Abstract
Baker’s yeast, S. cerevisiae, is an excellent model organism exploited for molecular genetic studies of the mechanisms of genome stability in eukaryotes. Genetic peculiarities of commonly used yeast strains impact the processes of DNA replication, repair, and recombination (RRR). We compared the genomic DNA sequence variation of the five strains that are intensively used for RRR studies. We used yeast next-generation sequencing data to detect the extent and significance of variation in 183 RRR genes. We present a detailed analysis of the differences that were found even in closely related strains. Polymorphisms of common yeast strains should be considered when interpreting the outcomes of genome stability studies, especially in cases of discrepancies between laboratories describing the same phenomena.
1. Introduction
Molecular biology and genetic studies using a simple eukaryote, Saccharomyces cerevisiae yeast, had, have, and will immensely impact the DNA replication, repair, and recombination (RRR) field [1,2]. Yeast strains that were collected and stored by Emil Hansen in the 1880s were introduced in laboratory practice by Öjvind Winge 50 years later. These strains diverted to several groups of modern yeast strains [3,4]. The strains have similar genetic content but multiple single nucleotide changes, different Ty1 element distributions, and structural variations [5,6]. The evidence has accumulated that some variations affect DNA maintenance genes that might be critical for the performance of this machinery. One of the most known classic examples is the finding that a widely used control strain, W303, carries a mutated RAD5 gene, allele rad5-535 [7]. Rad5 is a multifunctional helicase/ubiquitin ligase. The mutation alters the conserved nucleotide binding motif 535GXGKT to 535RXGKT (changed amino acid is underlined). Strains with the rad5-535 allele are slightly sensitive to a mutagen MMS and show altered genetic interactions with soh2 mutants affecting the RNA polymerase mediator complex. Rad5 is implicated in template switch during replication [8], and in translesion DNA synthesis (TLS) [9,10]. Polymorphism in RAD5 might be a weighty modifier of these processes. Strain yJF1, whose derivatives are used to produce proteins for reconstitutions of replication fork in vitro, is the relative of W303 and possesses this allele [11]. Variation between the strains is a common reason for different rates of replication origin activation [12]. The yeast genome results from ancestral genome duplication [13], and many genes have paralogs that are under less stringent selection and evolve rapidly. Examples are RNR genes that have remained paralogs at various stages of diversification. As a result, some strains with deletion of the gene encoding a major subunit of RNR1 are inviable, e.g., W303, but some are viable, BY4741/S288C [14,15]. Another example is a polymorphism of strains explaining critical parameters connecting telomere length and longevity [16].
Before the next-generation sequencing (NGS) era, discrepancies in the results of DNA repair studies were attributed to unknown variations in the genetic background of the used strains. In one extreme case, it was proposed that unknown genetic differences may affect the interpretation of genetic experiments defining the arrangement of DNA polymerases at the fork [17,18]. Many classic and new yeast strains that are used for studies of DNA metabolism and genome stability have now been sequenced, and the genetic causes of phenotypic differences have been uncovered [19,20]. Here, we analyzed the differences in significant RRR genes among several common laboratory strains and LAN series that we have used for the studies of peculiarities of mutagenesis in diploids by a base analog and APOBEC deaminases with an emphasis on kataegis and the hypermutable fraction of yeast cells [21,22,23]. LAN series are close derivatives of the CG379 strain, which has been used for pioneer studies of the consequences of defective DNA polymerase proofreading, defects of mismatch repair (MMR), or their combination on mutation rates [24,25,26,27,28,29]. A variant of the CG379 strain called E134 [29,30] is one of the primary strains for modeling mutator phenotypes of cancer-associated mutations [31,32].
