Functional Characterization of Allelic Variations of Human Cytochrome P450 2C8 (V181I, I244V, I331T, and L361F)
Abstract
1. Introduction
2. Results
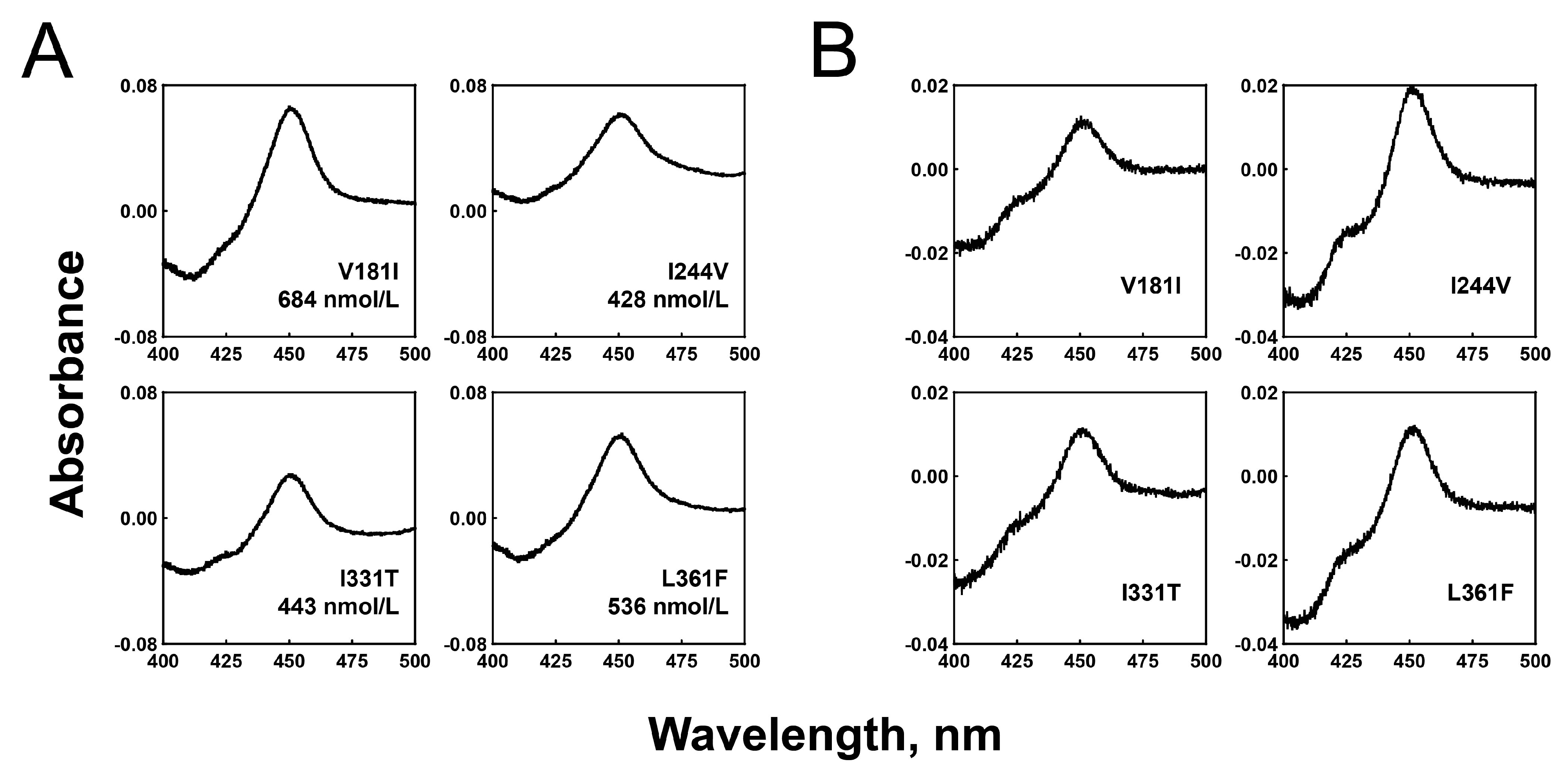
2.1. Expression and Purification of the Recombinant P450 2C8 Variant Enzymes
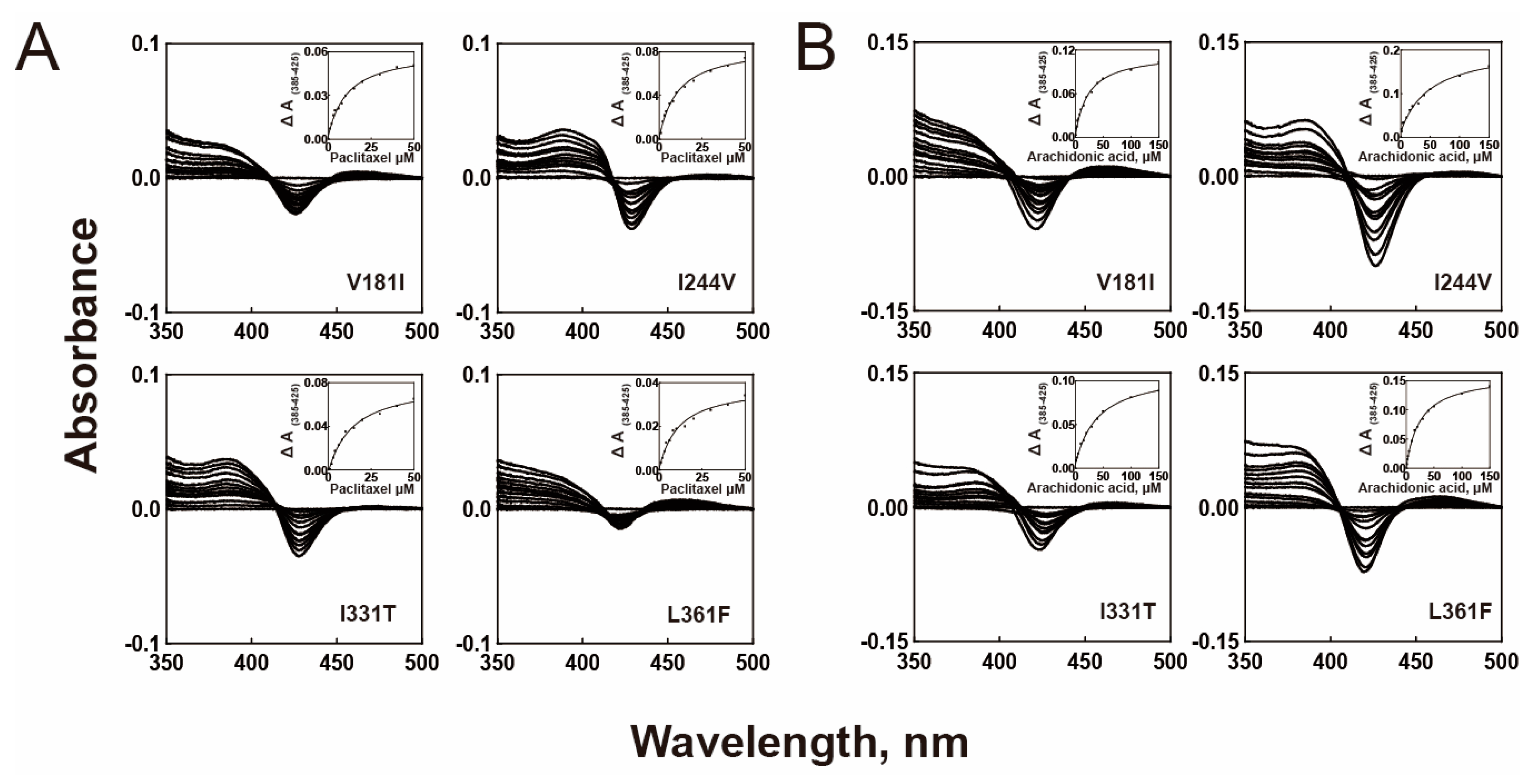
2.2. Substrate Binding Affinities
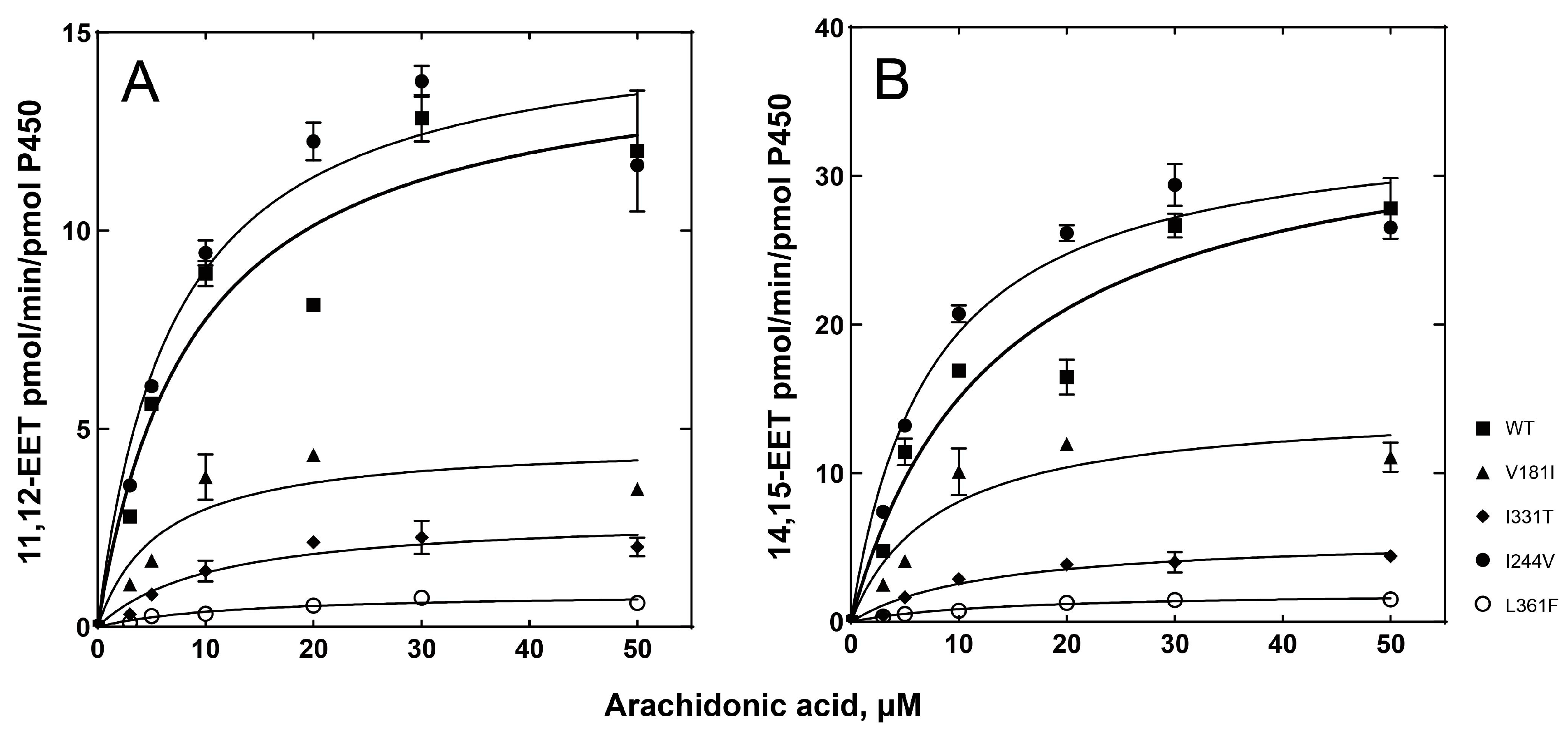
2.3. Catalytic Activities of the P450 2C8 Allelic Variants
3. Discussion
4. Materials and Methods
4.1. Chemicals
4.2. Construction of P450 2C8 Variants
4.3. Expression and Purification of P450 2C8 Variant Enzymes
4.4. Substrate Binding Assay
4.5. Catalytic Activity Assays
4.6. Docking Modeling
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Guengerich, F.P. A history of the roles of cytochrome P450 enzymes in the toxicity of drugs. Toxicol. Res. 2021, 37, 1–23. [Google Scholar] [PubMed]
- Rifkind, A.B.; Lee, C.; Chang, T.K.H.; Waxman, D.J. Arachidonic acid metabolism by human cytochrome P450s 2C8, 2C9, 2E1, and 1A2: Regioselective oxygenation and evidence for a role for CYP2C enzymes in arachidonic acid epoxygenation in human liver microsomes. Arch. Biochem. Biophys. 1995, 320, 380–389. [Google Scholar] [CrossRef]
- Goldstein, J.A.; de Morais, S.M.F. Biochemistry and molecular biology of the human CYP2C subfamily. Pharmacokinet. Genom. 1994, 4, 285–299. [Google Scholar] [CrossRef] [PubMed]
- Zanger, U.M.; Turpeinen, M.; Klein, K.; Schwab, M. Functional pharmacogenetics/genomics of human cytochromes P450 involved in drug biotransformation. Anal. Bioanal. Chem. 2008, 392, 1093–1108. [Google Scholar] [PubMed]
