Visual Detection of Stem-Loop Primer Amplification (SPA) Products without Denaturation Using Peroxidase-like DNA Machines (PxDM)
Abstract
1. Introduction
2. Results
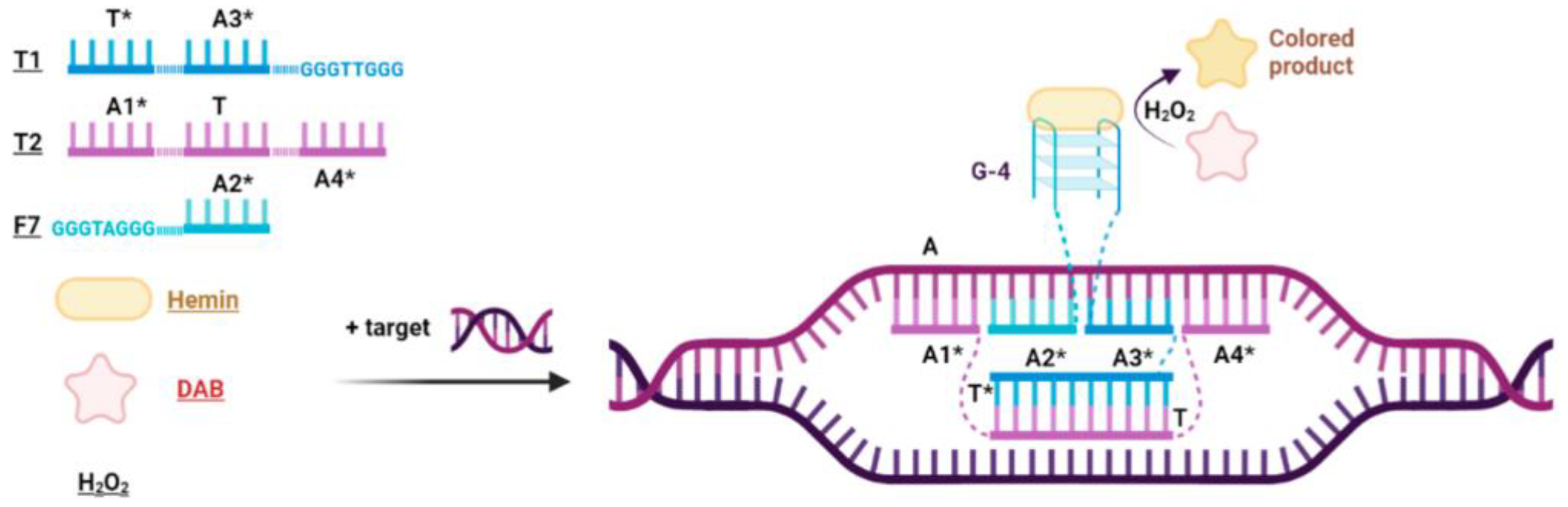
2.1. Design of Peroxidase-like DNA Machines (PxDM)
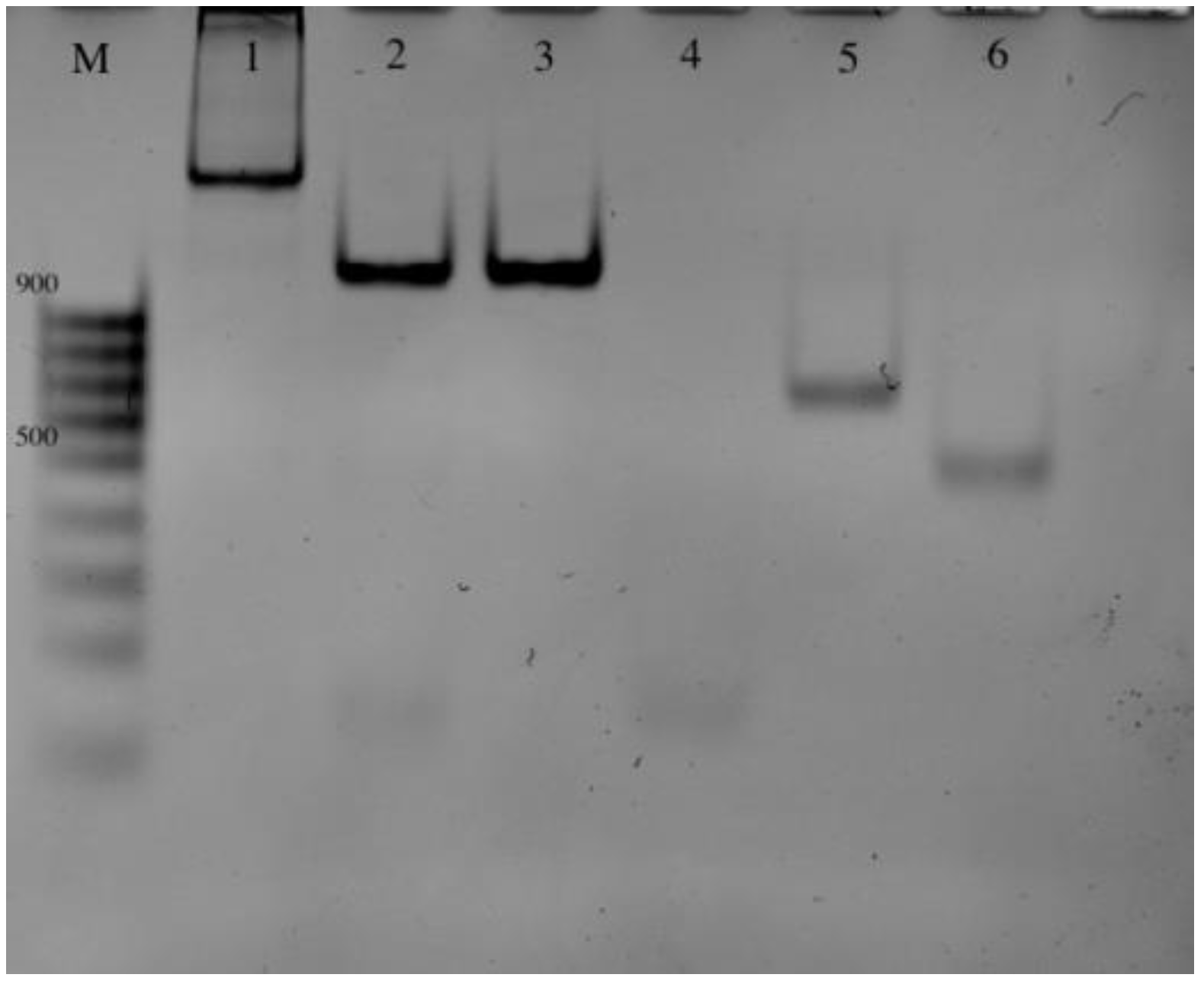
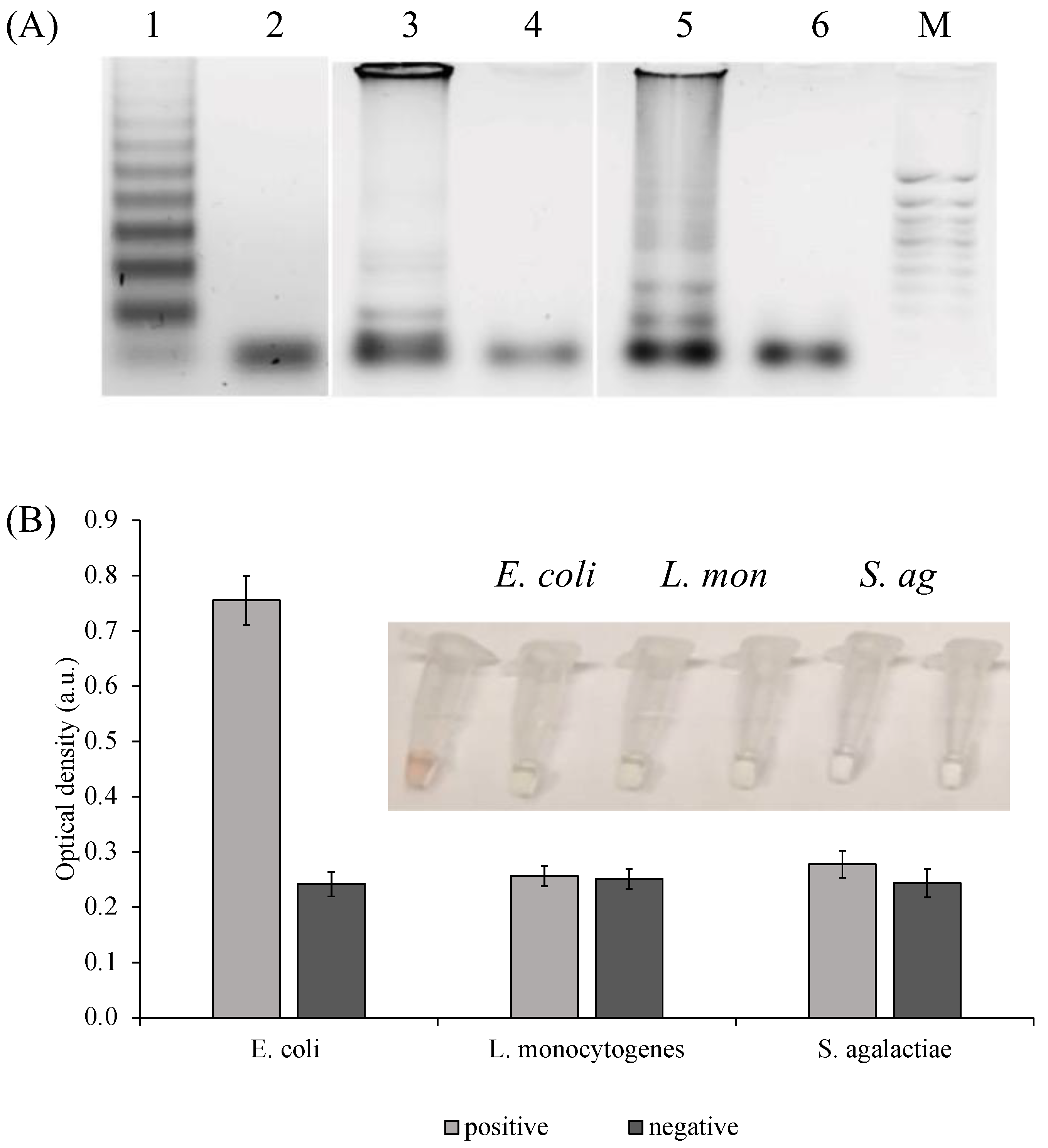
2.2. Amplification and Detection of the Target Fragments
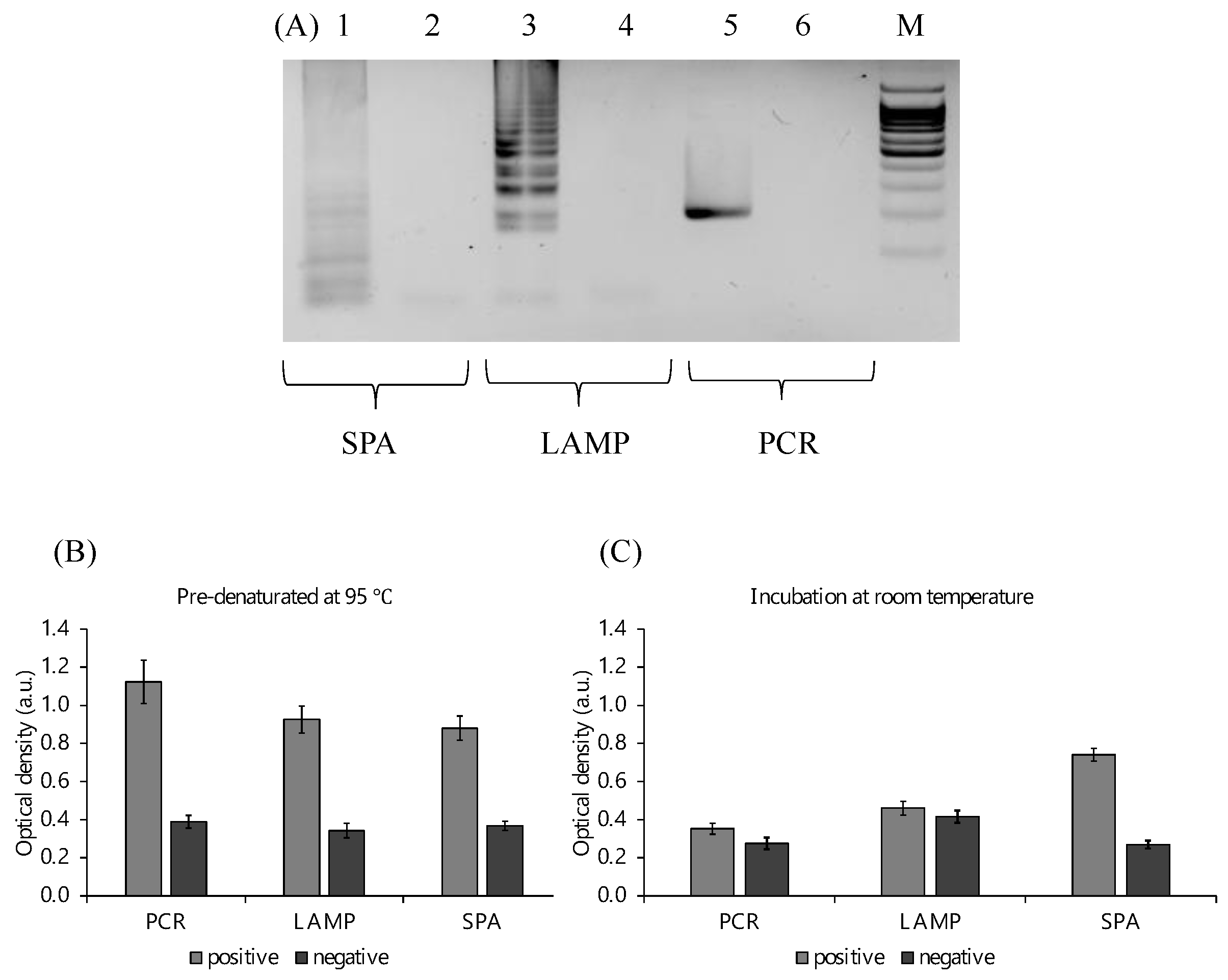
2.3. Optimization of the Peroxidase Reaction
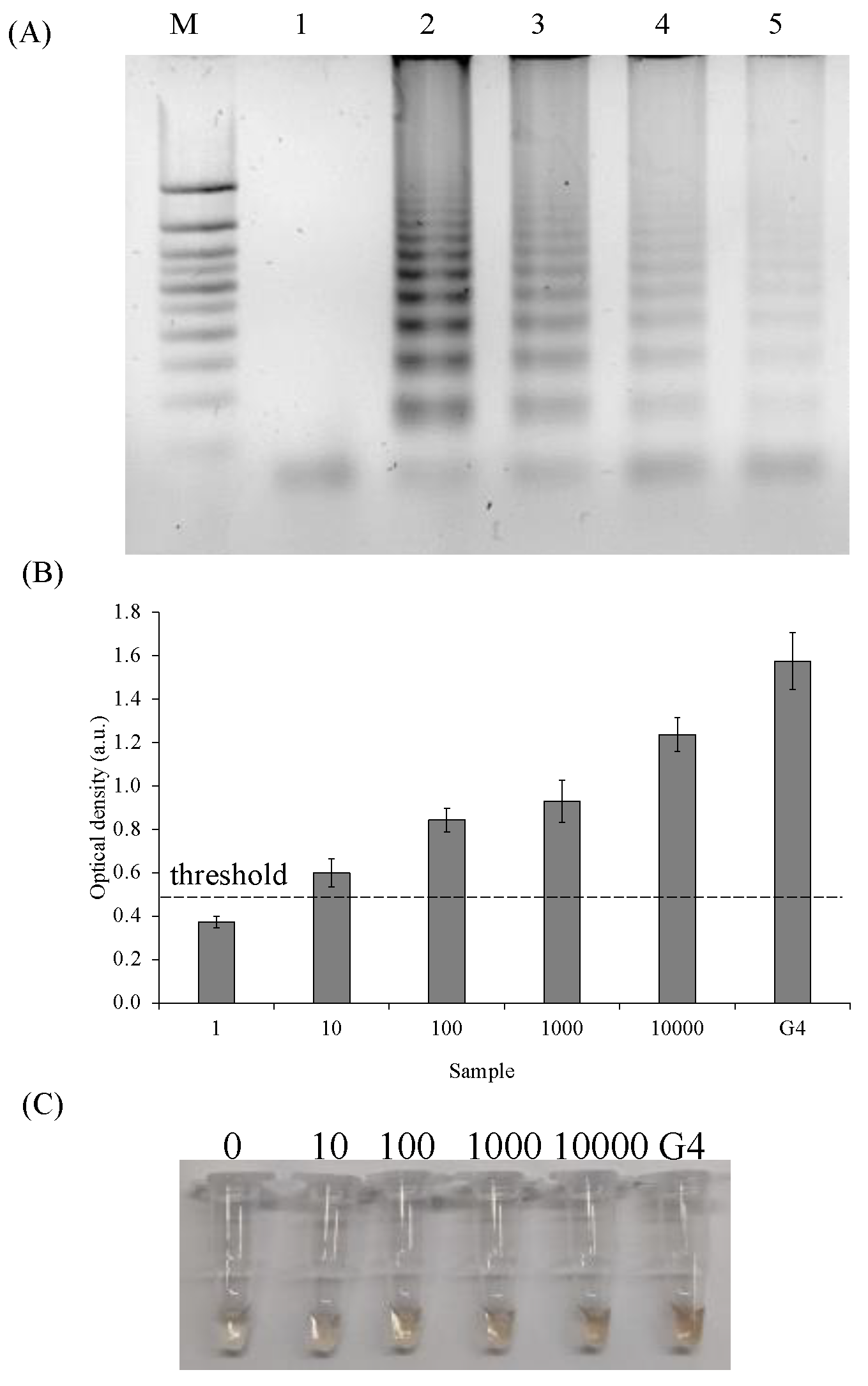
2.4. Limit of Detection (LOD) of the SPA/PxDM Assay
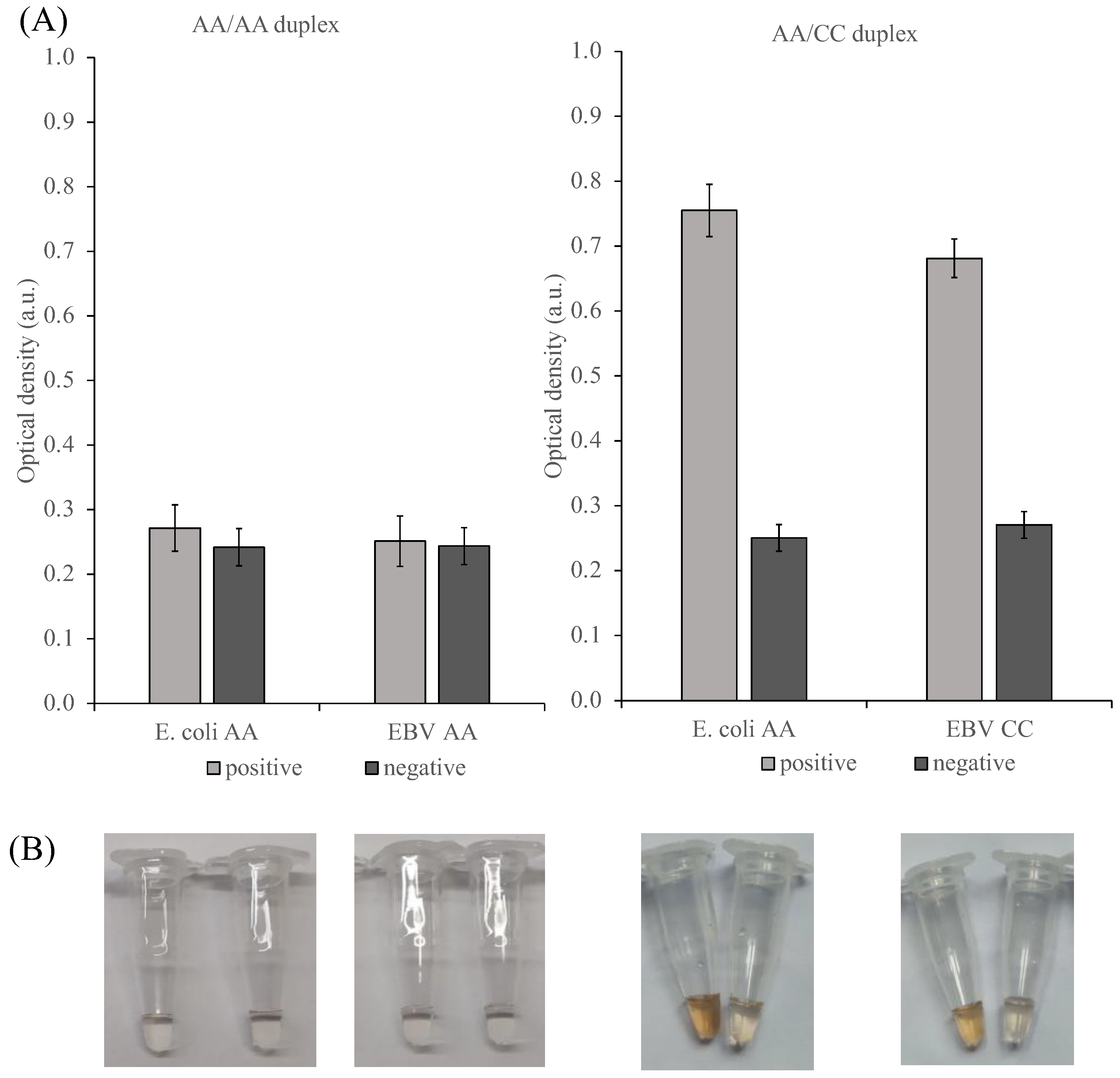
2.5. Selectivity of the SPA/PxDM Assay towards Different SPA Products and Single Nucleotide Mismatch
2.6. PxDM Assay Does Not Detect False Positives
2.7. Development of Diplex SPA and Detection of the SPA Products by PxDM Assay
3. Discussion
4. Materials and Methods
4.1. Materials
4.2. Cultivation of the Pathogens
4.3. DNA Fragments Preparation
4.4. Primer and Sensor Design
4.5. PCR Reaction
4.6. LAMP Reaction
4.7. SPA Reaction
4.8. Amplification Product Analysis
4.9. Optical Density Measurement/Colorimetric Detection/Visual Detection
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Lesens, O.; Desbiez, F.; Vidal, M. Clinical microbiology and infection: The official publication of the European Society of Clinical Microbiology and Infectious Diseases. Clin. Microbiol. Infect. 2011, 17, 1717–1726. [Google Scholar]