2. Results
2.1. Characteristics of Strains Examined
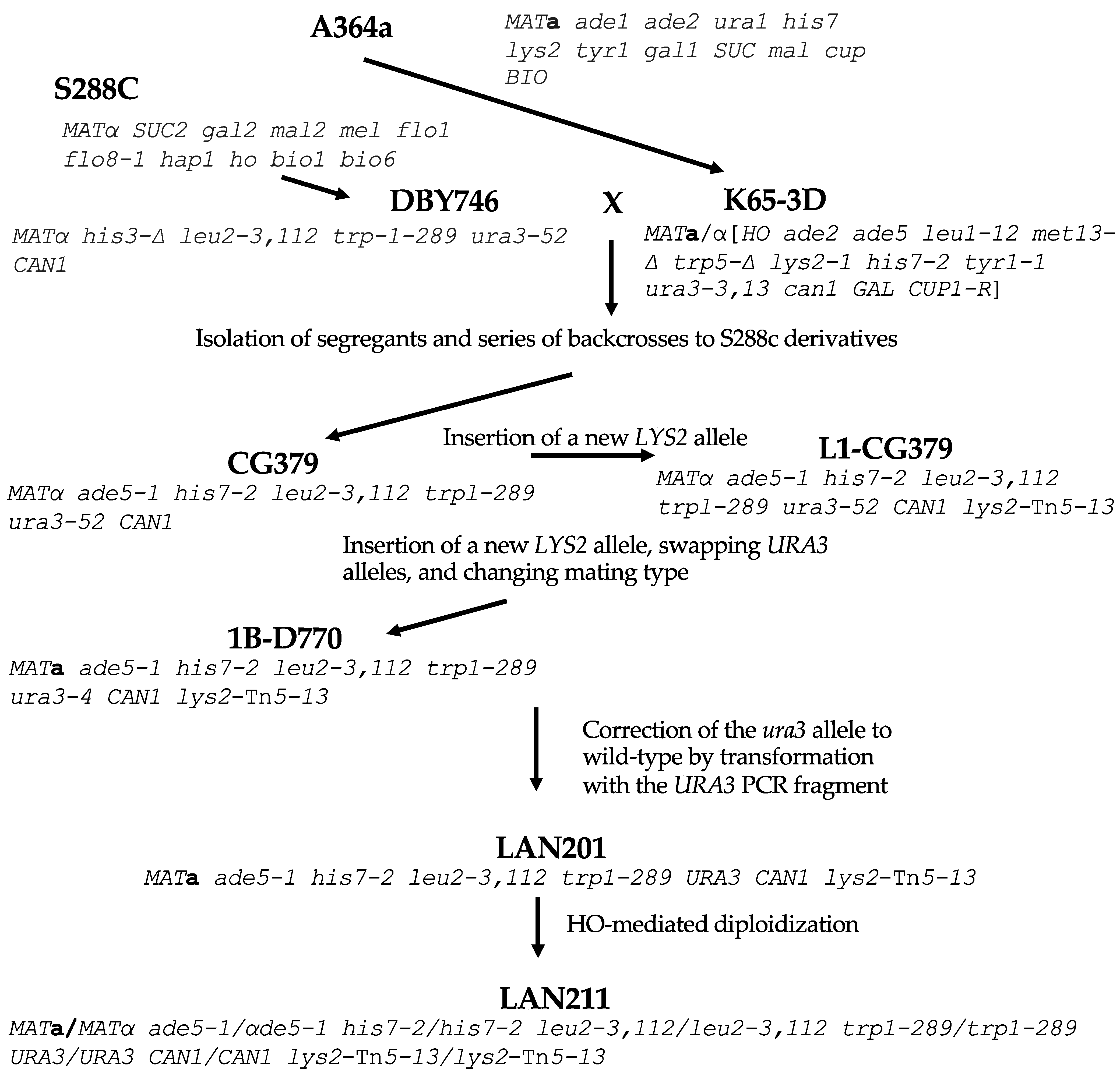
For our comparison, we analyzed the genome of the strain that is the closest relative of the first sequenced yeast strain, S288C, BY4742 (MATα his3-Δ1 leu2-Δ0 lys2-Δ0 ura3-Δ0). BY4742, and its sibling of a different mating type, BY4741, are widely used for various genetic studies, including genome instability analysis because of the availability of the library of systematic deletions [33,34]. Its genealogy is described in [35]. W303-1A and -1B are MATa and MATα haploids correspondingly, with the same genotype (ade2-1 ura3-1 his3-11, 15 trp1-1 leu2-3, 112 can1-100) but differ in mating type, originating from W303 diploid parent [36]. W303 is likely closely related to S288C although its history is not well-defined [37]. Less related is SK1, a homothallic strain (MATa/MATα [HO can1 gal2 cup] [38,39] that is widely used for studies of yeast metabolism and meiosis [40,41]. It is quite distant from S288C [42]. The known parts of the ancestry of LAN201 and LAN211 are summarized in Figure 1. They are derived from CG379 by a series of integration–excisions, one-step gene replacements, mating type switching by HO-containing plasmid, and crosses.
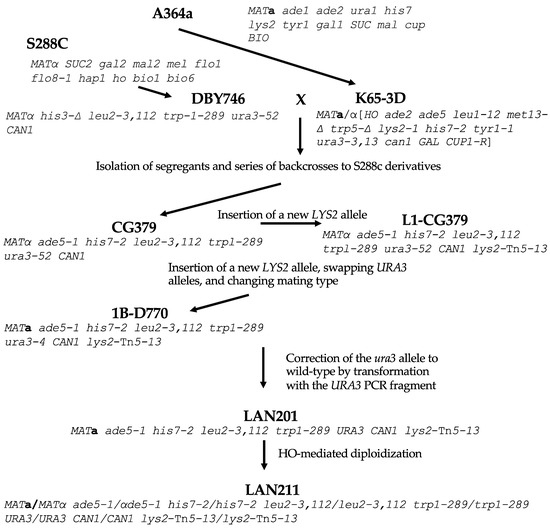
Figure 1.
The genealogy of LAN201 and LAN210 strains. The principal ancestors are A464a [43] and its derivative K65-3D (brackets in genotype are used to indicate the homozygous diploid genotype of homothallic HO strain) [44] on one side, and S288c [4] and its derivative DBY746, on the other [45]. Dr. Craig Giroux (NIEHS, USA) performed a series of backcrosses to find segregants with the desired markers. This yielded CG379, the basic strain for creating DNA polymerase mutations [24]. The lys2 allele with Tn5 insertion was introduced as described in [46], ura3-52 was replaced by the ura3-4 allele, and the strain was made diploid by HO endonuclease. Tetrad dissection gave segregant 1B-D770 with a changed mating type [30]. Next, ura3-4 was converted back to URA3 to create LAN201 [22], which gave rise to diploid LAN211 by auto-diploidization with the assistance of an HO-bearing plasmid.
2.2. DNA Sequence Variations in Genomes of the Yeast Strains
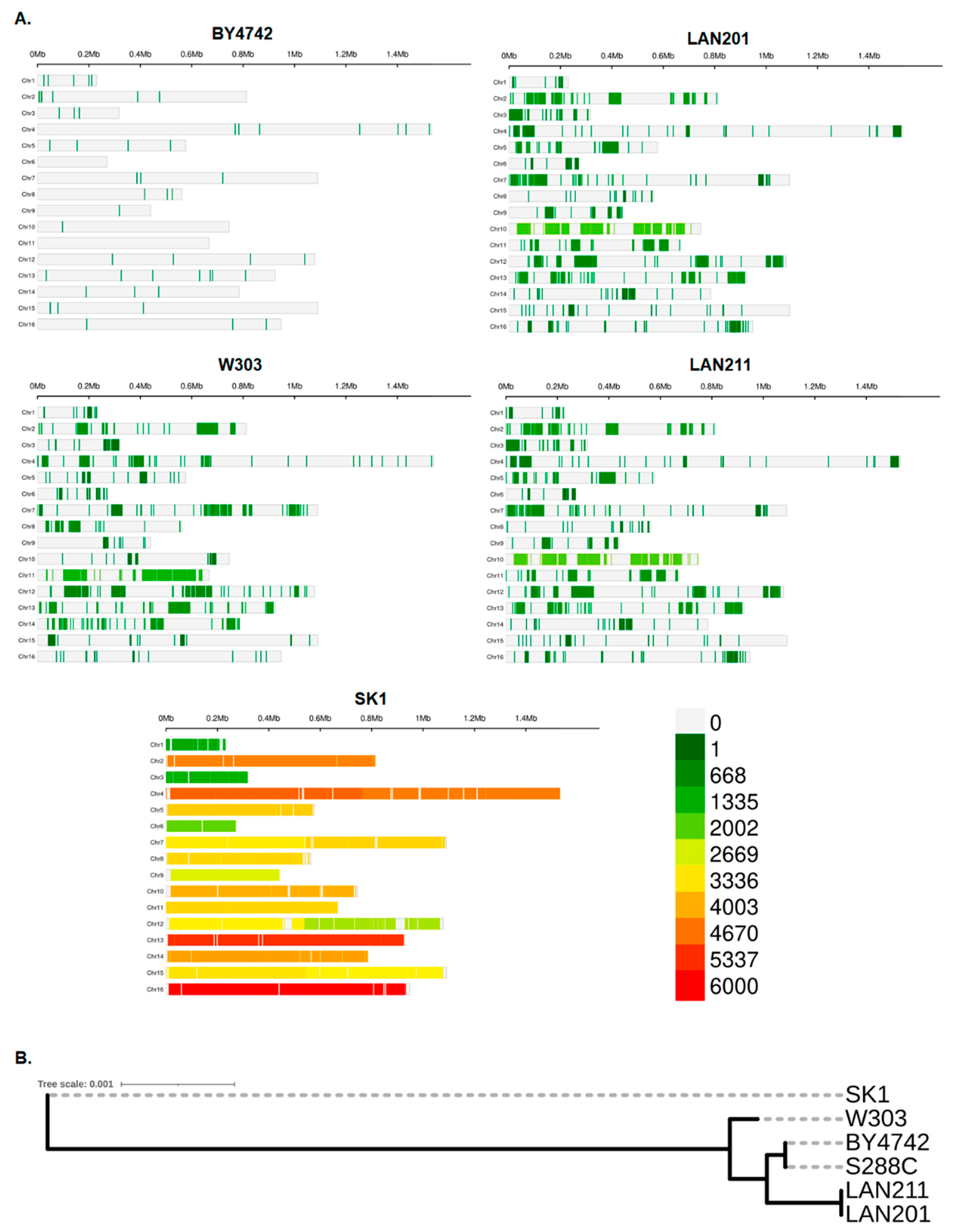
We compared whole genome sequencing data of the five strains (data sources are provided in Materials and Methods, Section 4.1, [22,33]) by the methodology described in Section 4.2 and Section 4.3. The results, expressed as differences from the canonical S288C genome, are summarized in Table 1, left half, and illustrated in Figure 2A. The closest to the S288C is the BY4742 strain, which has only 54 variants leading to amino acid changes, with a few variants per chromosome distributed relatively evenly (there is not a single chromosome without changes, Figure 2). The W303 and LAN series appear to have more differences from S288C, and the three strains are quite similar regarding the number of variants (around 2000). The pattern of mutation distribution in W303 differs from LANs in many cases. The most evident examples are large blocks with around 1000 mutations per Mb at different locations in chromosomes II, VII, and XI (Figure 2). LAN201 and its autodiploid LAN211 are virtually identical, both in pattern of mutation localization and in numbers of mutations. However, the transformation by HO-containing plasmid and diploidization (Figure 1) was not wholly benign and a few changes were accumulated. In the following paragraphs, we will refer to these strains under the umbrella name “LAN”. SK1 has overwhelming eight-fold more differences (Table 1). It is vividly demonstrated by a complete change of color in the panel describing this strain in Figure 2A. Panel Figure 2B illustrates a considerable distance between SK1 and the group of the other strains.

Table 1.
Genomic view of polymorphism of yeast strains: differences from S288C.
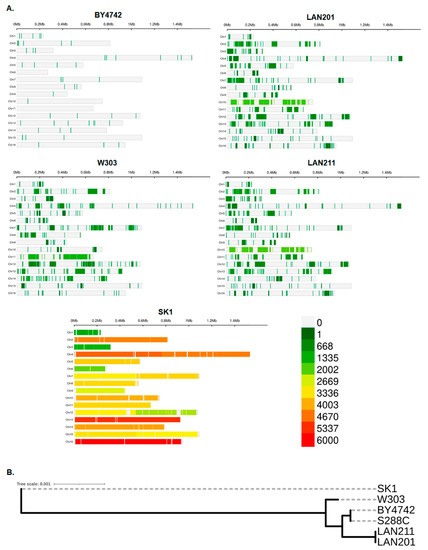
Figure 2.