- Hanioka, N.; Matsumoto, K.; Saito, Y.; Narimatsu, S. Functional characterization of CYP2C8.13 and CYP2C8.14: Catalytic activities toward paclitaxel. Basic Clin. Pharmacol. Toxicol. 2010, 107, 565–569. [Google Scholar] [CrossRef]
- Schoch, G.A.; Yano, J.K.; Wester, M.R.; Griffin, K.J.; Stout, C.D.; Johnson, E.F. Structure of human microsomal cytochrome P450 2C8. Evidence for a peripheral fatty acid binding site. J. Biol. Chem. 2004, 279, 9497–9503. [Google Scholar] [CrossRef]
- Shimada, T.; Yamazaki, H.; Mimura, M.; Inui, Y.; Guengerich, F.P. Interindividual variations in human liver cytochrome P-450 enzymes involved in the oxidation of drugs, carcinogens and toxic chemicals: Studies with liver microsomes of 30 Japanese and 30 Caucasians. J. Pharmacol. Exp. Ther. 1994, 270, 414. [Google Scholar]
- Zeldin, D.C.; Dubois, R.N.; Falck, J.R.; Capdevila, J.H. Molecular cloning, expression and characterization of an endogenous human cytochrome P450 arachidonic acid epoxygenase isoform. Arch. Biochem. Biophys. 1995, 322, 76–86. [Google Scholar] [CrossRef]
- Zhao, X.; Imig, J. Kidney CYP450 enzymes: Biological actions beyond drug metabolism. Curr. Drug Metab. 2003, 4, 73–84. [Google Scholar] [CrossRef]
- Yoshida, S.; Hirai, A.; Tamura, Y.; Yoshida, S. Possible involvement of arachidonic acid metabolites of cytochrome P450 monooxygenase pathway in vasopressin-stimulated glycogenolysis in isolated rat hepatocytes. Arch. Biochem. Biophys. 1990, 280, 346–351. [Google Scholar] [CrossRef]
