Iron Saturation Drives Lactoferrin Effects on Oxidative Stress and Neurotoxicity Induced by HIV-1 Tat
Abstract
1. Introduction
2. Results
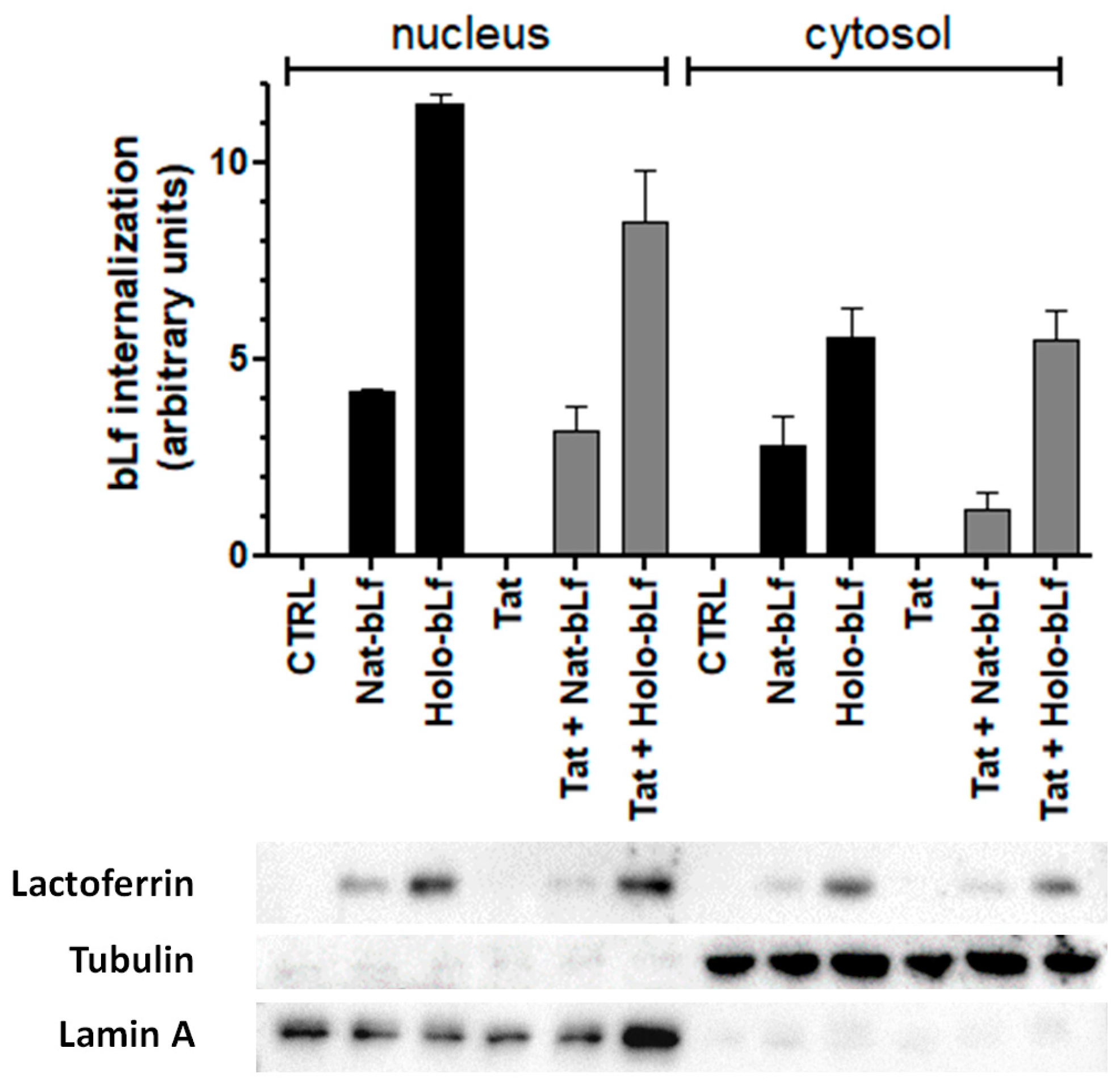
2.1. Native and Holo-Bovine Lactoferrin Are Internalized by U373 Cells
2.2. Bovine Lactoferrin Modulates Cell Antioxidant Response
2.3. Bovine Lactoferrin Potentiates Cell Defense against Intracellular Iron Overload
2.4. Bovine Lactoferrin Counteracts Tat-Mediated Lipid Peroxidation and DNA Damage in Astroglial Cells
2.5. Holo-bLf Exacerbates Tat-Induced Neutoxicity via System Xc−
3. Discussion
4. Materials and Methods
4.1. Reagents
4.2. Bovine Lactoferrin
4.3. Cell Cultures, Transfection and Treatments
4.4. Co-Cultures and Cell Viability Assay
4.5. Quantitative Real-Time Reverse Transcription-Polymerase Chain Reaction
4.6. Total and Nuclear Extracts
4.7. Western Blotting
4.8. Cytokine Analysis
4.9. Immunocytochemistry and Confocal Analysis
4.10. Measurements of Glutamate Concentration in Cell Supernatants
4.11. Measurements of Lipid Peroxidation
4.12. Statistical Analysis
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- UNIAIDS. 2021 Updated Information about HIV Prevalence. Available online: https://www.Unaids.Org/Sites/Default/Files/Media_asset/JC3032_AIDS_Data_book_2021_En.Pdf (accessed on 15 March 2023).
- Saylor, D.; Dickens, A.M.; Sacktor, N.; Haughey, N.; Slusher, B.; Pletnikov, M.; Mankowski, J.L.; Brown, A.; Volsky, D.J.; McArthur, J.C. HIV-Associated Neurocognitive Disorder—Pathogenesis and Prospects for Treatment. Nat. Rev. Neurol. 2016, 12, 234–248. [Google Scholar] [CrossRef] [PubMed]
- Jadhav, S.; Nema, V. HIV-Associated Neurotoxicity: The Interplay of Host and Viral Proteins. Mediat. Inflamm. 2021, 2021, 1267041. [Google Scholar] [CrossRef] [PubMed]
- Chauhan, A.; Turchan, J.; Pocernich, C.; Bruce-Keller, A.; Roth, S.; Butterfield, D.A.; Major, E.O.; Nath, A. Intracellular Human Immunodeficiency Virus Tat Expression in Astrocytes Promotes Astrocyte Survival but Induces Potent Neurotoxicity at Distant Sites via Axonal Transport. J. Biol. Chem. 2003, 278, 13512–13519. [Google Scholar] [CrossRef] [PubMed]
- Dickens, A.M.; Yoo, S.W.; Chin, A.C.; Xu, J.; Johnson, T.P.; Trout, A.L.; Hauser, K.F.; Haughey, N.J. Chronic Low-Level Expression of HIV-1 Tat Promotes a Neurodegenerative Phenotype with Aging. Sci. Rep. 2017, 7, 7748. [Google Scholar] [CrossRef]