Genome-wide overview of the strain’s differences. (A) View of individual chromosomes where regions with a sequence deviation from S288C are shown in a heatmap based on the number of single nucleotide polymorphisms (SNPs) within 1 Mb window size on the 16 chromosomes (the scale 0-6000 is on the bottom right). Analysis was performed with CMplot (Materials and Methods). (B) A simple genealogy tree illustrating the considerable evolutionary distance of SK1 from S288C, constructed by REALPHY v.1.13 as described in the Materials and Methods.
2.3. DNA Sequence Variations in Genes of the RRR Panel
Next, we compiled a list of 183 genes relevant to genome stability (arranged alphabetically with a short annotation in Supplementary Table S1). The panel includes genes encoding for DNA polymerases and accessory subunits; other proteins involved in DNA replication, repair, and recombination; nucleotide metabolism; chromatin remodeling; cell cycle; checkpoint; and others. We understand that the list might need to be updated because of so many intertwining processes in the cell, but we believe that we selected most of the important genes. Approximately one-third of the genes in our panel are essential for vegetative growth. Only five genes do not harbor non-synonymic or other significant changes: NTG1, RFA1, RIM1, UBC13, and HAM1. All other genes possess non-synonymous changes that are predicted (by methods described in Section 4.2) to exert a moderate to high impact on protein function, although they are primarily found in the genome of SK1 (Table 1, right half). The analysis of alterations in the gene of the panel revealed only 3 changes in BY4742, around 100 changes in W303-LANs, and more than 600 changes in SK1, averaging around 4 per gene. The information on the position and types of changes for RRR genes are summarized in Supplementary Table S2 and for all genes in Supplementary Table S3. Variations happen both in essential and non-essential genes.
2.4. DNA Sequence Variations in DNA Polymerase-Related Genes
DNA replication is a major characteristic of life. The variation in the genes for the apparatus of replication determines the properties of organisms. Due to our long-term interest in replication [47,48], we focused on polymorphisms of the DNA polymerase-related genes in the five strains (Figure 3). Non-synonymic changes and multiple mutations were prevalent in all the selected genes in SK1 irrespective of essentiality. On the contrary, BY4742 possessed almost no differences from S288C; the only affected essential genes were PSF3 (although multiple changes do not lead to amino acid changes in the protein, Figure 3B, Supplementary Table S2), a component of the GINS complex necessary for initiation of replication [49] and DPB2, a second subunit of pol ε [50]. Both genes are related to the leading DNA strand replication. LANs and W303 possess an intermediate number of variations. Table 2 lists the detected amino acid changes. Not all of the multiple changes listed in Figure 3 cause amino acid substitutions, but they can alter a gene’s transcription or mRNA stability (see details of these changes in Supplementary Table S2). These changes are inherited from CG379 because all the changes in the LAN strains listed in Table 2 are also present in another direct descendant of CG379, strain ySR128 (https://www.ncbi.nlm.nih.gov/nuccore/?term=ySR128 [ncbi.nlm.nih.gov] 29 March 2023) that is extensively used for studies of mutagenic effects of APOBEC deaminases and other agents preferentially damaging ssDNA [51,52,53]. W303 possessed the same amino acid change in Dpb2 as BY4742 and, in addition, a change in Rad5. This finding serves as an internal control for the accuracy of our analysis because the change of Rad5 in this strain was one of the examples of how yeast polymorphisms can affect parameters of genetic processes (Introduction, [7]). No amino acid changes were found in Dpb3, Psf1-3, Pol3, and Pol30, the proteins responsible for bulk replication of the yeast genome. However, another essential component of pol ε, Dpb2, varied in all strains. Conversely, a conditional non-essential Pol32 was stable in BY4742 and W303 but harbored several substitutions in LAN and SK1. Some of these substitutions were shared, alluding to common ancestors.

