Possible Pathogenesis and Prevention of Long COVID: SARS-CoV-2-Induced Mitochondrial Disorder
Abstract
:1. Introduction
2. SARS-CoV-2 and Long COVID
2.1. SARS-CoV-2 Non-Structural Proteins
2.2. Viral PAMPs
2.3. Long COVID
3. Altered Mitochondrial Function in SARS-CoV-2 Infection
3.1. Alteration of Mitochondrial Ca2+ Signaling
3.2. Alteration in Glycolysis–OXPHOS Equilibrium
3.3. Alteration of Mitochondrial Dynamics
3.4. Alteration of Autophagy and Apoptosis
3.5. Cell Death and mtDNA Release
4. Mitochondrial Redox Status and Inflammatory Response
4.1. Mitochondria Induce NLRP-3-Based Inflammatory Response
4.2. Activation of TLR7 and TLR9 Signaling
4.3. MAVS Signaling Regulates Inflammatory Responses
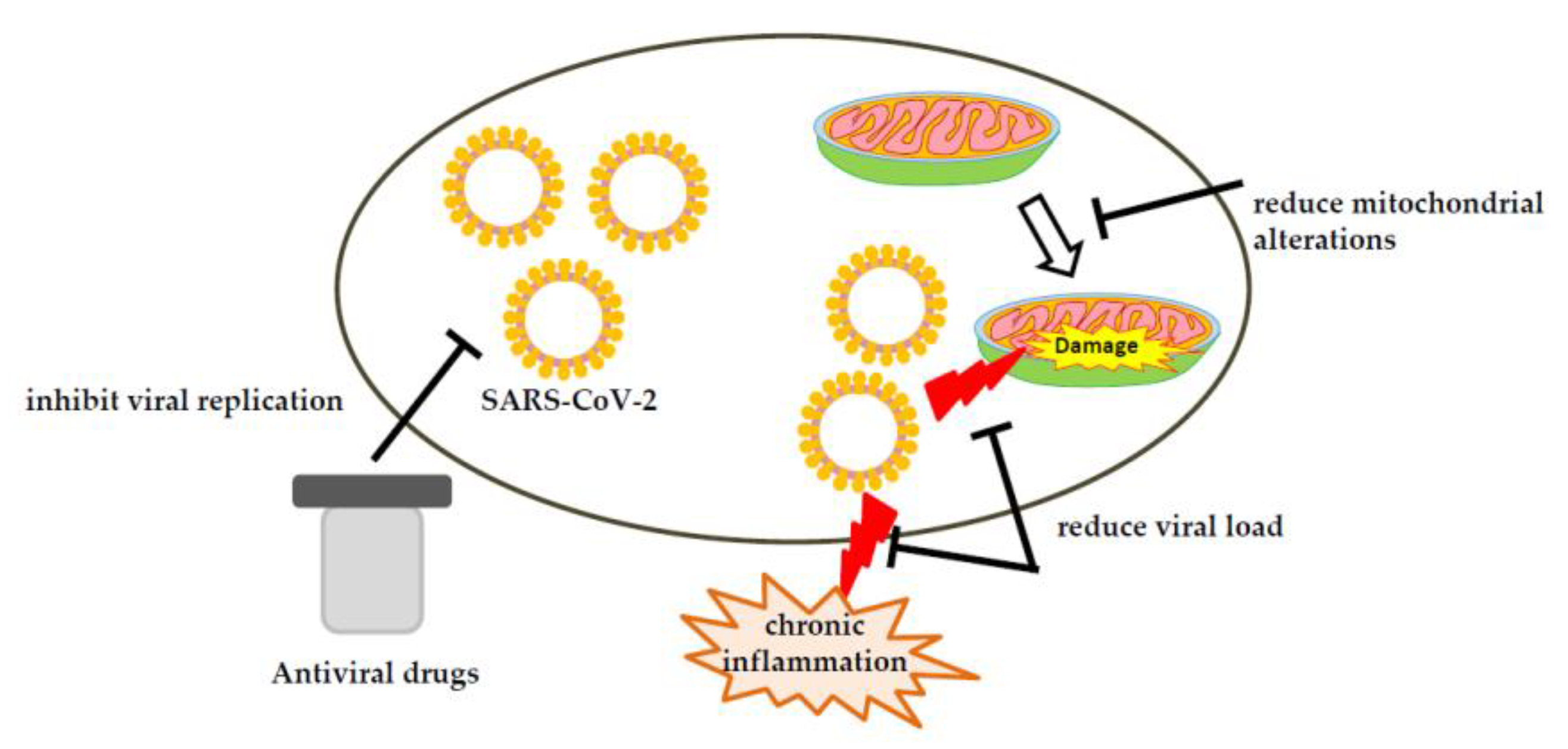
5. Therapeutic Strategies against COVID-19 Involving Mitochondria
6. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Xie, Y.; Wang, Z.; Liao, H.; Marley, G.; Wu, D.; Tang, W. Epidemiologic, clinical, and laboratory findings of the COVID-19 in the current pandemic: Systematic review and meta-analysis. BMC Infect. Dis 2020, 20, 640. [Google Scholar] [CrossRef] [PubMed]
- Chen, T.H.; Hsu, M.T.; Lee, M.Y.; Chou, C.K. Gastrointestinal Involvement in SARS-CoV-2 Infection. Viruses 2022, 14, 1188. [Google Scholar] [CrossRef] [PubMed]
- Van Kessel, S.A.M.; Olde Hartman, T.C.; Lucassen, P.; van Jaarsveld, C.H.M. Post-acute and long-COVID-19 symptoms in patients with mild diseases: A systematic review. Fam. Pract. 2022, 39, 159–167. [Google Scholar] [CrossRef] [PubMed]
- Department of Health and Human Services, Office of the Assistant Secretary for Health. National Research Action Plan on Long COVID; U.S. Department of Health & Human Services: Washington, DC, USA, 2022; Volume 20201. [Google Scholar]
- National Center for Immunization and Respiratory Diseases (NCIRD), Division of Viral Diseases. Long COVID or Post-COVID Conditions. Available online: https://www.cdc.gov/coronavirus/2019-ncov/long-term-effects/index.html (accessed on 16 December 2022).