- Mahony, J.B.; Blackhouse, G.; Babwah, J.; Smieja, M.; Buracond, S.; Chong, S.; Ciccotelli, W.; O’Shea, T.; Alnakhli, D.; Griffiths-Turner, M. Cost analysis of multiplex PCR testing for diagnosing respiratory virus infections. J. Clin. Microbiol. 2009, 47, 2812–2817. [Google Scholar] [CrossRef]
- Mori, Y.; Kanda, H.; Notomi, T. Loop-mediated isothermal amplification (LAMP): Recent progress in research and development. J. Infect. Chemother. 2013, 19, 404–411. [Google Scholar] [CrossRef] [PubMed]
- Tomita, N.; Mori, Y.; Kanda, H.; Notomi, T. Loop-mediated isothermal amplification (LAMP) of gene sequences and simple visual detection of products. Nat. Protoc. 2008, 3, 877–882. [Google Scholar] [CrossRef] [PubMed]
- Gorbenko, D.A.; Shkodenko, L.A.; Rubel, M.S.; Slita, A.V.; Nikitina, E.V.; Martens, E.A.; Kolpashchikov, D.M. DNA nanomachine for visual detection of structured RNA and double stranded DNA. Chem. Commun. 2022, 58, 5395–5398. [Google Scholar] [CrossRef]
- Jiang, H.-X.; Liang, Z.-Z.; Ma, Y.-H.; Kong, D.-M.; Hong, Z.-Y. G-quadruplex fluorescent probe-mediated real-time rolling circle amplification strategy for highly sensitive microRNA detection. Anal. Chim. Acta 2016, 943, 114–122. [Google Scholar] [CrossRef]