Figure 3.
DNA polymerases and accessory protein-encoding genes: differences from S288C. Essential genes are highlighted in light blue. One gene, POL32, is highlighted yellow because yeast with the deletion of the gene is viable at 30 °C but non-viable at 13 °C (cold-sensitivity) [54]. Deleting the POL32 ortholog in fission yeast S. pombe (cdc27) is also lethal [55]. Triangles indicate two variants of the corresponding types (color-coded) in the respective strain/gene combination. Multiple changes include upstream and downstream alterations (5′- and 3′-UTRs), synonymous changes, and missense mutations. (A) SNPs and indels: all changes, including promoter, transcription start, and termination zone sequences. (B) SNPs and indels in coding regions.

Table 2.
Non-synonymous amino acid differences of the five strains from S288C in DNA polymerases and accessory proteins.
2.5. Genetic Specifics of LAN Strains
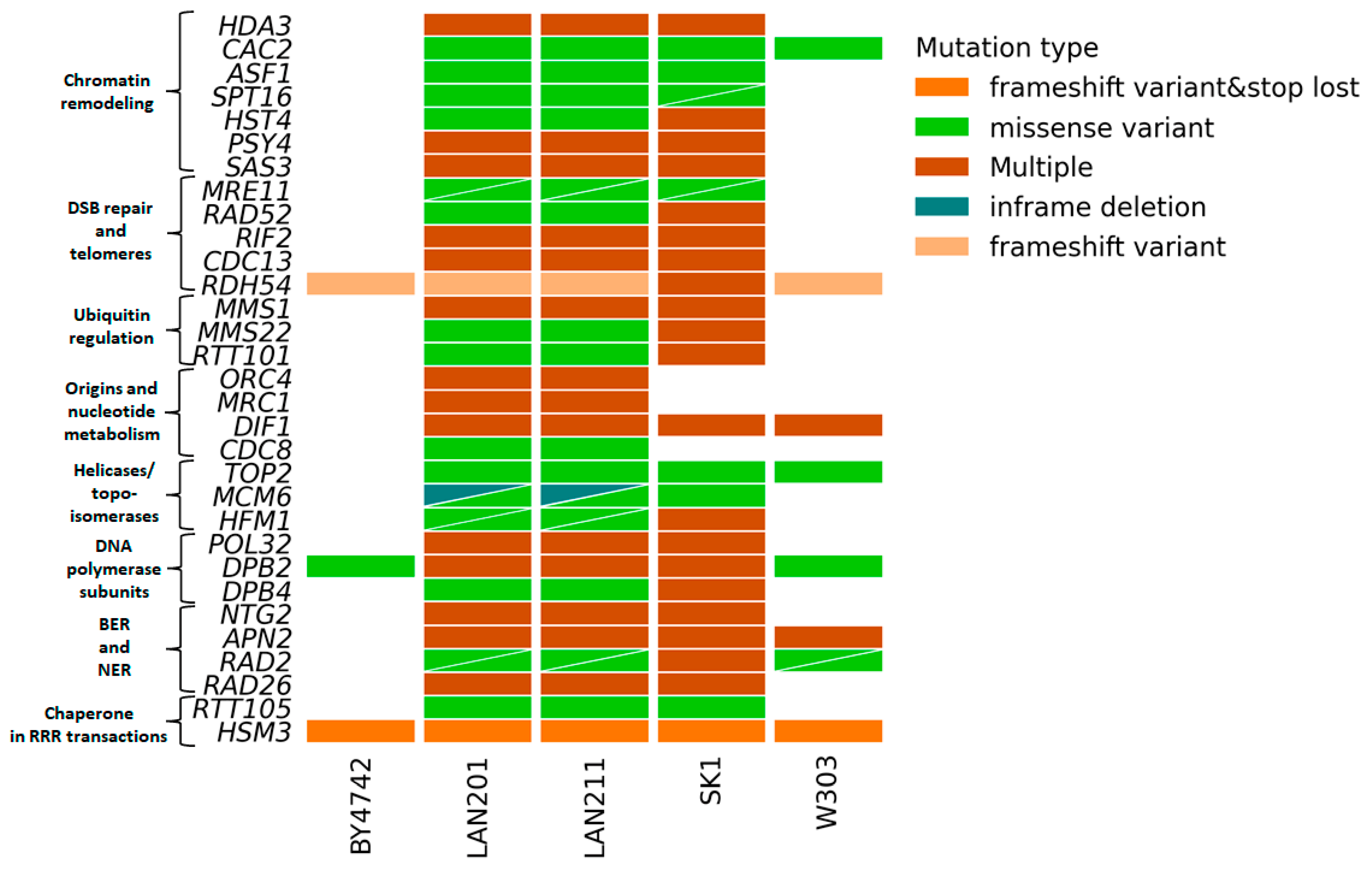
We were particularly interested in the status of RRR genes in LAN strains because of the extensive work performed by us and others with these strains or their close relatives (Introduction). Figure 4 illustrates the main differences between these strains from S288C and others (details of the analysis results are in Supplementary Table S2). The figure shows what genes are changed in the LAN strains and whether the same genes are polymorphic in other strains. Only a few genes that are variable in LANs are also variable in BY4742 and W303. In contrast, most genes that are variable in LANs are also variable in SK1. The ubiquitous variants across the panel are in the two non-essential genes: RDH54, involved in recombination, and HSM3, a chaperone involved in DNA mismatch repair, and in one essential gene, DPB2 discussed in Section 2.4. LAN strains carry variants in all genome stability pathways; the most prominent are in genes participating in chromatin remodeling, DSB repair, DNA polymerases, and cell cycle control. The significance of these variations has to be determined, but the results of our study might help with interpreting and comparing the results of studies performed on different strain backgrounds.
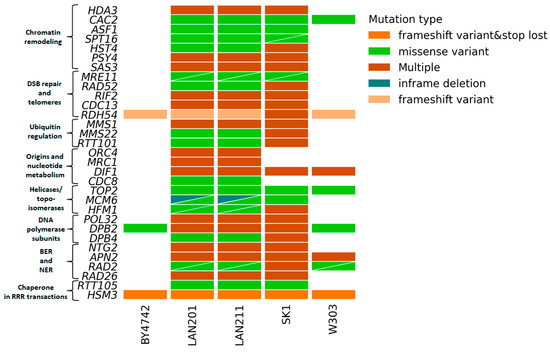
Figure 4.