- Falck, J.R.; Manna, S.; Moltz, J.; Chacos, N.; Capdevila, J. Epoxyeicosatrienoic acids stimulate glucagon and insulin release from isolated rat pancreatic islets. Biochem. Biophys. Res. Commun. 1983, 114, 743–749. [Google Scholar] [CrossRef] [PubMed]
- Dai, D.; Zeldin, D.C.; Blaisdell, J.A.; Chanas, B.; Coulter, S.J.; Ghanayem, B.I.; Goldstein, J.A. Polymorphisms in human CYP2C8 decrease metabolism of the anticancer drug paclitaxel and arachidonic acid. Pharm. Genom. 2001, 11, 597–607. [Google Scholar] [CrossRef]
- Gaedigk, A.; Boone, E.C.; Scherer, S.E.; Lee, S.-b.; Numanagić, I.; Sahinalp, C.; Smith, J.D.; McGee, S.; Radhakrishnan, A.; Qin, X.; et al. CYP2C8, CYP2C9, and CYP2C19 characterization using next-generation sequencing and haplotype analysis: A GeT-RM Collaborative Project. J. Mol. Diagn. 2022, 24, 337–350. [Google Scholar] [CrossRef]
- Evans, W.E.; Relling, M.V. Clinical pharmacokinetics-pharmacodynamics of anticancer drugs. Clin. Pharmacokinet. 1989, 16, 327–336. [Google Scholar] [CrossRef]
- Zorsky, P.E.; Perkins, J.B. Optimizing high-dose therapy using pharmacokinetic principles. Semin. Oncol. 1993, 20, 2–18. [Google Scholar] [PubMed]
- Schoch, G.A.; Yano, J.K.; Sansen, S.; Dansette, P.M.; Stout, C.D.; Johnson, E.F. Determinants of cytochrome P450 2C8 substrate binding: Structures of complexes with montelukast, troglitazone, felodipine, and 9-cis-retinoic acid. J. Biol. Chem. 2008, 283, 17227–17237. [Google Scholar] [CrossRef]