- Kovtunov, E.A.; Shkodenko, L.A.; Goncharova, E.A.; Nedorezova, D.D.; Sidorenko, S.V.; Koshel, E.I.; Kolpashchikov, D.M. Towards Point of Care Diagnostics: Visual Detection of Meningitis Pathogens Directly from Cerebrospinal Fluid. ChemistrySelect 2020, 5, 14572–14577. [Google Scholar] [CrossRef]
- Mori, Y.; Hirano, T.; Notomi, T.; Lesens, O.; Desbiez, F.; Vidal, M.; Wang, D.-G.; Brewster, J.D.; Paul, M.; Tomasula, P.M.; et al. Loop-mediated isothermal amplification (LAMP): A rapid, accurate, and cost-effective diagnostic method for infectious diseases. Clin. Microbiol. Infect. 2011, 30, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Chen, H.; Liu, K.; Li, Z.; Wang, P. Point of care testing for infectious diseases. Clin. Chim. Acta 2019, 493, 138–147. [Google Scholar] [CrossRef] [PubMed]
- Luo, G.; Yi, T.; Wang, Q.; Guo, B.; Fang, L.; Zhang, G.; Guo, X. Stem-loop-primer assisted isothermal amplification enabling high-specific and ultrasensitive nucleic acid detection. Biosens. Bioelectron. 2021, 184, 113239. [Google Scholar] [CrossRef] [PubMed]
- Mori, Y.; Hirano, T.; Notomi, T. Sequence specific visual detection of LAMP reactions by addition of cationic polymers. BMC Biotechnol. 2006, 6, 3. [Google Scholar] [CrossRef]