Variations predicted to alter gene function in 183-gene RRR panel in LAN strain series. The analysis was performed as in Figure 3. Triangles indicate two variants of the corresponding types (color-coded) in the respective strain/gene combination.
3. Discussion
The genetic background of yeast strains plays an insidious or treacherous role in the analysis of genome instability. In addition to numerous known examples of how a single nucleotide change in the genome completely changes the phenotype of the gene under study (some of them are listed in the Introduction), we have our own story. Our present examination of genomes was instigated in part by our recent work on genome-wide analysis of yeast clones that evolved under stress imposed by a deletion of the N-terminal, catalytic active containing half of Pol2, which was performed in LAN201 and E134 strain derivatives [57]. The deletion created in haploid strains confers a severe growth defect, but miserably growing cells yield healthy colonies with time. NGS analysis of the DNA of these fast growers revealed many different genomic variants and recurrent mutations in the essential CDC28 gene encoding for a catalytic subunit of cyclin-dependent kinase (CDK), a master regulator of the cell cycle. The genetic analysis confirmed that single nucleotide changes in several sites of the gene acquired by the healthy growers cause rescue of the slow growth phenotype (changes L62F, L84V, L86H, and I236N). These changes are located on the surfaces of Cdc28 interacting with cyclins and a Cks1 subunit of CDK. Thus, combining deleterious and benign mutations in two essential genes might provide growth advantages. We did not see any significant changes in the CDC28 gene in the examined strains. However, CKS1 has several mutations leading to non-synonymic amino acid changes that are predicted to have a moderate impact on protein function in the SK1 strain (Supplementary Table S2). Thus, the effect of these mutations may be similar to the effect of CDC28 alleles because they affect the same CDK complex.
Our work revealed a wide diversity of yeast strains commonly used to analyze genome stability. The genetic consequences of only a few variants have been examined. Many more studies are needed to be performed using structural modeling and genetic and biochemical approaches. Our study will serve as a guidebook for such endeavors and help to better interpret past and future genetic findings.
4. Materials and Methods
4.1. Sources of Raw Data
The NCBI sources and references to genomic data are in Table 3.

Table 3.
Yeast genomics data source.
4.2. Data Processing
The raw reads of the five yeast strains were filtered by AfterQC (v0.9.7) [58], and adapters, primers sequences, and low-quality nucleotides (Q < 20) were removed. Reads shorter than 35 nucleotides after trimming were discarded. The raw and filtered sequences were explored with FastQC to calculate and visualize sequence quality metrics [59]. The resulting filtered reads were then aligned to the S288c reference (R64-3-1) genome using bwa-mem (v0.7.17) with default parameters [60]. Samtools (v.1.17) [61] and picard (v2.27.5) were used to index the reference and create a dictionary. The mapped reads were sorted with samtools (v1.17) [61]. Duplicated reads were marked with Picard MarkDuplicates [62]. Alignment statistics were assessed using picard and samtools.
The alignment results were used to identify SNPs and indels within each genome by GATK HaplotypeCaller (v4.3.0.0) with sample_ploidy set to 1 or 2 depending on yeast strain ploidy [63]. GATK VariantFiltration was used to filter the vcf files with the following parameters: “QD < 2.0” –filter-name “QD2” -filter “QUAL < 30.0” –filter-name “QUAL30” -filter SOR > 3.0” –filter-name “SOR3” -filter “FS > 60.0” –filter-name “FS60” -filter “MQ < 40.0” –filter-name “MQ40” for snps filtration and QD < 2.0” –filter-name “QD2” -filter “QUAL < 30.0” –filter-name “QUAL30” -filter “FS > 200.0” –filter-name “FS200” for indel filtration. The variant call set was filtered by excluding multiallelic variants, thresholds DP < 10 for read depth, and allele depth AD < 5 for the alternative allele by bcftools (v1.17) [61]. Variants were annotated using snpEFF (v5.1) [64] and Ensembl Variant Effect Predictor (VEP) (v109) that uses multiple criteria to connect sequence variants to the predicted effect on gene function [65].
4.3. Oher Methods
SNP-based phylogenetic trees from whole genome sequence data were reconstructed using REALPHY by aligning the reads to S288C reference sequences with default parameters (v.1.13) [66]. The Interactive Tree Of Life (https://itol.embl.de) [67] was used to display of phylogenetic tree.
The number of SNPs within a sliding window of a 1 Mb size and SNP density plot was carried out with CMplot [61]. Genomic information was visualized with CoMut [68].
The summary of all genomic differences (not only RRR-panel) in our five strains is in Supplementary Table S3. The results for genes that we might have inadvertently not covered above can be found in this Table.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/ijms24097795/s1.
Author Contributions
Conceptualization, A.G.L. and Y.I.P.; methodology, software, and validation, A.S.Z., A.G.L. and Y.I.P.; formal analysis, A.S.Z.; investigation, A.S.Z., A.G.L. and Y.I.P.; resources, A.S.Z. and Y.I.P.; data curation, A.S.Z.; writing—original draft preparation, A.S.Z. and Y.I.P.; writing—review and editing, A.S.Z., A.G.L. and Y.I.P.; visualization, A.S.Z., A.G.L.,and Y.I.P.; supervision, A.G.L. and Y.I.P.; project administration, Y.I.P.; funding acquisition, A.S.Z. and Y.I.P. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by NE DHHS, grant number 2023-46; FPBCC Pilot 2022 award to YIP from The Fred and Pamela Buffett Cancer Center’s National Cancer Institute Cancer Support Grant P50 CA127297. ASZ was funded by Saint-Petersburg State University project #94031363.
Institutional Review Board Statement
Not applicable.
Data Availability Statement
Non-applicable.
Acknowledgments
We are thankful to Dmitry Gordenin (NIEHS, USA), Tom Petes (Duke University, USA), Linda Reha-Krantz (University of Alberta, Canada), and Polina Shcherbakova (UNMC, Omaha, USA) for the information on genealogy of yeast strains. We are grateful to Elizabeth Moore for the expert assistance in the maintenance of the YIP laboratory at UNMC and to the secretarial and administrative personnel of Eppley Institute for expert help with the submission of grants.
Conflicts of Interest
The authors declare no conflict of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript; or in the decision to publish the results.