- Domanski, T.L.; He, Y.Q.; Scott, E.E.; Wang, Q.; Halpert, J.R. The role of cytochrome 2B1 substrate recognition site residues 115, 294, 297, 298, and 362 in the oxidation of steroids and 7-alkoxycoumarins. Arch. Biochem. Biophys. 2001, 394, 21–28. [Google Scholar] [CrossRef]
- Lee, S.G.; Kim, V.; Lee, G.H.; Kim, C.; Jeong, E.; Guengerich, F.P.; Kim, D. Hydroxylation and lyase reactions of steroids catalyzed by mouse cytochrome P450 17A1 (Cyp17a1). J. Inorg. Biochem. 2023, 240, 112085. [Google Scholar] [CrossRef]
- Lee, I.-S.; Kim, D. Polymorphic metabolism by functional alterations of human cytochrome P450 enzymes. Arch. Pharm. Res. 2011, 34, 1799–1816. [Google Scholar] [PubMed]
- Bahadur, N.; Leathart, J.B.S.; Mutch, E.; Steimel-Crespi, D.; Dunn, S.A.; Gilissen, R.; Houdt, J.V.; Hendrickx, J.; Mannens, G.; Bohets, H.; et al. CYP2C8 polymorphisms in Caucasians and their relationship with paclitaxel 6α-hydroxylase activity in human liver microsomes. Biochem. Pharmacol. 2002, 64, 1579–1589. [Google Scholar] [CrossRef]
- Hichiya, H.; Tanaka-Kagawa, T.; Soyama, A.; Jinno, H.; Koyano, S.; Katori, N.; Matsushima, E.; Uchiyama, S.; Tokunaga, H.; Kimura, H.; et al. Functional characterization of five novel CYP2C8 variants, G171S, R186X, R186G, K247R, and K383N, found in a Japanese population. Drug Metab. Dispos. 2005, 33, 630. [Google Scholar] [CrossRef]
- Nakajima, M.; Yamamoto, T.; Nunoya, K.; Yokoi, T.; Nagashima, K.; Inoue, K.; Funae, Y.; Shimada, N.; Kamataki, T.; Kuroiwa, Y. Role of human cytochrome P4502A6 in C-oxidation of nicotine. Drug Metab. Dispos. 1996, 24, 1212. [Google Scholar] [PubMed]