- Varona, M.; Anderson, J.L. Visual detection of single-nucleotide polymorphisms using molecular beacon loop-mediated isothermal amplification with centrifuge-free DNA extraction. Anal. Chem. 2019, 91, 6991–6995. [Google Scholar] [CrossRef] [PubMed]
- Phillips, E.A.; Moehling, T.J.; Bhadra, S.; Ellington, A.D.; Linnes, J.C. Strand displacement probes combined with isothermal nucleic acid amplification for instrument-free detection from complex samples. Anal. Chem. 2018, 90, 6580–6586. [Google Scholar] [CrossRef]
- Wang, D.-G.; Brewster, J.D.; Paul, M.; Tomasula, P.M. Two methods for increased specificity and sensitivity in loop-mediated isothermal amplification. Molecules 2015, 20, 6048–6059. [Google Scholar] [CrossRef] [PubMed]
- Gao, X.; Sun, B.; Guan, Y. Pullulan reduces the non-specific amplification of loop-mediated isothermal amplification (LAMP). Anal. Bioanal. Chem. 2019, 411, 1211–1218. [Google Scholar] [CrossRef] [PubMed]
- Park, K.S. Nucleic acid aptamer-based methods for diagnosis of infections. Biosens. Bioelectron. 2018, 102, 179–188. [Google Scholar] [CrossRef]
- Travascio, P.; Li, Y.; Sen, D. DNA-enhanced peroxidase activity of a DNA-aptamer-hemin complex. Chem. Biol. 1998, 5, 505–517. [Google Scholar] [CrossRef] [PubMed]
- Long, Y.; Zhou, C.; Wang, C.; Cai, H.; Yin, C.; Yang, Q.; Xiao, D. Ultrasensitive visual detection of HIV DNA biomarkers via a multi-amplification nanoplatform. Sci. Rep. 2016, 6, 23949. [Google Scholar] [CrossRef]
