Characterization of A-to-I Editing in Pigs under a Long-Term High-Energy Diet
Abstract
1. Introduction
2. Results
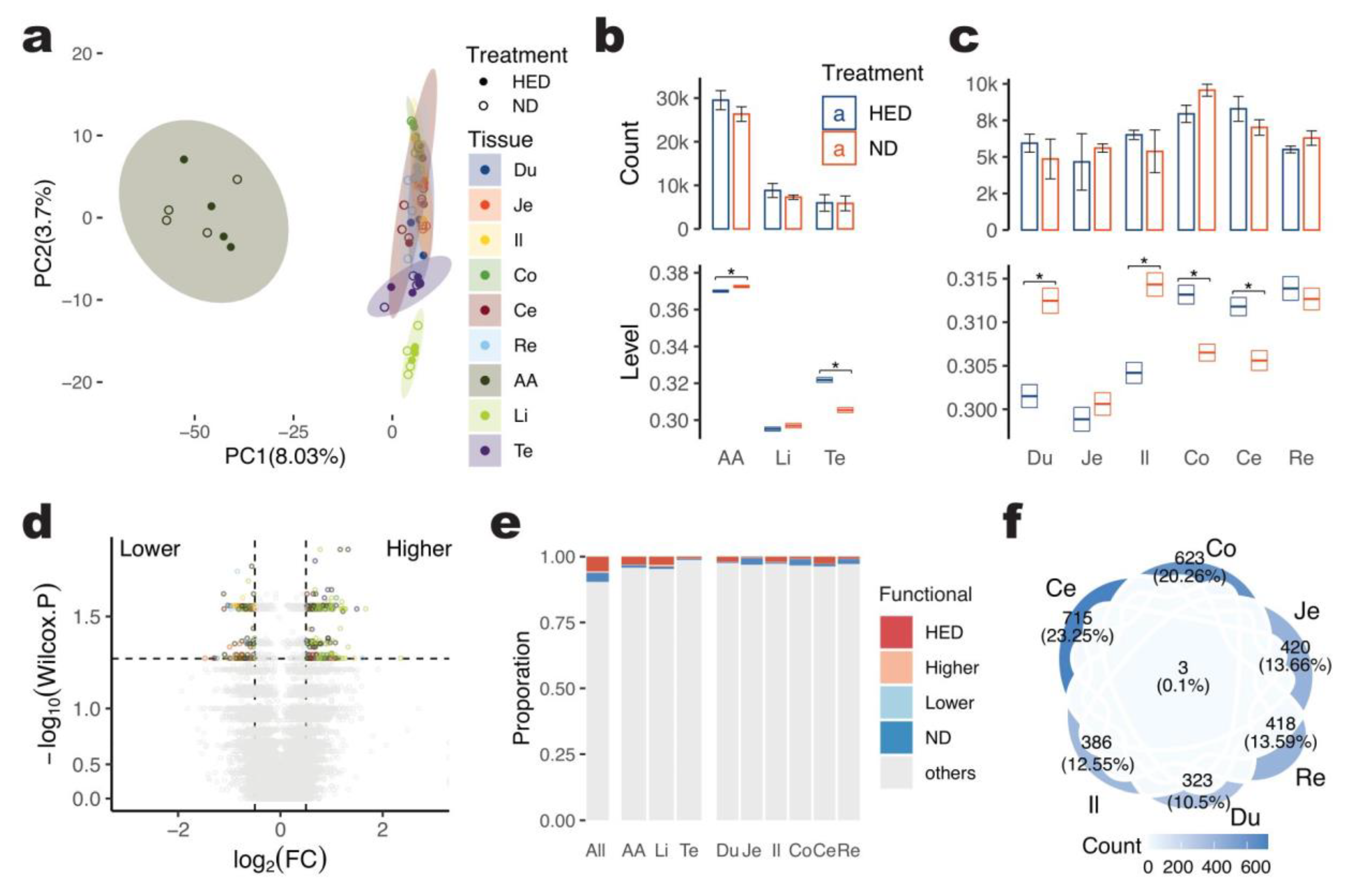
2.1. A-to-I Editing across Pig Genome
2.2. Functional Aspects of A-to-I Editing across Pig Tissues
2.3. Changes in A-to-I Editing for the Long-Term High-Energy Diet of Pigs
2.4. Gene Enrichment of Functional A-to-I Editing Sites
2.4.1. Functional A-to-I Editing Sites Involved in Aspects of the Living Process
2.4.2. Functional A-to-I Editing Sites Involved in Tissue’s Specific Function
2.4.3. Two Functional A-to-I Editing Sites Shared Nine Tissues
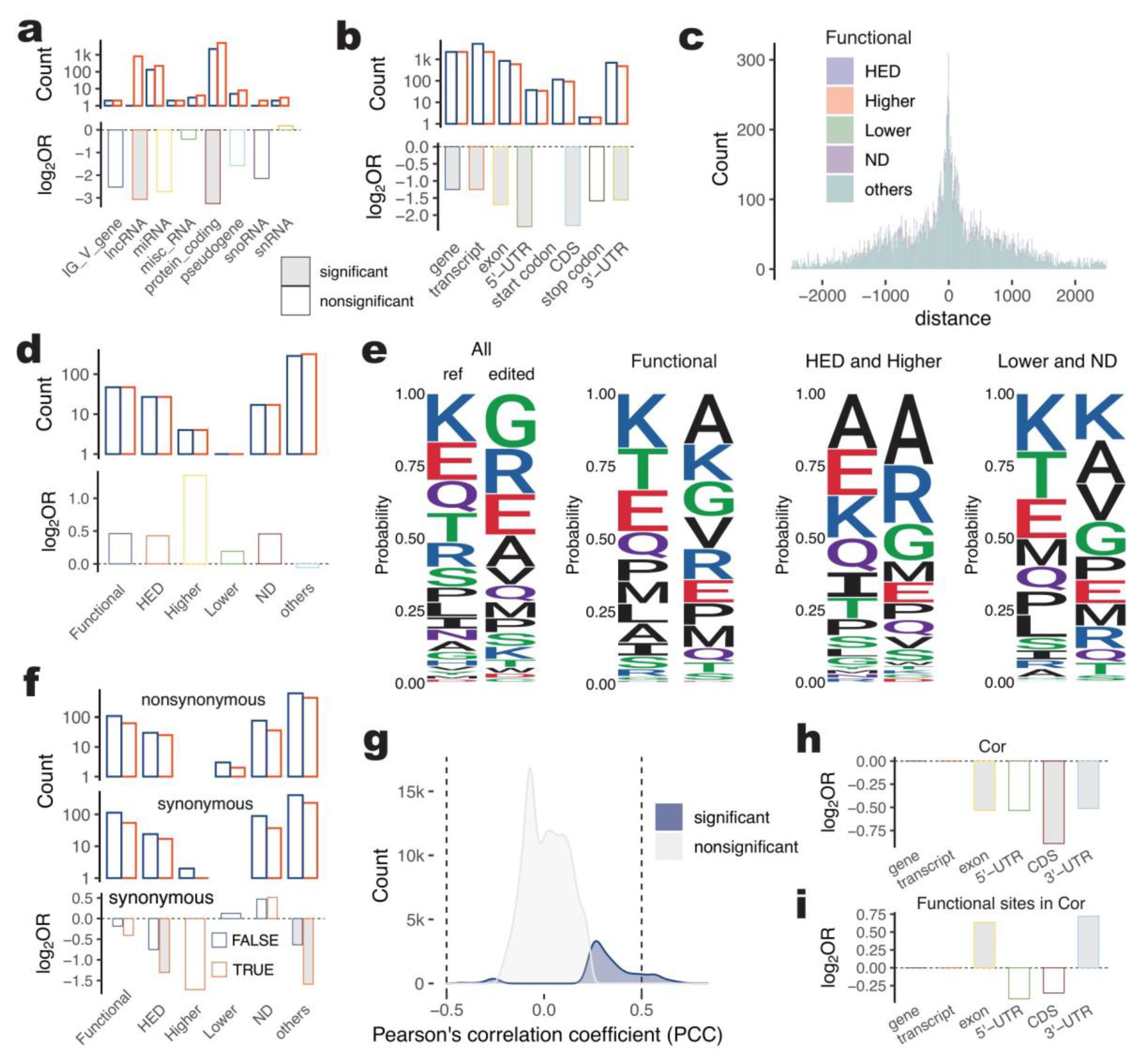
2.5. Characterization of Functional A-to-I Sites in Gene Regions
2.5.1. Functional A-to-I Editing Sites Not Enriched in the Gene Body (Non-Intron)
2.5.2. Functional A-to-I Editing Was Not Significantly Related to Alternative Splicing for HED Pig
2.5.3. Potential Protein-Coding Change of Functional A-to-I Editing
2.5.4. Gene Expression-Related Functional A-to-I Editing Enriched in 3′-UTRs
3. Discussion
4. Materials and Methods
4.1. Data and Samples
4.2. Data Preprocessing
4.3. Variation Calling
4.4. Identification of RNA Editing Sites
4.5. Filtration of RNA Editing Sites
4.6. Gene Annotation and Functional Enrichment
4.7. Principal Component Analysis (PCA)
4.8. Differential Expression Genes/Transcripts between Two Treatments
4.9. Differential Editing Sites between Two Treatments
4.10. Definitions of Treatment-Specific Editing Sites
4.11. Correlation between Gene/Transcript Expression and RNA Editing Level
4.12. Protein Coding Change of A-to-I Editing Sites
4.13. Plotting Gene Regions
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Pace, N.R. The universal nature of biochemistry. Proc. Natl. Acad. Sci. USA 2001, 98, 805–808. [Google Scholar] [CrossRef] [PubMed]
- Smith, E.; Morowitz, H.J. Universality in intermediary metabolism. Proc. Natl. Acad. Sci. USA 2004, 101, 13168–13173. [Google Scholar] [CrossRef] [PubMed]
- Xu, S.-S.; Zhang, X.-L.; Liu, S.-S.; Feng, S.-T.; Xiang, G.-M.; Xu, C.-J.; Fan, Z.-Y.; Xu, K.; Wang, N.; Wang, Y. Multi-omic analysis in a metabolic syndrome porcine model implicates arachidonic acid metabolism disorder as a risk factor for atherosclerosis. Front. Nutr. 2022, 9, 60. [Google Scholar] [CrossRef]