- Twomey, R.; DeMars, J.; Franklin, K.; Culos-Reed, S.N.; Weatherald, J.; Wrightson, J.G. Chronic Fatigue and Postexertional Malaise in People Living with Long COVID: An Observational Study. Phys. Ther. 2022, 102, pzac005. [Google Scholar] [CrossRef]
- Singh, I.; Joseph, P.; Heerdt, P.M.; Cullinan, M.; Lutchmansingh, D.D.; Gulati, M.; Possick, J.D.; Systrom, D.M.; Waxman, A.B. Persistent Exertional Intolerance After COVID-19: Insights from Invasive Cardiopulmonary Exercise Testing. Chest 2022, 161, 54–63. [Google Scholar] [CrossRef]
- Schwendinger, F.; Knaier, R.; Radtke, T.; Schmidt-Trucksass, A. Low Cardiorespiratory Fitness Post-COVID-19: A Narrative Review. Sports Med. 2023, 53, 51–74. [Google Scholar] [CrossRef] [PubMed]
- Bornstein, S.R.; Voit-Bak, K.; Donate, T.; Rodionov, R.N.; Gainetdinov, R.R.; Tselmin, S.; Kanczkowski, W.; Muller, G.M.; Achleitner, M.; Wang, J.; et al. Chronic post-COVID-19 syndrome and chronic fatigue syndrome: Is there a role for extracorporeal apheresis? Mol. Psychiatry 2022, 27, 34–37. [Google Scholar] [CrossRef]
- Gabanella, F.; Barbato, C.; Corbi, N.; Fiore, M.; Petrella, C.; de Vincentiis, M.; Greco, A.; Ferraguti, G.; Corsi, A.; Ralli, M.; et al. Exploring Mitochondrial Localization of SARS-CoV-2 RNA by Padlock Assay: A Pilot Study in Human Placenta. Int. J. Mol. Sci. 2022, 23, 2100. [Google Scholar] [CrossRef]
- Perlman, S.; Netland, J. Coronaviruses post-SARS: Update on replication and pathogenesis. Nat. Rev. Microbiol. 2009, 7, 439–450. [Google Scholar]
- Wu, K.E.; Fazal, F.M.; Parker, K.R.; Zou, J.; Chang, H.Y. RNA-GPS Predicts SARS-CoV-2 RNA Residency to Host Mitochondria and Nucleolus. Cell Syst. 2020, 11, 102–108.e3. [Google Scholar] [CrossRef]
- Chen, Y.; Zhou, Z.; Min, W. Mitochondria, Oxidative Stress and Innate Immunity. Front. Physiol. 2018, 9, 1487. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ramachandran, K.; Maity, S.; Muthukumar, A.R.; Kandala, S.; Tomar, D.; Abd El-Aziz, T.M.; Allen, C.; Sun, Y.; Venkatesan, M.; Madaris, T.R.; et al. SARS-CoV-2 infection enhances mitochondrial PTP complex activity to perturb cardiac energetics. iScience 2022, 25, 103722. [Google Scholar] [CrossRef] [PubMed]
- Dela Cruz, C.S.; Kang, M.J. Mitochondrial dysfunction and damage associated molecular patterns (DAMPs) in chronic inflammatory diseases. Mitochondrion 2018, 41, 37–44. [Google Scholar] [CrossRef] [PubMed]
- Grazioli, S.; Pugin, J. Mitochondrial Damage-Associated Molecular Patterns: From Inflammatory Signaling to Human Diseases. Front. Immunol. 2018, 9, 832. [Google Scholar] [PubMed] [Green Version]
- Marchi, S.; Guilbaud, E.; Tait, S.W.G.; Yamazaki, T.; Galluzzi, L. Mitochondrial control of inflammation. Nat. Rev. Immunol. 2023, 23, 159–173. [Google Scholar] [PubMed]
- Li, F. Structure, Function, and Evolution of Coronavirus Spike Proteins. Annu. Rev. Virol. 2016, 3, 237–261. [Google Scholar] [CrossRef] [Green Version]
- Subissi, L.; Posthuma, C.C.; Collet, A.; Zevenhoven-Dobbe, J.C.; Gorbalenya, A.E.; Decroly, E.; Snijder, E.J.; Canard, B.; Imbert, I. One severe acute respiratory syndrome coronavirus protein complex integrates processive RNA polymerase and exonuclease activities. Proc. Natl. Acad. Sci. USA 2014, 111, E3900–E3909. [Google Scholar] [CrossRef] [Green Version]
- Brian, D.A.; Baric, R.S. Coronavirus genome structure and replication. In Coronavirus Replication and Reverse Genetics; Current Topics in Microbiology and Immunology; Springer: Berlin/Heidelberg, Germany, 2005; Volume 287, pp. 1–30. [Google Scholar]
- Chang, L.J.; Chen, T.H. NSP16 2′-O-MTase in Coronavirus Pathogenesis: Possible Prevention and Treatments Strategies. Viruses 2021, 13, 538. [Google Scholar] [CrossRef]
- Kesheh, M.M.; Hosseini, P.; Soltani, S.; Zandi, M. An overview on the seven pathogenic human coronaviruses. Rev. Med. Virol. 2022, 32, e2282. [Google Scholar] [CrossRef]
- Farrag, M.A.; Amer, H.M.; Bhat, R.; Hamed, M.E.; Aziz, I.M.; Mubarak, A.; Dawoud, T.M.; Almalki, S.G.; Alghofaili, F.; Alnemare, A.K.; et al. SARS-CoV-2: An Overview of Virus Genetics, Transmission, and Immunopathogenesis. Int. J. Environ. Res. Public Health 2021, 18, 6312. [Google Scholar] [CrossRef]
- Zhang, Y.; Chen, Y.; Li, Y.; Huang, F.; Luo, B.; Yuan, Y.; Xia, B.; Ma, X.; Yang, T.; Yu, F.; et al. The ORF8 protein of SARS-CoV-2 mediates immune evasion through down-regulating MHC-Iota. Proc. Natl. Acad. Sci. USA 2021, 118, e2024202118. [Google Scholar] [CrossRef]
- Azad, G.K.; Khan, P.K. Variations in Orf3a protein of SARS-CoV-2 alter its structure and function. Biochem. Biophys. Rep. 2021, 26, 100933. [Google Scholar] [CrossRef] [PubMed]
- Castano-Rodriguez, C.; Honrubia, J.M.; Gutierrez-Alvarez, J.; DeDiego, M.L.; Nieto-Torres, J.L.; Jimenez-Guardeno, J.M.; Regla-Nava, J.A.; Fernandez-Delgado, R.; Verdia-Baguena, C.; Queralt-Martin, M.; et al. Role of Severe Acute Respiratory Syndrome Coronavirus Viroporins E, 3a, and 8a in Replication and Pathogenesis. mBio 2018, 9, e02325-17. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Siu, K.L.; Yuen, K.S.; Castano-Rodriguez, C.; Ye, Z.W.; Yeung, M.L.; Fung, S.Y.; Yuan, S.; Chan, C.P.; Yuen, K.Y.; Enjuanes, L.; et al. Severe acute respiratory syndrome coronavirus ORF3a protein activates the NLRP3 inflammasome by promoting TRAF3-dependent ubiquitination of ASC. FASEB J. 2019, 33, 8865–8877. [Google Scholar] [CrossRef]
- Yue, Y.; Nabar, N.R.; Shi, C.S.; Kamenyeva, O.; Xiao, X.; Hwang, I.Y.; Wang, M.; Kehrl, J.H. SARS-Coronavirus Open Reading Frame-3a drives multimodal necrotic cell death. Cell Death Dis. 2018, 9, 904. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Konno, Y.; Kimura, I.; Uriu, K.; Fukushi, M.; Irie, T.; Koyanagi, Y.; Sauter, D.; Gifford, R.J.; Consortium, U.-C.; Nakagawa, S.; et al. SARS-CoV-2 ORF3b Is a Potent Interferon Antagonist Whose Activity Is Increased by a Naturally Occurring Elongation Variant. Cell Rep. 2020, 32, 108185. [Google Scholar] [CrossRef]