- Deng, M.; Feng, S.; Luo, F.; Wang, S.; Sun, X.; Zhou, X.; Zhang, X.-L. Visual detection of rpoB mutations in rifampin-resistant Mycobacterium tuberculosis strains by use of an asymmetrically split peroxidase DNAzyme. J. Clin. Microbiol. 2012, 50, 3443–3450. [Google Scholar] [CrossRef]
- Darius, A.K.L.; Ling, N.J.; Mahesh, U. Visual detection of DNA from salmonella and mycobacterium using split DNAzymes. Mol. Biosyst. 2010, 6, 792–794. [Google Scholar] [CrossRef]
- Shimron, S.; Elbaz, J.; Henning, A.; Willner, I. Ion-induced DNAzyme switches. Chem. Commun. 2010, 46, 3250–3252. [Google Scholar] [CrossRef] [PubMed]
- Cox, A.J.; Bengtson, H.N.; Rohde, K.H.; Kolpashchikov, D.M. DNA nanotechnology for nucleic acid analysis: Multifunctional molecular DNA machine for RNA detection. Chem. Commun. 2016, 52, 14318–14321. [Google Scholar] [CrossRef]
- Nedorezova, D.D.; Fakhardo, A.F.; Nemirich, D.V.; Bryushkova, E.A.; Kolpashchikov, D.M. Towards DNA Nanomachines for Cancer Treatment: Achieving Selective and Efficient Cleavage of Folded RNA. Angew. Chem. Int. Ed. 2019, 58, 4654–4658. [Google Scholar] [CrossRef] [PubMed]
- Wadle, S.; Lehnert, M.; Rubenwolf, S.; Zengerle, R.; Von Stetten, F. Real-time PCR probe optimization using design of experiments approach. Biomol. Detect. Quantif. 2016, 7, 1–8. [Google Scholar] [CrossRef]