- Xu, S.-S.; Wang, N.; Huang, L.; Zhang, X.-L.; Feng, S.-T.; Liu, S.-S.; Wang, Y.; Liu, Z.-G.; Wang, B.-Y.; Wu, T.-W. Changes in the Mucosa-Associated Microbiome and Transcriptome across Gut Segments Are Associated with Obesity in a Metabolic Syndrome Porcine Model. Microbiol. Spectr. 2022, 10, e0071722. [Google Scholar] [CrossRef] [PubMed]
- Covello, P.S.; Gray, M.W. RNA editing in plant mitochondria. Nature 1989, 341, 662–666. [Google Scholar] [CrossRef]
- Sommer, B.; Kohler, M.; Sprengel, R.; Seeburg, P.H. RNA editing in brain controls a determinant of ion flow in glutamate-gated channels. Cell 1991, 67, 11–19. [Google Scholar] [CrossRef]
- Nishikura, K. Functions and regulation of RNA editing by ADAR deaminases. Annu. Rev. Biochem. 2010, 79, 321–349. [Google Scholar] [CrossRef]
- Eisenberg, E. Proteome Diversification by RNA Editing. Methods Mol. Biol. 2021, 2181, 229–251. [Google Scholar] [CrossRef]
- Nakano, M.; Fukami, T.; Gotoh, S.; Nakajima, M. A-to-I RNA Editing Up-regulates Human Dihydrofolate Reductase in Breast Cancer. J. Biol. Chem. 2017, 292, 4873–4884. [Google Scholar] [CrossRef]
- Rueter, S.M.; Dawson, T.R.; Emeson, R.B. Regulation of alternative splicing by RNA editing. Nature 1999, 399, 75–80. [Google Scholar] [CrossRef]
- Levanon, E.Y.; Eisenberg, E.; Yelin, R.; Nemzer, S.; Hallegger, M.; Shemesh, R.; Fligelman, Z.Y.; Shoshan, A.; Pollock, S.R.; Sztybel, D.; et al. Systematic identification of abundant A-to-I editing sites in the human transcriptome. Nat. Biotechnol. 2004, 22, 1001–1005. [Google Scholar] [CrossRef] [PubMed]
- Park, E.; Williams, B.; Wold, B.J.; Mortazavi, A. RNA editing in the human ENCODE RNA-seq data. Genome Res. 2012, 22, 1626–1633. [Google Scholar] [CrossRef] [PubMed]
- Hwang, T.; Park, C.K.; Leung, A.K.; Gao, Y.; Hyde, T.M.; Kleinman, J.E.; Rajpurohit, A.; Tao, R.; Shin, J.H.; Weinberger, D.R. Dynamic regulation of RNA editing in human brain development and disease. Nat. Neurosci. 2016, 19, 1093–1099. [Google Scholar] [CrossRef] [PubMed]
- Han, L.; Diao, L.; Yu, S.; Xu, X.; Li, J.; Zhang, R.; Yang, Y.; Werner, H.M.J.; Eterovic, A.K.; Yuan, Y.; et al. The Genomic Landscape and Clinical Relevance of A-to-I RNA Editing in Human Cancers. Cancer Cell. 2015, 28, 515–528. [Google Scholar] [CrossRef]