- Finkel, Y.; Mizrahi, O.; Nachshon, A.; Weingarten-Gabbay, S.; Morgenstern, D.; Yahalom-Ronen, Y.; Tamir, H.; Achdout, H.; Stein, D.; Israeli, O.; et al. The coding capacity of SARS-CoV-2. Nature 2021, 589, 125–130. [Google Scholar] [CrossRef]
- Gordon, D.E.; Jang, G.M.; Bouhaddou, M.; Xu, J.; Obernier, K.; White, K.M.; O’Meara, M.J.; Rezelj, V.V.; Guo, J.Z.; Swaney, D.L.; et al. A SARS-CoV-2 protein interaction map reveals targets for drug repurposing. Nature 2020, 583, 459–468. [Google Scholar] [CrossRef]
- Hachim, A.; Kavian, N.; Cohen, C.A.; Chin, A.W.H.; Chu, D.K.W.; Mok, C.K.P.; Tsang, O.T.Y.; Yeung, Y.C.; Perera, R.; Poon, L.L.M.; et al. ORF8 and ORF3b antibodies are accurate serological markers of early and late SARS-CoV-2 infection. Nat. Immunol. 2020, 21, 1293–1301. [Google Scholar] [CrossRef]
- Lee, J.G.; Huang, W.; Lee, H.; van de Leemput, J.; Kane, M.A.; Han, Z. Characterization of SARS-CoV-2 proteins reveals Orf6 pathogenicity, subcellular localization, host interactions and attenuation by Selinexor. Cell Biosci. 2021, 11, 58. [Google Scholar] [CrossRef]
- Miorin, L.; Kehrer, T.; Sanchez-Aparicio, M.T.; Zhang, K.; Cohen, P.; Patel, R.S.; Cupic, A.; Makio, T.; Mei, M.; Moreno, E.; et al. SARS-CoV-2 Orf6 hijacks Nup98 to block STAT nuclear import and antagonize interferon signaling. Proc. Natl. Acad. Sci. USA 2020, 117, 28344–28354. [Google Scholar] [CrossRef]
- Kopecky-Bromberg, S.A.; Martinez-Sobrido, L.; Frieman, M.; Baric, R.A.; Palese, P. Severe acute respiratory syndrome coronavirus open reading frame (ORF) 3b, ORF 6, and nucleocapsid proteins function as interferon antagonists. J. Virol. 2007, 81, 548–557. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, J.Y.; Liao, C.H.; Wang, Q.; Tan, Y.J.; Luo, R.; Qiu, Y.; Ge, X.Y. The ORF6, ORF8 and nucleocapsid proteins of SARS-CoV-2 inhibit type I interferon signaling pathway. Virus Res. 2020, 286, 198074. [Google Scholar] [CrossRef] [PubMed]
- Kato, K.; Ikliptikawati, D.K.; Kobayashi, A.; Kondo, H.; Lim, K.; Hazawa, M.; Wong, R.W. Overexpression of SARS-CoV-2 protein ORF6 dislocates RAE1 and NUP98 from the nuclear pore complex. Biochem. Biophys. Res. Commun. 2021, 536, 59–66. [Google Scholar] [CrossRef]
- Xia, H.; Cao, Z.; Xie, X.; Zhang, X.; Chen, J.Y.; Wang, H.; Menachery, V.D.; Rajsbaum, R.; Shi, P.Y. Evasion of Type I Interferon by SARS-CoV-2. Cell Rep. 2020, 33, 108234. [Google Scholar] [CrossRef] [PubMed]
- Cao, Z.; Xia, H.; Rajsbaum, R.; Xia, X.; Wang, H.; Shi, P.Y. Ubiquitination of SARS-CoV-2 ORF7a promotes antagonism of interferon response. Cell. Mol. Immunol. 2021, 18, 746–748. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Z.; Huang, C.; Zhou, Z.; Huang, Z.; Su, L.; Kang, S.; Chen, X.; Chen, Q.; He, S.; Rong, X.; et al. Structural insight reveals SARS-CoV-2 ORF7a as an immunomodulating factor for human CD14(+) monocytes. iScience 2021, 24, 102187. [Google Scholar] [CrossRef]
- Liu, D.X.; Fung, T.S.; Chong, K.K.; Shukla, A.; Hilgenfeld, R. Accessory proteins of SARS-CoV and other coronaviruses. Antiviral Res. 2014, 109, 97–109. [Google Scholar]
- Chen, C.Y.; Ping, Y.H.; Lee, H.C.; Chen, K.H.; Lee, Y.M.; Chan, Y.J.; Lien, T.C.; Jap, T.S.; Lin, C.H.; Kao, L.S.; et al. Open reading frame 8a of the human severe acute respiratory syndrome coronavirus not only promotes viral replication but also induces apoptosis. J. Infect. Dis. 2007, 196, 405–415. [Google Scholar] [CrossRef] [Green Version]
- Wong, H.H.; Fung, T.S.; Fang, S.; Huang, M.; Le, M.T.; Liu, D.X. Accessory proteins 8b and 8ab of severe acute respiratory syndrome coronavirus suppress the interferon signaling pathway by mediating ubiquitin-dependent rapid degradation of interferon regulatory factor 3. Virology 2018, 515, 165–175. [Google Scholar] [CrossRef]
- Lei, X.; Dong, X.; Ma, R.; Wang, W.; Xiao, X.; Tian, Z.; Wang, C.; Wang, Y.; Li, L.; Ren, L.; et al. Activation and evasion of type I interferon responses by SARS-CoV-2. Nat. Commun. 2020, 11, 3810. [Google Scholar] [CrossRef]
- Lin, X.; Fu, B.; Yin, S.; Li, Z.; Liu, H.; Zhang, H.; Xing, N.; Wang, Y.; Xue, W.; Xiong, Y.; et al. ORF8 contributes to cytokine storm during SARS-CoV-2 infection by activating IL-17 pathway. iScience 2021, 24, 102293. [Google Scholar] [CrossRef]
- Shi, C.S.; Qi, H.Y.; Boularan, C.; Huang, N.N.; Abu-Asab, M.; Shelhamer, J.H.; Kehrl, J.H. SARS-coronavirus open reading frame-9b suppresses innate immunity by targeting mitochondria and the MAVS/TRAF3/TRAF6 signalosome. J. Immunol. 2014, 193, 3080–3089. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kreimendahl, S.; Rassow, J. The Mitochondrial Outer Membrane Protein Tom70-Mediator in Protein Traffic, Membrane Contact Sites and Innate Immunity. Int. J. Mol. Sci. 2020, 21, 7262. [Google Scholar] [CrossRef] [PubMed]
- Singh, K.K.; Chaubey, G.; Chen, J.Y.; Suravajhala, P. Decoding SARS-CoV-2 hijacking of host mitochondria in COVID-19 pathogenesis. Am. J. Physiol. Cell. Physiol. 2020, 319, C258–C267. [Google Scholar] [CrossRef] [PubMed]
- Dominguez Andres, A.; Feng, Y.; Campos, A.R.; Yin, J.; Yang, C.C.; James, B.; Murad, R.; Kim, H.; Deshpande, A.J.; Gordon, D.E.; et al. SARS-CoV-2 ORF9c Is a Membrane-Associated Protein that Suppresses Antiviral Responses in Cells. bioRxiv 2020. [Google Scholar] [CrossRef]
- Gordon, D.E.; Hiatt, J.; Bouhaddou, M.; Rezelj, V.V.; Ulferts, S.; Braberg, H.; Jureka, A.S.; Obernier, K.; Guo, J.Z.; Batra, J.; et al. Comparative host-coronavirus protein interaction networks reveal pan-viral disease mechanisms. Science 2020, 370, eabe9403. [Google Scholar] [PubMed]
- Jiang, H.W.; Zhang, H.N.; Meng, Q.F.; Xie, J.; Li, Y.; Chen, H.; Zheng, Y.X.; Wang, X.N.; Qi, H.; Zhang, J.; et al. SARS-CoV-2 Orf9b suppresses type I interferon responses by targeting TOM70. Cell. Mol. Immunol. 2020, 17, 998–1000. [Google Scholar] [CrossRef]