- Park, H.-G.; Lim, Y.-R.; Eun, C.-Y.; Han, S.; Han, J.-S.; Cho, K.S.; Chun, Y.-J.; Kim, D. Candida albicans NADPH-P450 reductase: Expression, purification, and characterization of recombinant protein. Biochem. Biophys. Res. Commun. 2010, 396, 534–538. [Google Scholar] [CrossRef] [PubMed]
- Lee, R.; Kim, V.; Chun, Y.; Kim, D. Structure-Functional Analysis of Human Cytochrome P450 2C8 Using Directed Evolution. Pharmaceutics 2021, 13, 1429. [Google Scholar] [CrossRef]
- Han, S.; Choi, S.; Chun, Y.-J.; Yun, C.-H.; Lee, C.H.; Shin, H.J.; Na, H.S.; Chung, M.W.; Kim, D. Functional characterization of allelic variants of polymorphic human cytochrome P450 2A6 (CYP2A6*5, *7, *8, *18, *19, and *35). Biol. Pharm. Bull. 2012, 35, 394–399. [Google Scholar] [CrossRef]
- Jeong, D.; Park, H.-G.; Lim, Y.-R.; Lee, Y.; Kim, V.; Cho, M.-A.; Kim, D. Terfenadine metabolism of human cytochrome P450 2J2 containing genetic variations (G312R, P351L and P115L). Drug Metab. Pharmacokinet. 2018, 33, 61–66. [Google Scholar] [CrossRef]
- Yun, C.-H.; Miller, G.P.; Guengerich, F.P. Rate-determining steps in phenacetin oxidations by human cytochrome P450 1A2 and selected mutants. Biochemistry 2000, 39, 11319–11329. [Google Scholar] [CrossRef] [PubMed]
- Morris, G.M.; Goodsell, D.S.; Halliday, R.S.; Huey, R.; Hart, W.E.; Belew, R.K.; Olson, A.J. Automated docking using a Lamarckian genetic algorithm and an empirical binding free energy function. J. Comp. Chem. 1998, 19, 1639–1662. [Google Scholar] [CrossRef]