- Bakhtiarizadeh, M.R.; Salehi, A.; Rivera, R.M. Genome-wide identification and analysis of A-to-I RNA editing events in bovine by transcriptome sequencing. PLoS ONE 2018, 13, e0193316. [Google Scholar] [CrossRef]
- Yang, L.; Li, L.; Kyei, B.; Guo, J.; Zhan, S.; Zhao, W.; Song, Y.; Zhong, T.; Wang, L.; Xu, L.; et al. Systematic analyses reveal RNA editing events involved in skeletal muscle development of goat (Capra hircus). Funct. Integr. Genom. 2020, 20, 633–643. [Google Scholar] [CrossRef]
- Li, L.; Xu, X.; Xiao, M.; Huang, C.; Cao, J.; Zhan, S.; Guo, J.; Zhong, T.; Wang, L.; Yang, L. The Profiles and Functions of RNA Editing Sites Associated with High-Altitude Adaptation in Goats. Int. J. Mol. Sci. 2023, 24, 3115. [Google Scholar] [CrossRef]
- Zhang, Y.; Han, D.; Dong, X.; Wang, J.; Chen, J.; Yao, Y.; Darwish, H.Y.A.; Liu, W.; Deng, X. Genome-wide profiling of RNA editing sites in sheep. J. Anim. Sci. Biotechnol. 2019, 10, 31. [Google Scholar] [CrossRef]
- Roux, P.F.; Fresard, L.; Boutin, M.; Leroux, S.; Klopp, C.; Djari, A.; Esquerre, D.; Martin, P.G.; Zerjal, T.; Gourichon, D.; et al. The Extent of mRNA Editing Is Limited in Chicken Liver and Adipose, but Impacted by Tissular Context, Genotype, Age, and Feeding as Exemplified with a Conserved Edited Site in COG3. G3 2015, 6, 321–335. [Google Scholar] [CrossRef]
- Martinez-Montes, A.M.; Fernandez, A.; Perez-Montarelo, D.; Alves, E.; Benitez, R.M.; Nunez, Y.; Ovilo, C.; Ibanez-Escriche, N.; Folch, J.M.; Fernandez, A.I. Using RNA-Seq SNP data to reveal potential causal mutations related to pig production traits and RNA editing. Anim. Genet. 2017, 48, 151–165. [Google Scholar] [CrossRef]
- Yang, Y.; Zhu, M.; Fan, X.; Yao, Y.; Yan, J.; Tang, Y.; Liu, S.; Li, K.; Tang, Z. Developmental atlas of the RNA editome in Sus scrofa skeletal muscle. DNA Res. 2019, 26, 261–272. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Feng, X.; Tang, Z.; Li, S.C. Genome-Wide Investigation and Functional Analysis of Sus scrofa RNA Editing Sites across Eleven Tissues. Genes 2019, 10, 327. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Zhang, L.; Yue, J.; Wei, X.; Wang, L.; Liu, X.; Gao, H.; Hou, X.; Zhao, F.; Yan, H.; et al. Genome-wide identification of RNA editing in seven porcine tissues by matched DNA and RNA high-throughput sequencing. J. Anim. Sci. Biotechnol. 2019, 10, 24. [Google Scholar] [CrossRef] [PubMed]