- Mastrantonio, R.; D’Ezio, V.; Colasanti, M.; Persichini, T. Nrf2-Mediated System Xc− Activation in Astroglial Cells Is Involved in HIV-1 Tat-Induced Neurotoxicity. Mol. Neurobiol. 2019, 56, 3796–3806. [Google Scholar] [CrossRef]
- Dong, X.; Wang, Y.; Qin, Z. Molecular Mechanisms of Excitotoxicity and Their Relevance to Pathogenesis of Neurodegenerative Diseases. Acta Pharmacol. Sin. 2009, 30, 379–387. [Google Scholar] [CrossRef]
- Mastrantonio, R.; Cervelli, M.; Pietropaoli, S.; Mariottini, P.; Colasanti, M.; Persichini, T. HIV-Tat Induces the Nrf2/ARE Pathway through NMDA Receptor-Elicited Spermine Oxidase Activation in Human Neuroblastoma Cells. PLoS ONE 2016, 11, e0149802. [Google Scholar] [CrossRef]
- Lee Mosley, R.; Benner, E.J.; Kadiu, I.; Thomas, M.; Boska, M.D.; Hasan, K.; Laurie, C.; Gendelman, H.E. Neuroinflammation, Oxidative Stress, and the Pathogenesis of Parkinson’s Disease. Clin. Neurosci. Res. 2006, 6, 261–281. [Google Scholar] [CrossRef]
- Yu, P.; Chang, Y.-Z. Brain Iron Metabolism and Regulation. In Brain Iron Metabolism and CNS Diseases; Chang, Y.-Z., Ed.; Advances in Experimental Medicine and Biology; Springer: Singapore, 2019; Volume 1173, pp. 33–44. ISBN 9789811395888. [Google Scholar]
- Donovan, A.; Lima, C.A.; Pinkus, J.L.; Pinkus, G.S.; Zon, L.I.; Robine, S.; Andrews, N.C. The Iron Exporter Ferroportin/Slc40a1 Is Essential for Iron Homeostasis. Cell Metab. 2005, 1, 191–200. [Google Scholar] [CrossRef]
- Campos-Escamilla, C. The Role of Transferrins and Iron-Related Proteins in Brain Iron Transport: Applications to Neurological Diseases. In Advances in Protein Chemistry and Structural Biology; Elsevier: Amsterdam, The Netherlands, 2021; Volume 123, pp. 133–162. ISBN 978-0-12-822087-0. [Google Scholar]
- Duck, K.A.; Simpson, I.A.; Connor, J.R. Regulatory Mechanisms for Iron Transport across the Blood-Brain Barrier. Biochem. Biophys. Res. Commun. 2017, 494, 70–75. [Google Scholar] [CrossRef]
- Jeong, S.Y.; David, S. Glycosylphosphatidylinositol-Anchored Ceruloplasmin Is Required for Iron Efflux from Cells in the Central Nervous System. J. Biol. Chem. 2003, 278, 27144–27148. [Google Scholar] [CrossRef] [PubMed]
- Ganz, T.; Nemeth, E. Iron Homeostasis in Host Defence and Inflammation. Nat. Rev. Immunol. 2015, 15, 500–510. [Google Scholar] [CrossRef] [PubMed]