References
- Feldmann, H. Yeast: Molecular and Cell Biology, 2nd ed.; Wiley-Blackwell: Weinheim, Germany, 2012; p. 444. [Google Scholar]
- Lisby, M.; Mortensen, U.H. Editorial: 3Rs tightly intertwined to maintain genome stability. FEMS Yeast Res. 2017, 17, fox003. [Google Scholar] [CrossRef]
- Louis, E.J. Historical Evolution of Laboratory Strains of Saccharomyces cerevisiae. Cold Spring Harb. Protoc. 2016, 2016. [Google Scholar] [CrossRef]
- Mortimer, R.K.; Johnston, J.R. Genealogy of principal strains of the yeast genetic stock center. Genetics 1986, 113, 35–43. [Google Scholar] [CrossRef] [PubMed]
- Schacherer, J.; Ruderfer, D.M.; Gresham, D.; Dolinski, K.; Botstein, D.; Kruglyak, L. Genome-wide analysis of nucleotide-level variation in commonly used Saccharomyces cerevisiae strains. PLoS ONE 2007, 2, e322. [Google Scholar] [CrossRef]
- Marsit, S.; Leducq, J.B.; Durand, É.; Marchant, A.; Filteau, M.; Landry, C.R. Evolutionary biology through the lens of budding yeast comparative genomics. Nat. Rev. Genet. 2017, 18, 581–598. [Google Scholar] [CrossRef] [PubMed]
- Fan, H.Y.; Cheng, K.K.; Klein, H.L. Mutations in the RNA polymerase II transcription machinery suppress the hyperrecombination mutant hpr1 delta of Saccharomyces cerevisiae. Genetics 1996, 142, 749–759. [Google Scholar] [CrossRef]
- Blastyák, A.; Pintér, L.; Unk, I.; Prakash, L.; Prakash, S.; Haracska, L. Yeast Rad5 protein required for postreplication repair has a DNA helicase activity specific for replication fork regression. Mol. Cell 2007, 28, 167–175. [Google Scholar] [CrossRef]
- Gallo, D.; Kim, T.; Szakal, B.; Saayman, X.; Narula, A.; Park, Y.; Branzei, D.; Zhang, Z.; Brown, G.W. Rad5 Recruits Error-Prone DNA Polymerases for Mutagenic Repair of ssDNA Gaps on Undamaged Templates. Mol. Cell 2019, 73, 900–914.e9. [Google Scholar] [CrossRef]
- Xu, X.; Lin, A.; Zhou, C.; Blackwell, S.R.; Zhang, Y.; Wang, Z.; Feng, Q.; Guan, R.; Hanna, M.D.; Chen, Z.; et al. Involvement of budding yeast Rad5 in translesion DNA synthesis through physical interaction with Rev1. Nucleic Acids Res. 2016, 44, 5231–5245. [Google Scholar] [CrossRef]
- Yeeles, J.T.; Deegan, T.D.; Janska, A.; Early, A.; Diffley, J.F. Regulated eukaryotic DNA replication origin firing with purified proteins. Nature 2015, 519, 431–435. [Google Scholar] [CrossRef]
- Joshi, I.; Peng, J.; Alvino, G.; Kwan, E.; Feng, W. Exceptional origin activation revealed by comparative analysis in two laboratory yeast strains. PLoS ONE 2022, 17, e0263569. [Google Scholar] [CrossRef]
- Wolfe, K.H. Origin of the Yeast Whole-Genome Duplication. PLoS Biol. 2015, 13, e1002221. [Google Scholar] [CrossRef]
- Gupta, A.; Sharma, S.; Reichenbach, P.; Marjavaara, L.; Nilsson, A.K.; Lingner, J.; Chabes, A.; Rothstein, R.; Chang, M. Telomere length homeostasis responds to changes in intracellular dNTP pools. Genetics 2013, 193, 1095–1105. [Google Scholar] [CrossRef]
- Cerritelli, S.M.; El Hage, A. RNases H1 and H2: Guardians of the stability of the nuclear genome when supply of dNTPs is limiting for DNA synthesis. Curr. Genet. 2020, 66, 1073–1084. [Google Scholar] [CrossRef]
- Kwan, E.X.; Foss, E.; Kruglyak, L.; Bedalov, A. Natural polymorphism in BUL2 links cellular amino acid availability with chronological aging and telomere maintenance in yeast. PLoS Genet. 2011, 7, e1002250. [Google Scholar] [CrossRef]
- Johnson, R.E.; Klassen, R.; Prakash, L.; Prakash, S. A major role of DNA polymerase δ in replication of both the leading and lagging DNA strands. Mol. Cell 2015, 59, 163–175. [Google Scholar] [CrossRef]
- Burgers, P.M.J.; Gordenin, D.; Kunkel, T.A. Who is leading the replication Fork, Pol ε or Pol δ? Mol. Cell 2016, 61, 492–493. [Google Scholar] [CrossRef]
- Dowell, R.D.; Ryan, O.; Jansen, A.; Cheung, D.; Agarwala, S.; Danford, T.; Bernstein, D.A.; Rolfe, P.A.; Heisler, L.E.; Chin, B.; et al. Genotype to phenotype: A complex problem. Science 2010, 328, 469. [Google Scholar] [CrossRef]
- Chin, B.L.; Ryan, O.; Lewitter, F.; Boone, C.; Fink, G.R. Genetic variation in Saccharomyces cerevisiae: Circuit diversification in a signal transduction network. Genetics 2012, 192, 1523–1532. [Google Scholar] [CrossRef]
- Lada, A.G.; Kliver, S.F.; Dhar, A.; Polev, D.E.; Masharsky, A.E.; Rogozin, I.B.; Pavlov, Y.I. Disruption of transcriptional coactivator Sub1 leads to genome-wide re-distribution of clustered mutations Induced by APOBEC in active yeast genes. PLoS Genet. 2015, 11, e1005217. [Google Scholar] [CrossRef]
- Lada, A.G.; Stepchenkova, E.I.; Waisertreiger, I.S.; Noskov, V.N.; Dhar, A.; Eudy, J.D.; Boissy, R.J.; Hirano, M.; Rogozin, I.B.; Pavlov, Y.I. Genome-wide mutation avalanches induced in diploid yeast cells by a base analog or an APOBEC deaminase. PLoS Genet. 2013, 9, e1003736. [Google Scholar] [CrossRef] [PubMed]
- Lada, A.G.; Stepchenkova, E.I.; Zhuk, A.S.; Kliver, S.F.; Rogozin, I.B.; Polev, D.E.; Dhar, A.; Pavlov, Y.I. Recombination Is responsible for the increased recovery of drug-resistant mutants with hypermutated genomes in resting yeast diploids expressing APOBEC deaminases. Front. Genet. 2017, 8, 202. [Google Scholar] [CrossRef] [PubMed]