- Garrett, S.; Rosenthal, J.J. RNA editing underlies temperature adaptation in K+ channels from polar octopuses. Science 2012, 335, 848–851. [Google Scholar] [CrossRef]
- Yap, H.-Y.Y.; Chooi, Y.-H.; Firdaus-Raih, M.; Fung, S.-Y.; Ng, S.-T.; Tan, C.-S.; Tan, N.-H. The genome of the Tiger Milk mushroom, Lignosus rhinocerotis, provides insights into the genetic basis of its medicinal properties. BMC Genom. 2014, 15, 635. [Google Scholar] [CrossRef] [PubMed]
- Hou, N.; Du, X.; Wu, S. Advances in pig models of human diseases. Anim. Model. Exp. Med. 2022, 5, 141–152. [Google Scholar] [CrossRef]
- Rodriguez, R.R.; Gonzalez-Bulnes, A.; Garcia-Contreras, C.; Elena Rodriguez-Rodriguez, A.; Astiz, S.; Vazquez-Gomez, M.; Luis Pesantez, J.; Isabel, B.; Salido-Ruiz, E.; Gonzalez, J.; et al. The Iberian pig fed with high-fat diet: A model of renal disease in obesity and metabolic syndrome. Int. J. Obes. 2020, 44, 457–465. [Google Scholar] [CrossRef] [PubMed]
- Wang, P.; Lu, Z.; He, M.; Shi, B.; Lei, X.; Shan, A. The Effects of Endoplasmic-Reticulum-Resident Selenoproteins in a Nonalcoholic Fatty Liver Disease Pig Model Induced by a High-Fat Diet. Nutrients 2020, 12, 692. [Google Scholar] [CrossRef] [PubMed]
- Walters, E.M.; Prather, R.S. Advancing swine models for human health and diseases. Mo. Med. 2013, 110, 212–215. [Google Scholar]
- Liu, Z.Z.; Xia, J.H.; Xin, L.L.; Wang, Z.G.; Qian, L.; Wu, S.G.; Yang, S.L.; Li, K. Swine leukocyte antigen class II genes (SLA-DRA, SLA-DRB1, SLA-DQA, SLA-DQB1) polymorphism and genotyping in Guizhou minipigs. Genet. Mol. Res. 2015, 14, 15256–15266. [Google Scholar] [CrossRef]
- Zhang, L.; Ren, J.; Shi, P.; Lu, D.; Zhao, C.; Su, Y.; Zhang, L.; Huang, J. The Immunological Regulation Roles of Porcine beta-1, 4 Galactosyltransferase V (B4GALT5) in PRRSV Infection. Front. Cell. Infect. Microbiol. 2018, 8, 48. [Google Scholar] [CrossRef] [PubMed]
- Bae, B.; Miura, P. Emerging Roles for 3′ UTRs in Neurons. Int. J. Mol. Sci. 2020, 21, 3413. [Google Scholar] [CrossRef]