- Mancinelli, R.; Rosa, L.; Cutone, A.; Lepanto, M.S.; Franchitto, A.; Onori, P.; Gaudio, E.; Valenti, P. Viral Hepatitis and Iron Dysregulation: Molecular Pathways and the Role of Lactoferrin. Molecules 2020, 25, 1997. [Google Scholar] [CrossRef] [PubMed]
- Ward, R.J.; Zucca, F.A.; Duyn, J.H.; Crichton, R.R.; Zecca, L. The Role of Iron in Brain Ageing and Neurodegenerative Disorders. Lancet Neurol. 2014, 13, 1045–1060. [Google Scholar] [CrossRef]
- Valenti, P.; Antonini, G. Lactoferrin: Lactoferrin: An Important Host Defence against Microbial and Viral Attack. Cell. Mol. Life Sci. 2005, 62, 2576–2587. [Google Scholar] [CrossRef]
- Rosa, L.; Cutone, A.; Conte, M.P.; Campione, E.; Bianchi, L.; Valenti, P. An Overview on in Vitro and in Vivo Antiviral Activity of Lactoferrin: Its Efficacy against SARS-CoV-2 Infection. Biometals 2022, 2022, 1–10. [Google Scholar] [CrossRef]
- Kruzel, M.L.; Zimecki, M.; Actor, J.K. Lactoferrin in a Context of Inflammation-Induced Pathology. Front. Immunol. 2017, 8, 1438. [Google Scholar] [CrossRef]
- Lepanto, M.S.; Rosa, L.; Paesano, R.; Valenti, P.; Cutone, A. Lactoferrin in Aseptic and Septic Inflammation. Molecules 2019, 24, 1323. [Google Scholar] [CrossRef]
- Ianiro, G.; Rosa, L.; Bonaccorsi di Patti, M.C.; Valenti, P.; Musci, G.; Cutone, A. Lactoferrin: From the Structure to the Functional Orchestration of Iron Homeostasis. Biometals 2022, 2022, 1–26. [Google Scholar] [CrossRef]
- Cutone, A.; Frioni, A.; Berlutti, F.; Valenti, P.; Musci, G.; Bonaccorsi di Patti, M.C. Lactoferrin Prevents LPS-Induced Decrease of the Iron Exporter Ferroportin in Human Monocytes/Macrophages. Biometals 2014, 27, 807–813. [Google Scholar] [CrossRef]
- Cutone, A.; Rosa, L.; Lepanto, M.S.; Scotti, M.J.; Berlutti, F.; Bonaccorsi di Patti, M.C.; Musci, G.; Valenti, P. Lactoferrin Efficiently Counteracts the Inflammation-Induced Changes of the Iron Homeostasis System in Macrophages. Front. Immunol. 2017, 8, 705. [Google Scholar] [CrossRef] [PubMed]
- Cutone, A.; Lepanto, M.S.; Rosa, L.; Scotti, M.J.; Rossi, A.; Ranucci, S.; De Fino, I.; Bragonzi, A.; Valenti, P.; Musci, G.; et al. Aerosolized Bovine Lactoferrin Counteracts Infection, Inflammation and Iron Dysbalance in A Cystic Fibrosis Mouse Model of Pseudomonas Aeruginosa Chronic Lung Infection. Int. J. Mol. Sci. 2019, 20, 2128. [Google Scholar] [CrossRef] [PubMed]