- Yilmaz, L.S.; Loy, A.; Wright, E.S.; Wagner, M.; Noguera, D.R. Modeling formamide denaturation of probe-target hybrids for improved microarray probe design in microbial diagnostics. PLoS ONE 2012, 7, e43862. [Google Scholar] [CrossRef] [PubMed]
- Boissinot, K.; Huletsky, A.; Peytavi, R.; Turcotte, S.; Veillette, V.; Boissinot, M.; Picard, F.J.; Martel, E.A.; Bergeron, M.G. Rapid exonuclease digestion of PCR-amplified targets for improved microarray hybridization. Clin. Chem. 2007, 53, 2020–2023. [Google Scholar] [CrossRef]
- Gerasimova, Y.V.; Cornett, E.M.; Edwards, E.; Su, X.; Rohde, K.H.; Kolpashchikov, D.M. Deoxyribozyme cascade for visual detection of bacterial RNA. ChemBioChem 2013, 14, 2087–2090. [Google Scholar] [CrossRef]
- Lyalina, T.A.; Goncharova, E.A.; Prokofeva, N.Y.; Voroshilina, E.S.; Kolpashchikov, D.M. A DNA minimachine for selective and sensitive detection of DNA. Analyst 2019, 144, 416–420. [Google Scholar] [CrossRef]
- Wang, M.; Chen, D.; Wu, W.; Tang, H.; Kan, L.; Zong, Z.; Dou, X.; Ji, X.; Xiong, D.; Zhang, X. Analytical performance evaluation of five RT-PCR kits for severe acute respiratory syndrome coronavirus 2. J. Clin. Lab. Anal. 2021, 35, e23643. [Google Scholar] [CrossRef]
