo, S.; Lim, D.; Dong, Z.; Saunders, T.L.; Ma, P.X.; Marcelo, C.L.; Ritchie, H.H. Dentin Sialophosphoprotein: A Regulatory Protein for Dental Pulp Stem Cell Identity and Fate. Stem Cells Dev. 2014, 23, 2883–2894. [Google Scholar] [CrossRef]
- Liang, T.; Zhang, H.; Xu, Q.; Wang, S.; Qin, C.; Lu, Y. Mutant Dentin Sialophosphoprotein Causes Dentinogenesis Imperfecta. J. Dent. Res. 2019, 98, 912–919. [Google Scholar] [CrossRef]
- Lu, Y.; Ye, L.; Yu, S.; Zhang, S.; Xie, Y.; McKee, M.D.; Li, Y.C.; Kong, J.; Eick, J.D.; Dallas, S.L.; et al. Rescue of odontogenesis in Dmp1-deficient mice by targeted re-expression of DMP1 reveals roles for DMP1 in early odontogenesis and dentin apposition in vivo. Dev. Biol. 2007, 303, 191–201. [Google Scholar] [CrossRef]
- Ye, L.; MacDougall, M.; Zhang, S.; Xie, Y.; Zhang, J.; Li, Z.; Lu, Y.; Mishina, Y.; Feng, J.Q. Deletion of Dentin Matrix Protein-1 Leads to a Partial Failure of Maturation of Predentin into Dentin, Hypomineralization, and Expanded Cavities of Pulp and Root Canal during Postnatal Tooth Development. J. Biol. Chem. 2004, 279, 19141–19148. [Google Scholar] [CrossRef]
- Arora, S.; Cooper, P.R.; Ratnayake, J.T.; Friedlander, L.T.; Rizwan, S.B.; Seo, B.; Hussaini, H.M. A critical review of in vitro research methodologies used to study mineralization in human dental pulp cell cultures. Int. Endod. J. 2022, 55, 3–13. [Google Scholar] [CrossRef]
- Ching, H.S.; Luddin, N.; Rahman, I.A.; Ponnuraj, K.T. Expression of Odontogenic and Osteogenic Markers in DPSCs and SHED: A Review. Curr. Stem Cell Res. Ther. 2017, 12, 71–79. [Google Scholar] [CrossRef]
- Habel, N.; El-Hachem, N.; Soysouvanh, F.; Hadhiri-Bzioueche, H.; Giuliano, S.; Nguyen, S.; Horák, P.; Gay, A.-S.; Debayle, D.; Nottet, N.; et al. FBXO32 links ubiquitination to epigenetic reprograming of melanoma cells. Cell Death Differ. 2021, 28, 1837–1848. [Google Scholar] [CrossRef]
- Li, Y.; Gao, S.; Xue, W.; Ma, Y.; Meng, Y.; Zhang, D. mir-19a-3p Functions as an Oncogene by Regulating FBXO32 Expression in Multiple Myeloma. Balk. Med. J. 2021, 38, 43–49. [Google Scholar] [CrossRef]
- Mei, Z.; Zhang, D.; Hu, B.; Wang, J.; Shen, X.; Xiao, W. FBXO32 Targets c-Myc for Proteasomal Degradation and Inhibits c-Myc Activity. J. Biol. Chem. 2015, 290, 16202–16214. [Google Scholar] [CrossRef]
- Cai, E.Y.; Kufeld, M.N.; Schuster, S.; Arora, S.; Larkin, M.; Germanos, A.A.; Hsieh, A.C.; Beronja, S. Selective Translation of Cell Fate Regulators Mediates Tolerance to Broad Oncogenic Stress. Cell Stem Cell 2020, 27, 270–283.e7. [Google Scholar] [CrossRef]
- Liu, Q.; Fan, W.; He, Y.; Zhang, F.; Guan, X.; Deng, Q.; Lu, X.; He, H.; Huang, F. Effects of melatonin on the proliferation and differentiation of human dental pulp cells. Arch. Oral Biol. 2017, 83, 33–39. [Google Scholar] [CrossRef]