- Paesano, R.; Pacifici, E.; Benedetti, S.; Berlutti, F.; Frioni, A.; Polimeni, A.; Valenti, P. Safety and Efficacy of Lactoferrin versus Ferrous Sulphate in Curing Iron Deficiency and Iron Deficiency Anaemia in Hereditary Thrombophilia Pregnant Women: An Interventional Study. Biometals 2014, 27, 999–1006. [Google Scholar] [CrossRef] [PubMed]
- Lepanto, M.S.; Rosa, L.; Cutone, A.; Conte, M.P.; Paesano, R.; Valenti, P. Efficacy of Lactoferrin Oral Administration in the Treatment of Anemia and Anemia of Inflammation in Pregnant and Non-Pregnant Women: An Interventional Study. Front. Immunol. 2018, 9, 2123. [Google Scholar] [CrossRef] [PubMed]
- Suzuki, Y.A.; Lopez, V.; Lönnerdal, B. Lactoferrin: Mammalian Lactoferrin Receptors: Structure and Function. Cell. Mol. Life Sci. 2005, 62, 2560–2575. [Google Scholar] [CrossRef] [PubMed]
- Drake, J.W.; Holland, J.J. Mutation Rates among RNA Viruses. Proc. Natl. Acad. Sci. USA 1999, 96, 13910–13913. [Google Scholar] [CrossRef]
- Reshi, M.L.; Su, Y.-C.; Hong, J.-R. RNA Viruses: ROS-Mediated Cell Death. Int. J. Cell Biol. 2014, 2014, 467452. [Google Scholar] [CrossRef]
- Balakrishna Pillai, A.; JeanPierre, A.R.; Mariappan, V.; Ranganadin, P. Neutralizing the Free Radicals Could Alleviate the Disease Severity Following an Infection by Positive Strand RNA Viruses. Cell Stress Chaperones 2022, 27, 189–195. [Google Scholar] [CrossRef]
- Schmidt, S.M. The Role of Iron in Viral Infections. Front. Biosci. 2020, 25, 893–911. [Google Scholar] [CrossRef]
- Chang, H.-C.; Bayeva, M.; Taiwo, B.; Palella, F.J.; Hope, T.J.; Ardehali, H. Short Communication: High Cellular Iron Levels Are Associated with Increased HIV Infection and Replication. AIDS Res. Hum. Retrovir. 2015, 31, 305–312. [Google Scholar] [CrossRef]
- Drakesmith, H.; Prentice, A. Viral Infection and Iron Metabolism. Nat. Rev. Microbiol. 2008, 6, 541–552. [Google Scholar] [CrossRef] [PubMed]
- Liu, W.; Zhang, S.; Nekhai, S.; Liu, S. Depriving Iron Supply to the Virus Represents a Promising Adjuvant Therapeutic Against Viral Survival. Curr. Clin. Microbio. Rep. 2020, 7, 13–19. [Google Scholar] [CrossRef] [PubMed]
- Kumari, N.; Ammosova, T.; Diaz, S.; Lin, X.; Niu, X.; Ivanov, A.; Jerebtsova, M.; Dhawan, S.; Oneal, P.; Nekhai, S. Increased Iron Export by Ferroportin Induces Restriction of HIV-1 Infection in Sickle Cell Disease. Blood Adv. 2016, 1, 170–183. [Google Scholar] [CrossRef] [PubMed]