- Morrison, A.; Bell, J.B.; Kunkel, T.A.; Sugino, A. Eukaryotic DNA polymerase amino acid sequence required for 3′->5′ exonuclease activity. Proc. Natl. Acad. Sci. USA 1991, 88, 9473–9477. [Google Scholar] [CrossRef] [PubMed]
- Morrison, A.; Johnson, A.L.; Johnston, L.H.; Sugino, A. Pathway correcting DNA replication errors in Saccharomyces cerevisiae. EMBO J. 1993, 12, 1467–1473. [Google Scholar] [CrossRef] [PubMed]
- Morrison, A.; Sugino, A. The 3′-->5′ exonucleases of both DNA polymerases δ and ε participate in correcting errors of DNA replication in Saccharomyces cerevisiae. Mol. Gen. Genet. 1994, 242, 289–296. [Google Scholar] [CrossRef] [PubMed]
- Tran, H.T.; Degtyareva, N.P.; Gordenin, D.A.; Resnick, M.A. Genetic factors affecting the impact of DNA polymerase δ proofreading activity on mutation avoidance in yeast. Genetics 1999, 152, 47–59. [Google Scholar] [CrossRef] [PubMed]
- Tran, H.T.; Gordenin, D.A.; Resnick, M.A. The 3′-->5′ exonucleases of DNA polymerases δ and ε and the 5′-->3′ exonuclease Exo1 have major roles in postreplication mutation avoidance in Saccharomyces cerevisiae. Mol. Cell Biol. 1999, 19, 2000–2007. [Google Scholar] [CrossRef]
- Tran, H.T.; Keen, J.D.; Kricker, M.; Resnick, M.A.; Gordenin, D.A. Hypermutability of homonucleotide runs in mismatch repair and DNA polymerase proofreading yeast mutants. Mol Cell Biol 1997, 17, 2859–2865. [Google Scholar] [CrossRef]
- Shcherbakova, P.V.; Kunkel, T.A. Mutator phenotypes conferred by MLH1 overexpression and by heterozygosity for mlh1 mutations. Mol. Cell Biol. 1999, 19, 3177–3183. [Google Scholar] [CrossRef]
- Kane, D.P.; Shcherbakova, P.V. A common cancer-associated DNA polymerase ε mutation causes an exceptionally strong mutator phenotype, indicating fidelity defects distinct from loss of proofreading. Cancer Res. 2014, 74, 1895–1901. [Google Scholar] [CrossRef]
- Bulock, C.R.; Xing, X.; Shcherbakova, P.V. Mismatch repair and DNA polymerase δ proofreading prevent catastrophic accumulation of leading strand errors in cells expressing a cancer-associated DNA polymerase ε variant. Nucleic Acids Res. 2020, 48, 9124–9134. [Google Scholar] [CrossRef]
- Song, G.; Dickins, B.J.; Demeter, J.; Engel, S.; Gallagher, J.; Choe, K.; Dunn, B.; Snyder, M.; Cherry, J.M. AGAPE (Automated Genome Analysis PipelinE) for pan-genome analysis of Saccharomyces cerevisiae. PLoS ONE 2015, 10, e0120671. [Google Scholar] [CrossRef]
- Puddu, F.; Herzog, M.; Selivanova, A.; Wang, S.; Zhu, J.; Klein-Lavi, S.; Gordon, M.; Meirman, R.; Millan-Zambrano, G.; Ayestaran, I.; et al. Genome architecture and stability in the Saccharomyces cerevisiae knockout collection. Nature 2019, 573, 416–420. [Google Scholar] [CrossRef]
- Brachmann, C.B.; Davies, A.; Cost, G.J.; Caputo, E.; Li, J.; Hieter, P.; Boeke, J.D. Designer deletion strains derived from Saccharomyces cerevisiae S288C: A useful set of strains and plasmids for PCR-mediated gene disruption and other applications. Yeast 1998, 14, 115–132. [Google Scholar] [CrossRef]
- Thomas, B.J.; Rothstein, R. Elevated recombination rates in transcriptionally active DNA. Cell 1989, 56, 619–630. [Google Scholar] [CrossRef]
- Ralser, M.; Kuhl, H.; Ralser, M.; Werber, M.; Lehrach, H.; Breitenbach, M.; Timmermann, B. The Saccharomyces cerevisiae W303-K6001 cross-platform genome sequence: Insights into ancestry and physiology of a laboratory mutt. Open Biol. 2012, 2, 120093. [Google Scholar] [CrossRef]
- Kane, S.M.; Roth, R. Carbohydrate metabolism during ascospore development in yeast. J. Bacteriol. 1974, 118, 8–14. [Google Scholar] [CrossRef]
- Liti, G.; Carter, D.M.; Moses, A.M.; Warringer, J.; Parts, L.; James, S.A.; Davey, R.P.; Roberts, I.N.; Burt, A.; Koufopanou, V.; et al. Population genomics of domestic and wild yeasts. Nature 2009, 458, 337–341. [Google Scholar] [CrossRef]
- Börner, G.V.; Cha, R.S. Induction and analysis of synchronous meiotic yeast cultures. Cold Spring Harb. Protoc. 2015, 2015, 908–913. [Google Scholar] [CrossRef]
- Chakraborty, P.; Pankajam, A.V.; Dutta, A.; Nishant, K.T. Genome wide analysis of meiotic recombination in yeast: For a few SNPs more. IUBMB Life 2018, 70, 743–752. [Google Scholar] [CrossRef]
- Winzeler, E.A.; Castillo-Davis, C.I.; Oshiro, G.; Liang, D.; Richards, D.R.; Zhou, Y.; Hartl, D.L. Genetic diversity in yeast assessed with whole-genome oligonucleotide arrays. Genetics 2003, 163, 79–89. [Google Scholar] [CrossRef] [PubMed]
- Hartwell, L.H. Macromolecule synthesis in temperature-sensitive mutants of yeast. J. Bacteriol. 1967, 93, 1662–1670. [Google Scholar] [CrossRef] [PubMed]
- Klapholz, S.; Esposito, R.E. Isolation of SPO12-1 and SPO13-1 from a natural variant of yeast that undergoes a single meiotic division. Genetics 1980, 96, 567–588. [Google Scholar] [CrossRef] [PubMed]
- Botstein, D.; Falco, S.C.; Stewart, S.E.; Brennan, M.; Scherer, S.; Stinchcomb, D.T.; Struhl, K.; Davis, R.W. Sterile host yeasts (SHY): A eukaryotic system of biological containment for recombinant DNA experiments. Gene 1979, 8, 17–24. [Google Scholar] [CrossRef] [PubMed]
- Gordenin, D.A.; Proscyavichus, Y.Y.; Malkova, A.L.; Trofimova, M.V.; Peterzen, A. Yeast mutants with increased bacterial transposon Tn5 excision. Yeast 1991, 7, 37–50. [Google Scholar] [CrossRef]