- Li, J.; Deng, Q.; Fan, W.; Zeng, Q.; He, H.; Huang, F. Melatonin-induced suppression of DNA methylation promotes odontogenic differentiation in human dental pulp cells. Bioengineered 2020, 11, 829–840. [Google Scholar] [CrossRef]
- Deng, Q.; Liu, Q.; Zhang, H.; Fan, W.; Li, J.; Kang, J.; He, H.; Huang, F. Melatonin enhances hydrogen peroxide-induced apoptosis in human dental pulp cells. J. Dent. Sci. 2019, 14, 370–377. [Google Scholar] [CrossRef]
- Li, B.; Yu, F.; Wu, F.; Hui, T.; Liao, X.; Yin, B.; Wang, C.; Ye, L. EZH2 Impairs Human Dental Pulp Cell Mineralization via the Wnt/beta-Catenin Pathway. J. Dent. Res. 2018, 97, 571–579. [Google Scholar] [CrossRef]



| Gene | Species | Primer Sequence (5′–3′) | Amplicon Product Size (bp) | Accession No. |
|---|---|---|---|---|
| ALP | Human | F: AACATCAGGGACATTGACGTG R: GTATCTCGGTTTGAAGCTCTTCC | 159 | NM_000478.6 |
| DMP-1 | Human | F: CACTCAAGATTCAGGTGGCAG R: TCTGAGATGCGAGACTTCCTAAA | 75 | NM_001079911.3 |
| DSPP | Human | F: GCATTTGGGCAGTAGCATGG R: CACTGGCATTTAACTCATCCTGT | 132 | NM_014208.3 |
| FBXO32 | Human | F: GCCTTTGTGCCTACAACTGAA R: CTGCCCTTTGTCTGACAGAAT | 187 | NM_001242463.2 |
| BCL6 | Human | F: ACACATCTCGGCTCAATTTGC R: AGTGTCCACAACATGCTCCAT | 89 | NM_001130845.2 |
| DDIT3 | Human | F: GGAAACAGAGTGGTCATTCCC R: CTGCTTGAGCCGTTCATTCTC | 116 | NM_001195053.1 |
| ABHD2 | Human | F: TCTACTTCCAGGACTCGGGG R: TCACCCTTCCCATCTTCCCA | 138 | NM_007011.8 |
| S100A6 | Human | F: CTTCCACAAGTACTCCGGCA R: GCCAATGGTGAGCTCCTTCT | 91 | NM_014624.4 |
| NKD2 | Human | F: GAGGACCAGTGTCCCCTACAG R: CTCCGTCATCTGCGCTGAG | 91 | NM_001271082.2 |
| RHO | Human | F: CATGACCATCCCAGCGTTCT R: CTTGGACACGGTAGCAGAGG | 157 | NM_000539.3 |
| WFS1 | Human | F: GAGAATGTCGGCCAGGTCAA R: CGATGTAGTTCTTGGACTCGCT | 124 | NM_001145853.1 |
| GAPDH | Human | F: TCTCCTCTGACTTCAACAGCGACA R: CCCTGTTGCTGTAGCCAAATTCGT | 126 | NM_001256799.3 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Xu, K.; Liu, Q.; Huang, W.; Chu, Y.; Fan, W.; Liu, J.; He, Y.; Huang, F. Promotive Effect of FBXO32 on the Odontoblastic Differentiation of Human Dental Pulp Stem Cells. Int. J. Mol. Sci. 2023, 24, 7708. https://doi.org/10.3390/ijms24097708
Xu K, Liu Q, Huang W, Chu Y, Fan W, Liu J, He Y, Huang F. Promotive Effect of FBXO32 on the Odontoblastic Differentiation of Human Dental Pulp Stem Cells. International Journal of Molecular Sciences. 2023; 24(9):7708. https://doi.org/10.3390/ijms24097708
Chicago/Turabian StyleXu, Ke, Qin Liu, Wushuang Huang, Yanhao Chu, Wenguo Fan, Jiawei Liu, Yifan He, and Fang Huang. 2023. "Promotive Effect of FBXO32 on the Odontoblastic Differentiation of Human Dental Pulp Stem Cells" International Journal of Molecular Sciences 24, no. 9: 7708. https://doi.org/10.3390/ijms24097708
APA StyleXu, K., Liu, Q., Huang, W., Chu, Y., Fan, W., Liu, J., He, Y., & Huang, F. (2023). Promotive Effect of FBXO32 on the Odontoblastic Differentiation of Human Dental Pulp Stem Cells. International Journal of Molecular Sciences, 24(9), 7708. https://doi.org/10.3390/ijms24097708