- Mangino, G.; Famiglietti, M.; Capone, C.; Veroni, C.; Percario, Z.A.; Leone, S.; Fiorucci, G.; Lülf, S.; Romeo, G.; Agresti, C.; et al. HIV-1 Myristoylated Nef Treatment of Murine Microglial Cells Activates Inducible Nitric Oxide Synthase, NO2 Production and Neurotoxic Activity. PLoS ONE 2015, 10, e0130189. [Google Scholar] [CrossRef] [PubMed]
- Persichini, T.; Mastrantonio, R.; Matto, S.D.; Palomba, L.; Cantoni, O.; Colasanti, M. The Role of Arachidonic Acid in the Regulation of Nitric Oxide Synthase Isoforms by HIV Gp120 Protein in Astroglial Cells. Free. Radic. Biol. Med. 2014, 74, 14–20. [Google Scholar] [CrossRef] [PubMed]
- Capone, C.; Cervelli, M.; Angelucci, E.; Colasanti, M.; Macone, A.; Mariottini, P.; Persichini, T. A Role for Spermine Oxidase as a Mediator of Reactive Oxygen Species Production in HIV-Tat-Induced Neuronal Toxicity. Free. Radic. Biol. Med. 2013, 63, 99–107. [Google Scholar] [CrossRef]
- Madrid, R.; Janvier, K.; Hitchin, D.; Day, J.; Coleman, S.; Noviello, C.; Bouchet, J.; Benmerah, A.; Guatelli, J.; Benichou, S. Nef-Induced Alteration of the Early/Recycling Endosomal Compartment Correlates with Enhancement of HIV-1 Infectivity. J. Biol. Chem. 2005, 280, 5032–5044. [Google Scholar] [CrossRef]
- Wessling-Resnick, M. Crossing the Iron Gate: Why and How Transferrin Receptors Mediate Viral Entry. Annu. Rev. Nutr. 2018, 38, 431–458. [Google Scholar] [CrossRef]
- Cutone, A.; Rosa, L.; Bonaccorsi di Patti, M.C.; Iacovelli, F.; Conte, M.P.; Ianiro, G.; Romeo, A.; Campione, E.; Bianchi, L.; Valenti, P.; et al. Lactoferrin Binding to SARS-CoV-2 Spike Glycoprotein Blocks Pseudoviral Entry and Relieves Iron Protein Dysregulation in Several In Vitro Models. Pharmaceutics 2022, 14, 2111. [Google Scholar] [CrossRef]
- Cutone, A.; Colella, B.; Pagliaro, A.; Rosa, L.; Lepanto, M.S.; Bonaccorsi di Patti, M.C.; Valenti, P.; Di Bartolomeo, S.; Musci, G. Native and Iron-Saturated Bovine Lactoferrin Differently Hinder Migration in a Model of Human Glioblastoma by Reverting Epithelial-to-Mesenchymal Transition-like Process and Inhibiting Interleukin-6/STAT3 Axis. Cell. Signal. 2020, 65, 109461. [Google Scholar] [CrossRef]
- Rosa, L.; Cutone, A.; Lepanto, M.S.; Scotti, M.J.; Conte, M.P.; Paesano, R.; Valenti, P. Physico-Chemical Properties Influence the Functions and Efficacy of Commercial Bovine Lactoferrins. Biometals 2018, 31, 301–312. [Google Scholar] [CrossRef] [PubMed]