- Pancer, K.; Milewska, A.; Owczarek, K.; Dabrowska, A.; Kowalski, M.; Labaj, P.P.; Branicki, W.; Sanak, M.; Pyrc, K. The SARS-CoV-2 ORF10 is not essential in vitro or in vivo in humans. PLoS Pathog. 2020, 16, e1008959. [Google Scholar] [CrossRef]
- Seong, S.Y.; Matzinger, P. Hydrophobicity: An ancient damage-associated molecular pattern that initiates innate immune responses. Nat. Rev. Immunol. 2004, 4, 469–478. [Google Scholar] [CrossRef]
- Broz, P.; Dixit, V.M. Inflammasomes: Mechanism of assembly, regulation and signalling. Nat. Rev. Immunol. 2016, 16, 407–420. [Google Scholar] [CrossRef]
- Yu, J.; Nagasu, H.; Murakami, T.; Hoang, H.; Broderick, L.; Hoffman, H.M.; Horng, T. Inflammasome activation leads to Caspase-1-dependent mitochondrial damage and block of mitophagy. Proc. Natl. Acad. Sci. USA 2014, 111, 15514–15519. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zheng, M.; Karki, R.; Williams, E.P.; Yang, D.; Fitzpatrick, E.; Vogel, P.; Jonsson, C.B.; Kanneganti, T.D. TLR2 senses the SARS-CoV-2 envelope protein to produce inflammatory cytokines. Nat. Immunol. 2021, 22, 829–838. [Google Scholar] [CrossRef] [PubMed]
- Kircheis, R.; Planz, O. The Role of Toll-like Receptors (TLRs) and Their Related Signaling Pathways in Viral Infection and Inflammation. Int. J. Mol. Sci. 2023, 24, 6701. [Google Scholar] [CrossRef]
- Bell, J.K.; Askins, J.; Hall, P.R.; Davies, D.R.; Segal, D.M. The dsRNA binding site of human Toll-like receptor 3. Proc. Natl. Acad. Sci. USA 2006, 103, 8792–8797. [Google Scholar] [CrossRef] [Green Version]
- Zhang, Z.; Ohto, U.; Shibata, T.; Krayukhina, E.; Taoka, M.; Yamauchi, Y.; Tanji, H.; Isobe, T.; Uchiyama, S.; Miyake, K.; et al. Structural Analysis Reveals that Toll-like Receptor 7 Is a Dual Receptor for Guanosine and Single-Stranded RNA. Immunity 2016, 45, 737–748. [Google Scholar] [CrossRef] [Green Version]
- Kircheis, R.; Planz, O. Could a Lower Toll-like Receptor (TLR) and NF-kappaB Activation Due to a Changed Charge Distribution in the Spike Protein Be the Reason for the Lower Pathogenicity of Omicron? Int. J. Mol. Sci. 2022, 23, 5966. [Google Scholar] [CrossRef] [PubMed]
- Ahmad, M.S.; Shaik, R.A.; Ahmad, R.K.; Yusuf, M.; Khan, M.; Almutairi, A.B.; Alghuyaythat, W.K.Z.; Almutairi, S.B. “LONG COVID”: An insight. Eur. Rev. Med. Pharmacol. Sci. 2021, 25, 5561–5577. [Google Scholar]
- Doykov, I.; Hallqvist, J.; Gilmour, K.C.; Grandjean, L.; Mills, K.; Heywood, W.E. ’The long tail of COVID-19’—The detection of a prolonged inflammatory response after a SARS-CoV-2 infection in asymptomatic and mildly affected patients. F1000Research 2020, 9, 1349. [Google Scholar] [CrossRef]
- Marshall, M. The lasting misery of coronavirus long-haulers. Nature 2020, 585, 339–341. [Google Scholar] [CrossRef]
- Nabavi, N. Long covid: How to define it and how to manage it. BMJ 2020, 370, m3489. [Google Scholar] [CrossRef]
- Garg, P.; Arora, U.; Kumar, A.; Wig, N. The "post-COVID" syndrome: How deep is the damage? J. Med. Virol. 2021, 93, 673–674. [Google Scholar] [CrossRef] [PubMed]
- Visco, V.; Vitale, C.; Rispoli, A.; Izzo, C.; Virtuoso, N.; Ferruzzi, G.J.; Santopietro, M.; Melfi, A.; Rusciano, M.R.; Maglio, A.; et al. Post-COVID-19 Syndrome: Involvement and Interactions between Respiratory, Cardiovascular and Nervous Systems. J. Clin. Med. 2022, 11, 524. [Google Scholar] [CrossRef]
- Chudzik, M.; Babicki, M.; Kapusta, J.; Kaluzinska-Kolat, Z.; Kolat, D.; Jankowski, P.; Mastalerz-Migas, A. Long-COVID Clinical Features and Risk Factors: A Retrospective Analysis of Patients from the STOP-COVID Registry of the PoLoCOV Study. Viruses 2022, 14, 1755. [Google Scholar] [CrossRef] [PubMed]
- Xie, Y.; Xu, E.; Bowe, B.; Al-Aly, Z. Long-term cardiovascular outcomes of COVID-19. Nat. Med. 2022, 28, 583–590. [Google Scholar] [CrossRef] [PubMed]
- Leng, A.; Shah, M.; Ahmad, S.A.; Premraj, L.; Wildi, K.; Li Bassi, G.; Pardo, C.A.; Choi, A.; Cho, S.M. Pathogenesis Underlying Neurological Manifestations of Long COVID Syndrome and Potential Therapeutics. Cells 2023, 12, 816. [Google Scholar] [CrossRef]
- Budzynska, N.; Morys, J. Anxiety and Depression Levels and Coping Strategies among Polish Healthcare Workers during the COVID-19 Pandemic. Int. J. Environ. Res. Public Health 2023, 20, 3319. [Google Scholar] [CrossRef]
- Pfaff, E.R.; Madlock-Brown, C.; Baratta, J.M.; Bhatia, A.; Davis, H.; Girvin, A.; Hill, E.; Kelly, E.; Kostka, K.; Loomba, J.; et al. Coding long COVID: Characterizing a new disease through an ICD-10 lens. BMC Med. 2023, 21, 58. [Google Scholar] [CrossRef]
- Lledo, G.M.; Sellares, J.; Brotons, C.; Sans, M.; Anton, J.D.; Blanco, J.; Bassat, Q.; Sarukhan, A.; Miro, J.M.; de Sanjose, S.; et al. Post-acute COVID-19 syndrome: A new tsunami requiring a universal case definition. Clin. Microbiol. Infect. 2022, 28, 315–318. [Google Scholar] [CrossRef] [PubMed]
- Nalbandian, A.; Sehgal, K.; Gupta, A.; Madhavan, M.V.; McGroder, C.; Stevens, J.S.; Cook, J.R.; Nordvig, A.S.; Shalev, D.; Sehrawat, T.S.; et al. Post-acute COVID-19 syndrome. Nat. Med. 2021, 27, 601–615. [Google Scholar] [CrossRef]
- Gaebler, C.; Wang, Z.; Lorenzi, J.C.C.; Muecksch, F.; Finkin, S.; Tokuyama, M.; Cho, A.; Jankovic, M.; Schaefer-Babajew, D.; Oliveira, T.Y.; et al. Evolution of Antibody Immunity to SARS-CoV-2. bioRxiv 2021. [Google Scholar] [CrossRef]
- de Melo, G.D.; Lazarini, F.; Levallois, S.; Hautefort, C.; Michel, V.; Larrous, F.; Verillaud, B.; Aparicio, C.; Wagner, S.; Gheusi, G.; et al. COVID-19-related anosmia is associated with viral persistence and inflammation in human olfactory epithelium and brain infection in hamsters. Sci. Transl. Med. 2021, 13, eabf8396. [Google Scholar] [CrossRef]
- Weinstock, L.B.; Brook, J.B.; Walters, A.S.; Goris, A.; Afrin, L.B.; Molderings, G.J. Mast cell activation symptoms are prevalent in Long-COVID. Int. J. Infect. Dis. 2021, 112, 217–226. [Google Scholar] [CrossRef] [PubMed]