- Slotkin, W.; Nishikura, K. Adenosine-to-inosine RNA editing and human disease. Genome Med. 2013, 5, 105. [Google Scholar] [CrossRef] [PubMed]
- Hideyama, T.; Yamashita, T.; Suzuki, T.; Tsuji, S.; Higuchi, M.; Seeburg, P.H.; Takahashi, R.; Misawa, H.; Kwak, S. Induced loss of ADAR2 engenders slow death of motor neurons from Q/R site-unedited GluR2. J. Neurosci. 2010, 30, 11917–11925. [Google Scholar] [CrossRef] [PubMed]
- Galeano, F.; Tomaselli, S.; Locatelli, F.; Gallo, A. A-to-I RNA editing: The "ADAR" side of human cancer. Semin. Cell. Dev. Biol. 2012, 23, 244–250. [Google Scholar] [CrossRef] [PubMed]
- Stellos, K.; Gatsiou, A.; Stamatelopoulos, K.; Perisic Matic, L.; John, D.; Lunella, F.F.; Jae, N.; Rossbach, O.; Amrhein, C.; Sigala, F.; et al. Adenosine-to-inosine RNA editing controls cathepsin S expression in atherosclerosis by enabling HuR-mediated post-transcriptional regulation. Nat. Med. 2016, 22, 1140–1150. [Google Scholar] [CrossRef]
- Bernard, A.; Khrestchatisky, M. Assessing the extent of RNA editing in the TMII regions of GluR5 and GluR6 kainate receptors during rat brain development. J. Neurochem. 1994, 62, 2057–2060. [Google Scholar] [CrossRef]
- Levin, J.Z.; Yassour, M.; Adiconis, X.; Nusbaum, C.; Thompson, D.A.; Friedman, N.; Gnirke, A.; Regev, A. Comprehensive comparative analysis of strand-specific RNA sequencing methods. Nat. Methods 2010, 7, 709–715. [Google Scholar] [CrossRef]
- Zhao, S.; Zhang, Y.; Gordon, W.; Quan, J.; Xi, H.; Du, S.; von Schack, D.; Zhang, B. Comparison of stranded and non-stranded RNA-seq transcriptome profiling and investigation of gene overlap. BMC Genom. 2015, 16, 675. [Google Scholar] [CrossRef]
- Nguyen, T.A.; Heng, J.W.J.; Kaewsapsak, P.; Kok, E.P.L.; Stanojevic, D.; Liu, H.; Cardilla, A.; Praditya, A.; Yi, Z.; Lin, M.; et al. Direct identification of A-to-I editing sites with nanopore native RNA sequencing. Nat. Methods 2022, 19, 833–844. [Google Scholar] [CrossRef]
- Garalde, D.R.; Snell, E.A.; Jachimowicz, D.; Sipos, B.; Lloyd, J.H.; Bruce, M.; Pantic, N.; Admassu, T.; James, P.; Warland, A.; et al. Highly parallel direct RNA sequencing on an array of nanopores. Nat. Methods 2018, 15, 201–206. [Google Scholar] [CrossRef]