- Needs, S.H.; Osborn, H.M.I.; Edwards, A.D. Counting bacteria in microfluidic devices: Smartphone compatible ‘dip-and-test’ viable cell quantitation using resazurin amplified detection in microliter capillary arrays. J. Microbiol. Methods 2021, 187, 106199. [Google Scholar] [CrossRef]
- Bilotto, P.; Imre, A.M.; Dworschak, D.; Mears, L.L.E.; Valtiner, M. Visualization of Ion|Surface Binding and In Situ Evaluation of Surface Interaction Free Energies via Competitive Adsorption Isotherms. ACS Phys. Chem. Au 2021, 1, 45–53. [Google Scholar] [CrossRef] [PubMed]
- Available online: http://primerexplorer.jp/e (accessed on 29 July 2020).
- Okonechnikov, K.; Golosova, O.; Fursov, M.; the UGENE Team. Unipro UGENE: A unified bioinformatics toolkit. Bioinformatics 2012, 28, 1166–1167. [Google Scholar] [CrossRef] [PubMed]
- Markham, N.R.; Zuker, M. DINAMelt web server for nucleic acid melting prediction. Nucleic Acids Res. 2005, 33, W577–W581. [Google Scholar] [CrossRef] [PubMed]
- Zuker, M. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 2003, 31, 3406–3415. [Google Scholar] [CrossRef] [PubMed]
- Zadeh, J.N.; Steenberg, C.D.; Bois, J.S.; Wolfe, B.R.; Pierce, M.B.; Khan, A.R.; Dirks, R.M.; Pierce, N.A. NUPACK: Analysis and design of nucleic acid systems. J. Comput. Chem. 2011, 32, 170–173. [Google Scholar] [CrossRef] [PubMed]




Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Maltzeva, Y.I.; Gorbenko, D.A.; Nikitina, E.V.; Rubel, M.S.; Kolpashchikov, D.M. Visual Detection of Stem-Loop Primer Amplification (SPA) Products without Denaturation Using Peroxidase-like DNA Machines (PxDM). Int. J. Mol. Sci. 2023, 24, 7812. https://doi.org/10.3390/ijms24097812
Maltzeva YI, Gorbenko DA, Nikitina EV, Rubel MS, Kolpashchikov DM. Visual Detection of Stem-Loop Primer Amplification (SPA) Products without Denaturation Using Peroxidase-like DNA Machines (PxDM). International Journal of Molecular Sciences. 2023; 24(9):7812. https://doi.org/10.3390/ijms24097812
Chicago/Turabian StyleMaltzeva, Yulia I., Daria A. Gorbenko, Ekaterina V. Nikitina, Maria S. Rubel, and Dmitry M. Kolpashchikov. 2023. "Visual Detection of Stem-Loop Primer Amplification (SPA) Products without Denaturation Using Peroxidase-like DNA Machines (PxDM)" International Journal of Molecular Sciences 24, no. 9: 7812. https://doi.org/10.3390/ijms24097812
APA StyleMaltzeva, Y. I., Gorbenko, D. A., Nikitina, E. V., Rubel, M. S., & Kolpashchikov, D. M. (2023). Visual Detection of Stem-Loop Primer Amplification (SPA) Products without Denaturation Using Peroxidase-like DNA Machines (PxDM). International Journal of Molecular Sciences, 24(9), 7812. https://doi.org/10.3390/ijms24097812