- Afrin, L.B.; Weinstock, L.B.; Molderings, G.J. COVID-19 hyperinflammation and post-Covid-19 illness may be rooted in mast cell activation syndrome. Int. J. Infect. Dis. 2020, 100, 327–332. [Google Scholar] [CrossRef]
- Proal, A.D.; VanElzakker, M.B. Long COVID or Post-acute Sequelae of COVID-19 (PASC): An Overview of Biological Factors That May Contribute to Persistent Symptoms. Front. Microbiol. 2021, 12, 698169. [Google Scholar] [CrossRef]
- Cao, X.; Nguyen, V.; Tsai, J.; Gao, C.; Tian, Y.; Zhang, Y.; Carver, W.; Kiaris, H.; Cui, T.; Tan, W. The SARS-CoV-2 Spike protein induces long-term transcriptional perturbations of mitochondrial metabolic genes, causes cardiac fibrosis, and reduces myocardial contractile in obese mice. bioRxiv 2023. [Google Scholar] [CrossRef]
- Motta, C.S.; Torices, S.; da Rosa, B.G.; Marcos, A.C.; Alvarez-Rosa, L.; Siqueira, M.; Moreno-Rodriguez, T.; Matos, A.D.R.; Caetano, B.C.; Martins, J.; et al. Human Brain Microvascular Endothelial Cells Exposure to SARS-CoV-2 Leads to Inflammatory Activation through NF-kappaB Non-Canonical Pathway and Mitochondrial Remodeling. Viruses 2023, 15, 745. [Google Scholar] [CrossRef]
- Peluso, M.J.; Deeks, S.G.; Mustapic, M.; Kapogiannis, D.; Henrich, T.J.; Lu, S.; Goldberg, S.A.; Hoh, R.; Chen, J.Y.; Martinez, E.O.; et al. SARS-CoV-2 and Mitochondrial Proteins in Neural-Derived Exosomes of COVID-19. Ann. Neurol. 2022, 91, 772–781. [Google Scholar] [CrossRef]
- Piotrowicz, K.; Gasowski, J.; Michel, J.P.; Veronese, N. Post-COVID-19 acute sarcopenia: Physiopathology and management. Aging Clin. Exp. Res. 2021, 33, 2887–2898. [Google Scholar] [CrossRef] [PubMed]
- Nardacci, R.; Colavita, F.; Castilletti, C.; Lapa, D.; Matusali, G.; Meschi, S.; Del Nonno, F.; Colombo, D.; Capobianchi, M.R.; Zumla, A.; et al. Evidences for lipid involvement in SARS-CoV-2 cytopathogenesis. Cell Death Dis. 2021, 12, 263. [Google Scholar] [CrossRef]
- Cortese, M.; Lee, J.Y.; Cerikan, B.; Neufeldt, C.J.; Oorschot, V.M.J.; Kohrer, S.; Hennies, J.; Schieber, N.L.; Ronchi, P.; Mizzon, G.; et al. Integrative Imaging Reveals SARS-CoV-2-Induced Reshaping of Subcellular Morphologies. Cell Host Microbe 2020, 28, 853–866.e5. [Google Scholar] [CrossRef] [PubMed]
- Faizan, M.I.; Chaudhuri, R.; Sagar, S.; Albogami, S.; Chaudhary, N.; Azmi, I.; Akhtar, A.; Ali, S.M.; Kumar, R.; Iqbal, J.; et al. NSP4 and ORF9b of SARS-CoV-2 Induce Pro-Inflammatory Mitochondrial DNA Release in Inner Membrane-Derived Vesicles. Cells 2022, 11, 2969. [Google Scholar] [CrossRef] [PubMed]
- Singh, K.; Chen, Y.C.; Hassanzadeh, S.; Han, K.; Judy, J.T.; Seifuddin, F.; Tunc, I.; Sack, M.N.; Pirooznia, M. Network Analysis and Transcriptome Profiling Identify Autophagic and Mitochondrial Dysfunctions in SARS-CoV-2 Infection. Front. Genet. 2021, 12, 599261. [Google Scholar] [CrossRef]
- Mando, C.; Savasi, V.M.; Anelli, G.M.; Corti, S.; Serati, A.; Lisso, F.; Tasca, C.; Novielli, C.; Cetin, I. Mitochondrial and Oxidative Unbalance in Placentas from Mothers with SARS-CoV-2 Infection. Antioxidants 2021, 10, 1517. [Google Scholar] [CrossRef]
- Hartsell, E.M.; Gillespie, M.N.; Langley, R.J. Does acute and persistent metabolic dysregulation in COVID-19 point to novel biomarkers and future therapeutic strategies? Eur. Respir. J. 2022, 59, 2102417. [Google Scholar] [CrossRef] [PubMed]
- Pliss, A.; Kuzmin, A.N.; Prasad, P.N.; Mahajan, S.D. Mitochondrial Dysfunction: A Prelude to Neuropathogenesis of SARS-CoV-2. ACS Chem. Neurosci. 2022, 13, 308–312. [Google Scholar] [CrossRef]
- Zhu, L.; She, Z.G.; Cheng, X.; Qin, J.J.; Zhang, X.J.; Cai, J.; Lei, F.; Wang, H.; Xie, J.; Wang, W.; et al. Association of Blood Glucose Control and Outcomes in Patients with COVID-19 and Pre-existing Type 2 Diabetes. Cell Metab. 2020, 31, 1068–1077 e1063. [Google Scholar] [CrossRef]
- Garreta, E.; Prado, P.; Stanifer, M.L.; Monteil, V.; Marco, A.; Ullate-Agote, A.; Moya-Rull, D.; Vilas-Zornoza, A.; Tarantino, C.; Romero, J.P.; et al. A diabetic milieu increases ACE2 expression and cellular susceptibility to SARS-CoV-2 infections in human kidney organoids and patient cells. Cell Metab. 2022, 34, 857–873 e859. [Google Scholar] [CrossRef]
- Palaiodimos, L.; Kokkinidis, D.G.; Li, W.; Karamanis, D.; Ognibene, J.; Arora, S.; Southern, W.N.; Mantzoros, C.S. Severe obesity, increasing age and male sex are independently associated with worse in-hospital outcomes, and higher in-hospital mortality, in a cohort of patients with COVID-19 in the Bronx, New York. Metabolism 2020, 108, 154262. [Google Scholar] [CrossRef]
- Holder, K.; Reddy, P.H. The COVID-19 Effect on the Immune System and Mitochondrial Dynamics in Diabetes, Obesity, and Dementia. Neuroscientist 2021, 27, 331–339. [Google Scholar] [CrossRef]
- Mavrogiannaki, A.N.; Migdalis, I.N. Nonalcoholic Fatty liver disease, diabetes mellitus and cardiovascular disease: Newer data. Int. J. Endocrinol. 2013, 2013, 450639. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Costa, F.F.; Rosario, W.R.; Ribeiro Farias, A.C.; de Souza, R.G.; Duarte Gondim, R.S.; Barroso, W.A. Metabolic syndrome and COVID-19: An update on the associated comorbidities and proposed therapies. Diabetes Metab. Syndr. 2020, 14, 809–814. [Google Scholar] [CrossRef] [PubMed]
- Moreno Fernandez-Ayala, D.J.; Navas, P.; Lopez-Lluch, G. Age-related mitochondrial dysfunction as a key factor in COVID-19 disease. Exp. Gerontol. 2020, 142, 111147. [Google Scholar] [CrossRef]
- Chang, X.; Ismail, N.I.; Rahman, A.; Xu, D.; Chan, R.W.Y.; Ong, S.G.; Ong, S.B. Long COVID-19 and the Heart: Is Cardiac Mitochondria the Missing Link? Antioxid. Redox Signal. 2023, 38, 599–618. [Google Scholar] [CrossRef]
- Chen, X.; Cao, R.; Zhong, W. Host Calcium Channels and Pumps in Viral Infections. Cells 2019, 9, 94. [Google Scholar] [CrossRef] [Green Version]