- Pavlov, Y.I.; Zhuk, A.S.; Stepchenkova, E.I. DNA polymerases at the eukaryotic replication fork thirty years after: Connection to cancer. Cancers 2020, 12, 3489. [Google Scholar] [CrossRef]
- Pavlov, Y.I.; Shcherbakova, P.V. DNA polymerases at the eukaryotic fork—20 years later. Mutat. Res. 2010, 685, 45–53. [Google Scholar] [CrossRef]
- Muramatsu, S.; Hirai, K.; Tak, Y.S.; Kamimura, Y.; Araki, H. CDK-dependent complex formation between replication proteins Dpb11, Sld2, Pol ε, and GINS in budding yeast. Genes Dev. 2010, 24, 602–612. [Google Scholar] [CrossRef]
- Araki, H.; Hamatake, R.K.; Johnston, L.H.; Sugino, A. DPB2, the gene encoding DNA polymerase II subunit B, is required for chromosome replication in Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA 1991, 88, 4601–4605. [Google Scholar] [CrossRef]
- Roberts, S.A.; Sterling, J.; Thompson, C.; Harris, S.; Mav, D.; Shah, R.; Klimczak, L.J.; Kryukov, G.V.; Malc, E.; Mieczkowski, P.A.; et al. Clustered mutations in yeast and in human cancers can arise from damaged long single-strand DNA regions. Mol. Cell 2012, 46, 424–435. [Google Scholar] [CrossRef]
- Chan, K.; Roberts, S.A.; Klimczak, L.J.; Sterling, J.F.; Saini, N.; Malc, E.P.; Kim, J.; Kwiatkowski, D.J.; Fargo, D.C.; Mieczkowski, P.A.; et al. An APOBEC3A hypermutation signature is distinguishable from the signature of background mutagenesis by APOBEC3B in human cancers. Nat. Genet. 2015, 47, 1067–1072. [Google Scholar] [CrossRef]
- Hoopes, J.I.; Cortez, L.M.; Mertz, T.M.; Malc, E.P.; Mieczkowski, P.A.; Roberts, S.A. APOBEC3A and APOBEC3B preferentially deaminate the lagging strand template during DNA replication. Cell Rep. 2016, 14, 1273–1282. [Google Scholar] [CrossRef] [PubMed]
- Gerik, K.J.; Li, X.; Pautz, A.; Burgers, P.M. Characterization of the two small subunits of Saccharomyces cerevisiae DNA polymerase delta. J. Biol. Chem. 1998, 273, 19747–19755. [Google Scholar] [CrossRef]
- MacNeill, S.A.; Moreno, S.; Reynolds, N.; Nurse, P.; Fantes, P.A. The fission yeast Cdc1 protein, a homologue of the small subunit of DNA polymerase delta, binds to Pol3 and Cdc27. Embo J. 1996, 15, 4613–4628. [Google Scholar] [CrossRef]
- Rothstein, R.J. One-step gene disruption in yeast. Methods Enzymol. 1983, 101, 202–211. [Google Scholar] [CrossRef]
- Stepchenkova, E.I.; Zhuk, A.S.; Cui, J.; Tarakhovskaya, E.R.; Barbari, S.R.; Shcherbakova, P.V.; Polev, D.E.; Fedorov, R.; Poliakov, E.; Rogozin, I.B.; et al. Compensation for the absence of the catalytically active half of DNA polymerase ε in yeast by positively selected mutations in CDC28. Genetics 2021, 218, iyab060. [Google Scholar] [CrossRef]
- Chen, S.; Huang, T.; Zhou, Y.; Han, Y.; Xu, M.; Gu, J. AfterQC: Automatic filtering, trimming, error removing and quality control for fastq data. BMC Bioinform. 2017, 18, 80. [Google Scholar] [CrossRef]
- Andrews, S. FastQC: A Quality Control Tool for High Throughput Sequence Data. 2010. Available online: https://www.bioinformatics.babraham.ac.uk/projects/fastqc/ (accessed on 29 March 2023).
- Li, H.; Durbin, R. Fast and accurate short read alignment with Burrows–Wheeler transform. Bioinformatics 2009, 25, 1754–1760. [Google Scholar] [CrossRef]
- Danecek, P.; Bonfield, J.K.; Liddle, J.; Marshall, J.; Ohan, V.; Pollard, M.O.; Whitwham, A.; Keane, T.; McCarthy, S.A.; Davies, R.M.; et al. Twelve years of SAMtools and BCFtools. GigaScience 2021, 10, giab008. [Google Scholar] [CrossRef]
- Broad Institute. Picard Tools. Broad Institute, GitHub Repository. 2021. Available online: http://broadinstitute.github.io/picard/ (accessed on 29 March 2023).
- Poplin, R.; Ruano-Rubio, V.; DePristo, M.A.; Fennell, T.J.; Carneiro, M.O.; Van der Auwera, G.A.; Kling, D.E.; Gauthier, L.D.; Levy-Moonshine, A.; Roazen, D.; et al. Scaling accurate genetic variant discovery to tens of thousands of samples. bioRxiv 2018. [Google Scholar] [CrossRef]
- Cingolani, P.; Platts, A.; Wang, L.; Coon, M.; Nguyen, T.; Wang, L.; Land, S.J.; Lu, X.; Ruden, D.M. A program for annotating and predicting the effects of single nucleotide polymorphisms, SnpEff: SNPs in the genome of Drosophila melanogaster strain w1118; iso-2; iso-3. Fly 2012, 6, 80–92. [Google Scholar] [CrossRef] [PubMed]
- McLaren, W.; Gil, L.; Hunt, S.E.; Riat, H.S.; Ritchie, G.R.S.; Thormann, A.; Flicek, P.; Cunningham, F. The Ensembl Variant Effect Predictor. Genome Biol. 2016, 17, 122. [Google Scholar] [CrossRef] [PubMed]
- Bertels, F.; Silander, O.K.; Pachkov, M.; Rainey, P.B.; van Nimwegen, E. Automated reconstruction of whole-genome phylogenies from short-sequence reads. Mol. Biol. Evol. 2014, 31, 1077–1088. [Google Scholar] [CrossRef]
- Letunic, I.; Bork, P. Interactive Tree of Life (iTOL) v5: An online tool for phylogenetic tree display and annotation. Nucleic Acids Res. 2021, 49, W293–W296. [Google Scholar] [CrossRef]
- Crowdis, J.; He, M.X.; Reardon, B.; Van Allen, E.M. CoMut: Visualizing integrated molecular information with comutation plots. Bioinformatics 2020, 36, 4348–4349. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).