- Persichini, T.; Mariotto, S.; Suzuki, H.; Butturini, E.; Mastrantonio, R.; Cantoni, O.; Colasanti, M. Cross-Talk Between NO Synthase Isoforms in Neuro-Inflammation: Possible Implications in HIV-Associated Neurocognitive Disorders. Curr. Med. Chem. 2016, 23, 2706–2714. [Google Scholar] [CrossRef] [PubMed]
- Fogal, B.; Li, J.; Lobner, D.; McCullough, L.D.; Hewett, S.J. System Xc− Activity and Astrocytes Are Necessary for Interleukin-1β-Mediated Hypoxic Neuronal Injury. J. Neurosci. 2007, 27, 10094–10105. [Google Scholar] [CrossRef] [PubMed]
- Qin, S.; Colin, C.; Hinners, I.; Gervais, A.; Cheret, C.; Mallat, M. System Xc− and Apolipoprotein E Expressed by Microglia Have Opposite Effects on the Neurotoxicity of Amyloid-β Peptide 1–40. J. Neurosci. 2006, 26, 3345–3356. [Google Scholar] [CrossRef]
- Pietropaoli, S.; Leonetti, A.; Cervetto, C.; Venturini, A.; Mastrantonio, R.; Baroli, G.; Persichini, T.; Colasanti, M.; Maura, G.; Marcoli, M.; et al. Glutamate Excitotoxicity Linked to Spermine Oxidase Overexpression. Mol. Neurobiol. 2018, 55, 7259–7270. [Google Scholar] [CrossRef]
- Piani, D.; Fontana, A. Involvement of the Cystine Transport System Xc− in the Macrophage-Induced Glutamate-Dependent Cytotoxicity to Neurons. J. Immunol. 1994, 152, 3578–3585. [Google Scholar] [CrossRef]
- Gupta, S.; Knight, A.G.; Gupta, S.; Knapp, P.E.; Hauser, K.F.; Keller, J.N.; Bruce-Keller, A.J. HIV-Tat Elicits Microglial Glutamate Release: Role of NAPDH Oxidase and the Cystine–Glutamate Antiporter. Neurosci. Lett. 2010, 485, 233–236. [Google Scholar] [CrossRef]
- Ferrarese, C.; Aliprandi, A.; Tremolizzo, L.; Stanzani, L.; De Micheli, A.; Dolara, A.; Frattola, L. Increased Glutamate in CSF and Plasma of Patients with HIV Dementia. Neurology 2001, 57, 671–675. [Google Scholar] [CrossRef]
- Cassol, E.; Misra, V.; Dutta, A.; Morgello, S.; Gabuzda, D. Cerebrospinal Fluid Metabolomics Reveals Altered Waste Clearance and Accelerated Aging in HIV Patients with Neurocognitive Impairment. AIDS 2014, 28, 1579–1591. [Google Scholar] [CrossRef]
- Ernst, T.; Jiang, C.S.; Nakama, H.; Buchthal, S.; Chang, L. Lower Brain Glutamate Is Associated with Cognitive Deficits in HIV Patients: A New Mechanism for HIV-Associated Neurocognitive Disorder. J. Magn. Reson. Imaging 2010, 32, 1045–1053. [Google Scholar] [CrossRef]
- Albano, R.; Liu, X.; Lobner, D. Regulation of System Xc− in the SOD1-G93A Mouse Model of ALS. Exp. Neurol. 2013, 250, 69–73. [Google Scholar] [CrossRef] [PubMed]