- Han, Z.; Madara, J.J.; Herbert, A.; Prugar, L.I.; Ruthel, G.; Lu, J.; Liu, Y.; Liu, W.; Liu, X.; Wrobel, J.E.; et al. Calcium Regulation of Hemorrhagic Fever Virus Budding: Mechanistic Implications for Host-Oriented Therapeutic Intervention. PLoS Pathog. 2015, 11, e1005220. [Google Scholar] [CrossRef] [Green Version]
- Qu, Y.; Sun, Y.; Yang, Z.; Ding, C. Calcium Ions Signaling: Targets for Attack and Utilization by Viruses. Front. Microbiol. 2022, 13, 889374. [Google Scholar] [CrossRef]
- Saurav, S.; Tanwar, J.; Ahuja, K.; Motiani, R.K. Dysregulation of host cell calcium signaling during viral infections: Emerging paradigm with high clinical relevance. Mol. Asp. Med. 2021, 81, 101004. [Google Scholar]
- Lai, A.L.; Freed, J.H. SARS-CoV-2 Fusion Peptide has a Greater Membrane Perturbating Effect than SARS-CoV with Highly Specific Dependence on Ca(2). J. Mol. Biol. 2021, 433, 166946. [Google Scholar] [CrossRef]
- Straus, M.R.; Tang, T.; Lai, A.L.; Flegel, A.; Bidon, M.; Freed, J.H.; Daniel, S.; Whittaker, G.R. Ca(2+) Ions Promote Fusion of Middle East Respiratory Syndrome Coronavirus with Host Cells and Increase Infectivity. J. Virol. 2020, 94, e00426-20. [Google Scholar] [CrossRef]
- Kern, D.M.; Sorum, B.; Mali, S.S.; Hoel, C.M.; Sridharan, S.; Remis, J.P.; Toso, D.B.; Kotecha, A.; Bautista, D.M.; Brohawn, S.G. Cryo-EM structure of SARS-CoV-2 ORF3a in lipid nanodiscs. Nat. Struct. Mol. Biol. 2021, 28, 573–582. [Google Scholar] [CrossRef] [PubMed]
- Davies, J.P.; Almasy, K.M.; McDonald, E.F.; Plate, L. Comparative Multiplexed Interactomics of SARS-CoV-2 and Homologous Coronavirus Nonstructural Proteins Identifies Unique and Shared Host-Cell Dependencies. ACS Infect. Dis. 2020, 6, 3174–3189. [Google Scholar] [CrossRef] [PubMed]
- Ren, Y.; Shu, T.; Wu, D.; Mu, J.; Wang, C.; Huang, M.; Han, Y.; Zhang, X.Y.; Zhou, W.; Qiu, Y.; et al. The ORF3a protein of SARS-CoV-2 induces apoptosis in cells. Cell. Mol. Immunol. 2020, 17, 881–883. [Google Scholar] [CrossRef]
- Bojkova, D.; Klann, K.; Koch, B.; Widera, M.; Krause, D.; Ciesek, S.; Cinatl, J.; Munch, C. Proteomics of SARS-CoV-2-infected host cells reveals therapy targets. Nature 2020, 583, 469–472. [Google Scholar] [CrossRef]
- Nunn, A.V.W.; Guy, G.W.; Brysch, W.; Bell, J.D. Understanding Long COVID.; Mitochondrial Health and Adaptation-Old Pathways, New Problems. Biomedicines 2022, 10, 3113. [Google Scholar] [CrossRef]
- Qu, C.; Zhang, S.; Wang, W.; Li, M.; Wang, Y.; van der Heijde-Mulder, M.; Shokrollahi, E.; Hakim, M.S.; Raat, N.J.H.; Peppelenbosch, M.P.; et al. Mitochondrial electron transport chain complex III sustains hepatitis E virus replication and represents an antiviral target. FASEB J. 2019, 33, 1008–1019. [Google Scholar] [CrossRef] [PubMed]
- Singh, D.; Agusti, A.; Anzueto, A.; Barnes, P.J.; Bourbeau, J.; Celli, B.R.; Criner, G.J.; Frith, P.; Halpin, D.M.G.; Han, M.; et al. Global Strategy for the Diagnosis, Management, and Prevention of Chronic Obstructive Lung Disease: The GOLD science committee report 2019. Eur. Respir. J. 2019, 53, 1900164. [Google Scholar] [CrossRef]
- Shin, C.S.; Meng, S.; Garbis, S.D.; Moradian, A.; Taylor, R.W.; Sweredoski, M.J.; Lomenick, B.; Chan, D.C. LONP1 and mtHSP70 cooperate to promote mitochondrial protein folding. Nat. Commun. 2021, 12, 265. [Google Scholar] [CrossRef]
- Codo, A.C.; Davanzo, G.G.; Monteiro, L.B.; de Souza, G.F.; Muraro, S.P.; Virgilio-da-Silva, J.V.; Prodonoff, J.S.; Carregari, V.C.; de Biagi Junior, C.A.O.; Crunfli, F.; et al. Elevated Glucose Levels Favor SARS-CoV-2 Infection and Monocyte Response through a HIF-1alpha/Glycolysis-Dependent Axis. Cell Metab. 2020, 32, 498–499. [Google Scholar] [CrossRef]
- Soria-Castro, E.; Soto, M.E.; Guarner-Lans, V.; Rojas, G.; Perezpena-Diazconti, M.; Criales-Vera, S.A.; Manzano Pech, L.; Perez-Torres, I. The kidnapping of mitochondrial function associated with the SARS-CoV-2 infection. Histol. Histopathol. 2021, 36, 947–965. [Google Scholar]
- Starkov, A.A.; Fiskum, G. Regulation of brain mitochondrial H2O2 production by membrane potential and NAD(P)H redox state. J. Neurochem. 2003, 86, 1101–1107. [Google Scholar] [CrossRef] [PubMed]
- Green, D.R.; Reed, J.C. Mitochondria and apoptosis. Science 1998, 281, 1309–1312. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Wang, L.; Yan, S.; Yang, F.; Xiang, L.; Zhu, J.; Shen, B.; Gong, Z. Clinical characteristics of 25 death cases with COVID-19: A retrospective review of medical records in a single medical center, Wuhan, China. Int. J. Infect. Dis. 2020, 94, 128–132. [Google Scholar] [CrossRef] [PubMed]
- Lei, Y.; Zhang, J.; Schiavon, C.R.; He, M.; Chen, L.; Shen, H.; Zhang, Y.; Yin, Q.; Cho, Y.; Andrade, L.; et al. SARS-CoV-2 Spike Protein Impairs Endothelial Function via Downregulation of ACE 2. Circ. Res. 2021, 128, 1323–1326. [Google Scholar] [CrossRef]
- Shang, C.; Liu, Z.; Zhu, Y.; Lu, J.; Ge, C.; Zhang, C.; Li, N.; Jin, N.; Li, Y.; Tian, M.; et al. SARS-CoV-2 Causes Mitochondrial Dysfunction and Mitophagy Impairment. Front. Microbiol. 2021, 12, 780768. [Google Scholar] [CrossRef] [PubMed]
- Andrade Silva, M.; da Silva, A.; do Amaral, M.A.; Fragas, M.G.; Camara, N.O.S. Metabolic Alterations in SARS-CoV-2 Infection and Its Implication in Kidney Dysfunction. Front. Physiol. 2021, 12, 624698. [Google Scholar] [CrossRef] [PubMed]
- Moolamalla, S.T.R.; Balasubramanian, R.; Chauhan, R.; Priyakumar, U.D.; Vinod, P.K. Host metabolic reprogramming in response to SARS-CoV-2 infection: A systems biology approach. Microb. Pathog. 2021, 158, 105114. [Google Scholar] [CrossRef]
- Zhang, W.; Wang, G.; Xu, Z.G.; Tu, H.; Hu, F.; Dai, J.; Chang, Y.; Chen, Y.; Lu, Y.; Zeng, H.; et al. Lactate Is a Natural Suppressor of RLR Signaling by Targeting MAVS. Cell 2019, 178, 176–189.e15. [Google Scholar] [CrossRef]