- Liu, H.; Begik, O.; Lucas, M.C.; Ramirez, J.M.; Mason, C.E.; Wiener, D.; Schwartz, S.; Mattick, J.S.; Smith, M.A.; Novoa, E.M. Accurate detection of m(6)A RNA modifications in native RNA sequences. Nat. Commun. 2019, 10, 4079. [Google Scholar] [CrossRef] [PubMed]
- Picardi, E.; Pesole, G. REDItools: High-throughput RNA editing detection made easy. Bioinformatics 2013, 29, 1813–1814. [Google Scholar] [CrossRef] [PubMed]
- Pan, Z.; Yao, Y.; Yin, H.; Cai, Z.; Wang, Y.; Bai, L.; Kern, C.; Halstead, M.; Chanthavixay, G.; Trakooljul, N.; et al. Pig genome functional annotation enhances the biological interpretation of complex traits and human disease. Nat. Commun. 2021, 12, 5848. [Google Scholar] [CrossRef] [PubMed]

| Treatment | Du | Je | Il | Ce | Co | Re | AA | Li | Te |
|---|---|---|---|---|---|---|---|---|---|
| HED | 11,581 | 12,122 | 12,907 | 17,424 | 15,651 | 12,579 | 46,251 | 15,775 | 14,359 |
| ND | 12,171 | 11,025 | 12,610 | 15,161 | 17,843 | 13,148 | 44,508 | 13,206 | 13,457 |
| Tissue | Spe/Dif | HED | Higher | Lower | ND |
|---|---|---|---|---|---|
| All | 6041 | 3550 | 278 | 155 | 2191 |
| Du | 382 | 311 | 5 | 9 | 57 |
| Je | 486 | 60 | 7 | 25 | 394 |
| Il | 456 | 342 | 12 | 26 | 76 |
| Ce | 781 | 553 | 13 | 7 | 208 |
| Co | 698 | 161 | 20 | 21 | 496 |
| Re | 495 | 137 | 9 | 17 | 332 |
| AA | 2027 | 1423 | 77 | 46 | 481 |
| Li | 823 | 537 | 111 | 8 | 167 |
| Te | 231 | 120 | 28 | - | 83 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yang, L.; Huang, L.; Mu, Y.; Li, K. Characterization of A-to-I Editing in Pigs under a Long-Term High-Energy Diet. Int. J. Mol. Sci. 2023, 24, 7921. https://doi.org/10.3390/ijms24097921
Yang L, Huang L, Mu Y, Li K. Characterization of A-to-I Editing in Pigs under a Long-Term High-Energy Diet. International Journal of Molecular Sciences. 2023; 24(9):7921. https://doi.org/10.3390/ijms24097921
Chicago/Turabian StyleYang, Liu, Lei Huang, Yulian Mu, and Kui Li. 2023. "Characterization of A-to-I Editing in Pigs under a Long-Term High-Energy Diet" International Journal of Molecular Sciences 24, no. 9: 7921. https://doi.org/10.3390/ijms24097921
APA StyleYang, L., Huang, L., Mu, Y., & Li, K. (2023). Characterization of A-to-I Editing in Pigs under a Long-Term High-Energy Diet. International Journal of Molecular Sciences, 24(9), 7921. https://doi.org/10.3390/ijms24097921