- Melet, A.; Marques-Soares, C.; Schoch, G.A.; Macherey, A.-C.; Jaouen, M.; Dansette, P.M.; Sari, M.-A.; Johnson, E.F.; Mansuy, D. Analysis of human cytochrome P450 2C8 substrate specificity using a substrate pharmacophore and site-directed mutants. Biochemistry 2004, 43, 15379–15392. [Google Scholar] [CrossRef] [PubMed]
- Moodie, E.E.M.; Richardson, T.S.; Stephens, D.A. Demystifying optimal dynamic treatment regimes. Biometrics 2007, 63, 447–455. [Google Scholar] [CrossRef]



| P450 2C8 Variants | Substrates | |
|---|---|---|
| Paclitaxel | Arachidonic Acid | |
| WT | 10.2 ± 1.0 μM | 20.3 ± 1.4 μM |
| V181I | 10.8 ± 1.4 μM | 22.5 ± 2.1 μM |
| I331T | 13.7 ± 2.0 μM | 39.1 ± 3.8 μM |
| I244V | 10.4 ± 1.5 μM | 49.0 ± 9.5 μM |
| L361F | 10.8 ± 2.2 μM | 25.2 ± 1.7 μM |
| P450 2C8 Variants | 6α-Hydroxylation of Paclitaxel | |||
|---|---|---|---|---|
| kcat (min−1) | Km (µM) | kcat/Km | Relative Catalytic Efficiency | |
| WT | 0.81 ± 0.03 | 4.9 ± 0.8 | 0.164 ± 0.028 | 1 |
| V181I | 0.23 ± 0.01 | 12.8 ± 2.7 | 0.018 ± 0.004 | 0.11 |
| I331T | 0.47 ± 0.01 | 11.3 ± 1.2 | 0.042 ± 0.005 | 0.26 |
| I244V | 1.77 ± 0.09 | 8.6 ± 1.6 | 0.205 ± 0.039 | 1.25 |
| L361F | 0.12 ± 0.01 | 14.8 ± 2.5 | 0.008 ± 0.001 | 0.05 |
| P450 2C8 Variants | Arachdonic Acid Epoxidation | |||||||
|---|---|---|---|---|---|---|---|---|
| ω-9 Epoxidation of Arachidonic Acid | ω-6 Epoxidation of Arachidonic Acid | |||||||
| kcat (min−1) | Km (µM) | kcat/Km | Relative Catalytic Efficiency | kcat (min−1) | Km (µM) | kcat/Km | Relative Catalytic Efficiency | |
| WT | 14.6 ± 1.3 | 8.8 ± 2.4 | 1.65 ± 0.47 | 1 | 35.1 ± 3.4 | 13.3 ± 3.4 | 2.64 ± 0.73 | 1 |
| V181I | 4.7 ± 0.6 | 5.8 ± 2.5 | 0.81 ± 0.37 | 0.49 | 14.6 ± 1.8 | 8.0 ± 2.9 | 1.80 ± 0.68 | 0.68 |