- Giacomello, M.; Pyakurel, A.; Glytsou, C.; Scorrano, L. The cell biology of mitochondrial membrane dynamics. Nat. Rev. Mol. Cell Biol. 2020, 21, 204–224. [Google Scholar]
- Terman, A.; Kurz, T.; Navratil, M.; Arriaga, E.A.; Brunk, U.T. Mitochondrial turnover and aging of long-lived postmitotic cells: The mitochondrial-lysosomal axis theory of aging. Antioxid. Redox Signal. 2010, 12, 503–535. [Google Scholar] [CrossRef] [Green Version]
- De, R.; Sarkar, S.; Mazumder, S.; Debsharma, S.; Siddiqui, A.A.; Saha, S.J.; Banerjee, C.; Nag, S.; Saha, D.; Pramanik, S.; et al. Macrophage migration inhibitory factor regulates mitochondrial dynamics and cell growth of human cancer cell lines through CD74-NF-kappaB signaling. J. Biol. Chem. 2018, 293, 19740–19760. [Google Scholar] [CrossRef] [Green Version]
- Bach, D.; Pich, S.; Soriano, F.X.; Vega, N.; Baumgartner, B.; Oriola, J.; Daugaard, J.R.; Lloberas, J.; Camps, M.; Zierath, J.R.; et al. Mitofusin-2 determines mitochondrial network architecture and mitochondrial metabolism. A novel regulatory mechanism altered in obesity. J. Biol. Chem. 2003, 278, 17190–17197. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Smith, M.E.; Tippetts, T.S.; Brassfield, E.S.; Tucker, B.J.; Ockey, A.; Swensen, A.C.; Anthonymuthu, T.S.; Washburn, T.D.; Kane, D.A.; Prince, J.T.; et al. Mitochondrial fission mediates ceramide-induced metabolic disruption in skeletal muscle. Biochem. J. 2013, 456, 427–439. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Morita, M.; Prudent, J.; Basu, K.; Goyon, V.; Katsumura, S.; Hulea, L.; Pearl, D.; Siddiqui, N.; Strack, S.; McGuirk, S.; et al. mTOR Controls Mitochondrial Dynamics and Cell Survival via MTFP1. Mol. Cell 2017, 67, 922–935.e5. [Google Scholar] [PubMed] [Green Version]
- Morita, M.; Gravel, S.P.; Hulea, L.; Larsson, O.; Pollak, M.; St-Pierre, J.; Topisirovic, I. mTOR coordinates protein synthesis, mitochondrial activity and proliferation. Cell Cycle 2015, 14, 473–480. [Google Scholar] [CrossRef] [Green Version]
- Zemirli, N.; Pourcelot, M.; Ambroise, G.; Hatchi, E.; Vazquez, A.; Arnoult, D. Mitochondrial hyperfusion promotes NF-kappaB activation via the mitochondrial E3 ligase MULAN. FEBS J. 2014, 281, 3095–3112. [Google Scholar] [CrossRef] [Green Version]
- Galluzzi, L.; Vitale, I.; Abrams, J.M.; Alnemri, E.S.; Baehrecke, E.H.; Blagosklonny, M.V.; Dawson, T.M.; Dawson, V.L.; El-Deiry, W.S.; Fulda, S.; et al. Molecular definitions of cell death subroutines: Recommendations of the Nomenclature Committee on Cell Death 2012. Cell Death Differ. 2012, 19, 107–120. [Google Scholar]
- Kroemer, G.; Galluzzi, L.; Brenner, C. Mitochondrial membrane permeabilization in cell death. Physiol. Rev. 2007, 87, 99–163. [Google Scholar]
- Madeddu, E.; Maniga, B.; Zaffanello, M.; Fanos, V.; Marcialis, A. The SARS-CoV2 and mitochondria: The impact on cell fate. Acta Biomed. 2022, 93, e2022199. [Google Scholar] [PubMed]
- Cao, G.; Xing, J.; Xiao, X.; Liou, A.K.; Gao, Y.; Yin, X.M.; Clark, R.S.; Graham, S.H.; Chen, J. Critical role of calpain I in mitochondrial release of apoptosis-inducing factor in ischemic neuronal injury. J. Neurosci. 2007, 27, 9278–9293. [Google Scholar] [CrossRef] [Green Version]
- Wang, Y.; Kim, N.S.; Haince, J.F.; Kang, H.C.; David, K.K.; Andrabi, S.A.; Poirier, G.G.; Dawson, V.L.; Dawson, T.M. Poly(ADP-ribose) (PAR) binding to apoptosis-inducing factor is critical for PAR polymerase-1-dependent cell death (parthanatos). Sci. Signal. 2011, 4, ra20. [Google Scholar] [CrossRef] [Green Version]
- Wang, Y.; Kim, N.S.; Li, X.; Greer, P.A.; Koehler, R.C.; Dawson, V.L.; Dawson, T.M. Calpain activation is not required for AIF translocation in PARP-1-dependent cell death (parthanatos). J. Neurochem. 2009, 110, 687–696. [Google Scholar] [CrossRef]
- Kesavardhana, S.; Kanneganti, T.D. Mechanisms governing inflammasome activation, assembly and pyroptosis induction. Int. Immunol. 2017, 29, 201–210. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vandenabeele, P.; Galluzzi, L.; Vanden Berghe, T.; Kroemer, G. Molecular mechanisms of necroptosis: An ordered cellular explosion. Nat. Rev. Mol. Cell Biol. 2010, 11, 700–714. [Google Scholar] [PubMed]
- Redondo, N.; Zaldivar-Lopez, S.; Garrido, J.J.; Montoya, M. SARS-CoV-2 Accessory Proteins in Viral Pathogenesis: Knowns and Unknowns. Front. Immunol. 2021, 12, 708264. [Google Scholar] [CrossRef] [PubMed]
- Valdes-Aguayo, J.J.; Garza-Veloz, I.; Badillo-Almaraz, J.I.; Bernal-Silva, S.; Martinez-Vazquez, M.C.; Juarez-Alcala, V.; Vargas-Rodriguez, J.R.; Gaeta-Velasco, M.L.; Gonzalez-Fuentes, C.; Avila-Carrasco, L.; et al. Mitochondria and Mitochondrial DNA: Key Elements in the Pathogenesis and Exacerbation of the Inflammatory State Caused by COVID-19. Medicina 2021, 57, 928. [Google Scholar] [CrossRef] [PubMed]
- Dirican, E.; Savrun, S.T.; Aydin, I.E.; Gulbay, G.; Karaman, U. Analysis of mitochondrial DNA cytochrome-b (CYB) and ATPase-6 gene mutations in COVID-19 patients. J. Med. Virol. 2022, 94, 3138–3146. [Google Scholar] [CrossRef] [PubMed]
- Wu, Y.; Wang, X.H.; Li, X.H.; Song, L.Y.; Yu, S.L.; Fang, Z.C.; Liu, Y.Q.; Yuan, L.Y.; Peng, C.Y.; Zhang, S.Y.; et al. Common mtDNA variations at C5178a and A249d/T6392C/G10310A decrease the risk of severe COVID-19 in a Han Chinese population from Central China. Mil. Med. Res. 2021, 8, 57. [Google Scholar] [CrossRef]
- Riley, J.S.; Tait, S.W. Mitochondrial DNA in inflammation and immunity. EMBO Rep. 2020, 21, e49799. [Google Scholar] [CrossRef]
- Scozzi, D.; Cano, M.; Ma, L.; Zhou, D.; Zhu, J.H.; O’Halloran, J.A.; Goss, C.; Rauseo, A.M.; Liu, Z.; Sahu, S.K.; et al. Circulating mitochondrial DNA is an early indicator of severe illness and mortality from COVID-19. JCI Insight 2021, 6, 143299. [Google Scholar] [CrossRef]
- Andargie, T.E.; Tsuji, N.; Seifuddin, F.; Jang, M.K.; Yuen, P.S.; Kong, H.; Tunc, I.; Singh, K.; Charya, A.; Wilkins, K.; et al. Cell-free DNA maps COVID-19 tissue injury and risk of death and can cause tissue injury. JCI Insight 2021, 6, 147610. [Google Scholar] [CrossRef] [PubMed]