- D’Ezio, V.; Colasanti, M.; Persichini, T. Amyloid-β 25-35 Induces Neurotoxicity through the Up-Regulation of Astrocytic System Xc−. Antioxidants 2021, 10, 1685. [Google Scholar] [CrossRef] [PubMed]
- Iwamaru, Y.; Shimizu, Y.; Imamura, M.; Murayama, Y.; Endo, R.; Tagawa, Y.; Ushiki-Kaku, Y.; Takenouchi, T.; Kitani, H.; Mohri, S.; et al. Lactoferrin induces cell surface retention of prion protein and inhibits prion accumulation. J. Neurochem. 2008, 107, 636–646. [Google Scholar] [CrossRef] [PubMed]
- Schirmbeck, G.H.; Sizonenko, S.; Sanches, E.F. Neuroprotective Role of Lactoferrin during Early Brain Development and Injury through Lifespan. Nutrients 2022, 14, 2923. [Google Scholar] [CrossRef] [PubMed]
- Bradford, M.M. A Rapid and Sensitive Method for the Quantitation of Microgram Quantities of Protein Utilizing the Principle of Protein-Dye Binding. Anal. Biochem. 1976, 72, 248–254. [Google Scholar] [CrossRef]











Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ianiro, G.; D’Ezio, V.; Carpinelli, L.; Casella, C.; Bonaccorsi di Patti, M.C.; Rosa, L.; Valenti, P.; Colasanti, M.; Musci, G.; Cutone, A.; et al. Iron Saturation Drives Lactoferrin Effects on Oxidative Stress and Neurotoxicity Induced by HIV-1 Tat. Int. J. Mol. Sci. 2023, 24, 7947. https://doi.org/10.3390/ijms24097947
Ianiro G, D’Ezio V, Carpinelli L, Casella C, Bonaccorsi di Patti MC, Rosa L, Valenti P, Colasanti M, Musci G, Cutone A, et al. Iron Saturation Drives Lactoferrin Effects on Oxidative Stress and Neurotoxicity Induced by HIV-1 Tat. International Journal of Molecular Sciences. 2023; 24(9):7947. https://doi.org/10.3390/ijms24097947
Chicago/Turabian StyleIaniro, Giusi, Veronica D’Ezio, Ludovica Carpinelli, Cecilia Casella, Maria Carmela Bonaccorsi di Patti, Luigi Rosa, Piera Valenti, Marco Colasanti, Giovanni Musci, Antimo Cutone, and et al. 2023. "Iron Saturation Drives Lactoferrin Effects on Oxidative Stress and Neurotoxicity Induced by HIV-1 Tat" International Journal of Molecular Sciences 24, no. 9: 7947. https://doi.org/10.3390/ijms24097947
APA StyleIaniro, G., D’Ezio, V., Carpinelli, L., Casella, C., Bonaccorsi di Patti, M. C., Rosa, L., Valenti, P., Colasanti, M., Musci, G., Cutone, A., & Persichini, T. (2023). Iron Saturation Drives Lactoferrin Effects on Oxidative Stress and Neurotoxicity Induced by HIV-1 Tat. International Journal of Molecular Sciences, 24(9), 7947. https://doi.org/10.3390/ijms24097947