| I331T | 2.8 ± 0.3 | 10.7 ± 3.4 | 0.26 ± 0.09 | 0.16 | 5.8 ± 0.5 | 12.6 ± 3.1 | 0.46 ± 0.12 | 0.17 |
| I244V | 15.3 ± 1.0 | 7.0 ± 1.5 | 2.19 ± 0.47 | 1.33 | 34.0 ± 1.8 | 7.4 ± 1.3 | 4.57 ± 0.82 | 1.73 |
| L361F | 0.9 ± 0.1 | 12.9 ± 3.7 | 0.07 ± 0.02 | 0.04 | 2.0 ± 0.1 | 13.7 ± 2.2 | 0.15 ± 0.03 | 0.06 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lee, Y.-B.; Kim, V.; Lee, S.-G.; Lee, G.-H.; Kim, C.; Jeong, E.; Kim, D. Functional Characterization of Allelic Variations of Human Cytochrome P450 2C8 (V181I, I244V, I331T, and L361F). Int. J. Mol. Sci. 2023, 24, 8032. https://doi.org/10.3390/ijms24098032
Lee Y-B, Kim V, Lee S-G, Lee G-H, Kim C, Jeong E, Kim D. Functional Characterization of Allelic Variations of Human Cytochrome P450 2C8 (V181I, I244V, I331T, and L361F). International Journal of Molecular Sciences. 2023; 24(9):8032. https://doi.org/10.3390/ijms24098032
Chicago/Turabian StyleLee, Yoo-Bin, Vitchan Kim, Sung-Gyu Lee, Gyu-Hyeong Lee, Changmin Kim, Eunseo Jeong, and Donghak Kim. 2023. "Functional Characterization of Allelic Variations of Human Cytochrome P450 2C8 (V181I, I244V, I331T, and L361F)" International Journal of Molecular Sciences 24, no. 9: 8032. https://doi.org/10.3390/ijms24098032
APA StyleLee, Y.-B., Kim, V., Lee, S.-G., Lee, G.-H., Kim, C., Jeong, E., & Kim, D. (2023). Functional Characterization of Allelic Variations of Human Cytochrome P450 2C8 (V181I, I244V, I331T, and L361F). International Journal of Molecular Sciences, 24(9), 8032. https://doi.org/10.3390/ijms24098032