- Xie, J.H.; Li, Y.Y.; Jin, J. The essential functions of mitochondrial dynamics in immune cells. Cell. Mol. Immunol. 2020, 17, 712–721. [Google Scholar] [CrossRef] [PubMed]
- Dickson, K.B.; Zhou, J. Role of reactive oxygen species and iron in host defense against infection. Front. Biosci. Landmark Ed. 2020, 25, 1600–1616. [Google Scholar]
- Chen, T.H.; Koh, K.Y.; Lin, K.M.; Chou, C.K. Mitochondrial Dysfunction as an Underlying Cause of Skeletal Muscle Disorders. Int. J. Mol. Sci. 2022, 23, 12926. [Google Scholar] [CrossRef]
- Miller, B.; Silverstein, A.; Flores, M.; Cao, K.; Kumagai, H.; Mehta, H.H.; Yen, K.; Kim, S.J.; Cohen, P. Host mitochondrial transcriptome response to SARS-CoV-2 in multiple cell models and clinical samples. Sci. Rep. 2021, 11, 3. [Google Scholar] [CrossRef]
- Liu, Q.; Zhang, D.; Hu, D.; Zhou, X.; Zhou, Y. The role of mitochondria in NLRP3 inflammasome activation. Mol. Immunol. 2018, 103, 115–124. [Google Scholar] [PubMed]
- Burtscher, J.; Cappellano, G.; Omori, A.; Koshiba, T.; Millet, G.P. Mitochondria: In the Cross Fire of SARS-CoV-2 and Immunity. iScience 2020, 23, 101631. [Google Scholar] [CrossRef]
- He, Y.; Hara, H.; Nunez, G. Mechanism and Regulation of NLRP3 Inflammasome Activation. Trends Biochem. Sci. 2016, 41, 1012–1021. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhou, R.; Yazdi, A.S.; Menu, P.; Tschopp, J. A role for mitochondria in NLRP3 inflammasome activation. Nature 2011, 469, 221–225. [Google Scholar] [CrossRef]
- Volt, H.; Garcia, J.A.; Doerrier, C.; Diaz-Casado, M.E.; Guerra-Librero, A.; Lopez, L.C.; Escames, G.; Tresguerres, J.A.; Acuna-Castroviejo, D. Same molecule but different expression: Aging and sepsis trigger NLRP3 inflammasome activation, a target of melatonin. J. Pineal Res. 2016, 60, 193–205. [Google Scholar] [CrossRef]
- Zhou, Z.; Zhang, M.; Wang, Y.; Zheng, F.; Huang, Y.; Huang, K.; Yu, Q.; Cai, C.; Chen, D.; Tian, Y.; et al. Clinical characteristics of older and younger patients infected with SARS-CoV-2. Aging 2020, 12, 11296–11305. [Google Scholar] [CrossRef]
- Pan, P.; Shen, M.; Yu, Z.; Ge, W.; Chen, K.; Tian, M.; Xiao, F.; Wang, Z.; Wang, J.; Jia, Y.; et al. SARS-CoV-2 N protein promotes NLRP3 inflammasome activation to induce hyperinflammation. Nat. Commun. 2021, 12, 4664. [Google Scholar] [CrossRef] [PubMed]
- Shi, C.S.; Nabar, N.R.; Huang, N.N.; Kehrl, J.H. SARS-Coronavirus Open Reading Frame-8b triggers intracellular stress pathways and activates NLRP3 inflammasomes. Cell Death Discov. 2019, 5, 101. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nieto-Torres, J.L.; DeDiego, M.L.; Verdia-Baguena, C.; Jimenez-Guardeno, J.M.; Regla-Nava, J.A.; Fernandez-Delgado, R.; Castano-Rodriguez, C.; Alcaraz, A.; Torres, J.; Aguilella, V.M.; et al. Severe acute respiratory syndrome coronavirus envelope protein ion channel activity promotes virus fitness and pathogenesis. PLoS Pathog. 2014, 10, e1004077. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fung, S.Y.; Yuen, K.S.; Ye, Z.W.; Chan, C.P.; Jin, D.Y. A tug-of-war between severe acute respiratory syndrome coronavirus 2 and host antiviral defence: Lessons from other pathogenic viruses. Emerg. Microbes Infect. 2020, 9, 558–570. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Astuti, I.; Ysrafil. Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2): An overview of viral structure and host response. Diabetes Metab. Syndr. 2020, 14, 407–412. [Google Scholar] [CrossRef]
- Albornoz, E.A.; Amarilla, A.A.; Modhiran, N.; Parker, S.; Li, X.X.; Wijesundara, D.K.; Aguado, J.; Zamora, A.P.; McMillan, C.L.D.; Liang, B.; et al. SARS-CoV-2 drives NLRP3 inflammasome activation in human microglia through spike protein. Mol. Psychiatry 2022. [Google Scholar] [CrossRef]
- Su, C.M.; Wang, L.; Yoo, D. Activation of NF-kappaB and induction of proinflammatory cytokine expressions mediated by ORF7a protein of SARS-CoV-2. Sci. Rep. 2021, 11, 13464. [Google Scholar] [CrossRef]
- Chen, I.Y.; Moriyama, M.; Chang, M.F.; Ichinohe, T. Severe Acute Respiratory Syndrome Coronavirus Viroporin 3a Activates the NLRP3 Inflammasome. Front. Microbiol. 2019, 10, 50. [Google Scholar] [CrossRef] [Green Version]
- Kasuga, Y.; Zhu, B.; Jang, K.J.; Yoo, J.S. Innate immune sensing of coronavirus and viral evasion strategies. Exp. Mol. Med. 2021, 53, 723–736. [Google Scholar] [CrossRef]
- Ebermeyer, T.; Cognasse, F.; Berthelot, P.; Mismetti, P.; Garraud, O.; Hamzeh-Cognasse, H. Platelet Innate Immune Receptors and TLRs: A Double-Edged Sword. Int. J. Mol. Sci. 2021, 22, 7894. [Google Scholar] [CrossRef] [PubMed]
- Costa, T.J.; Potje, S.R.; Fraga-Silva, T.F.C.; da Silva-Neto, J.A.; Barros, P.R.; Rodrigues, D.; Machado, M.R.; Martins, R.B.; Santos-Eichler, R.A.; Benatti, M.N.; et al. Mitochondrial DNA and TLR9 activation contribute to SARS-CoV-2-induced endothelial cell damage. Vascul. Pharmacol. 2022, 142, 106946. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Chen, M.; Cao, H.; Zhu, Y.; Zheng, J.; Zhou, H. Extraordinary GU-rich single-strand RNA identified from SARS coronavirus contributes an excessive innate immune response. Microbes Infect. 2013, 15, 88–95. [Google Scholar] [CrossRef] [PubMed]
- Salvi, V.; Nguyen, H.O.; Sozio, F.; Schioppa, T.; Gaudenzi, C.; Laffranchi, M.; Scapini, P.; Passari, M.; Barbazza, I.; Tiberio, L.; et al. SARS-CoV-2-associated ssRNAs activate inflammation and immunity via TLR7/8. JCI Insight 2021, 6, 150542. [Google Scholar] [CrossRef] [PubMed]
- Bortolotti, D.; Gentili, V.; Rizzo, S.; Schiuma, G.; Beltrami, S.; Strazzabosco, G.; Fernandez, M.; Caccuri, F.; Caruso, A.; Rizzo, R. TLR3 and TLR7 RNA Sensor Activation during SARS-CoV-2 Infection. Microorganisms 2021, 9, 1820. [Google Scholar] [CrossRef]
- Cervantes-Barragan, L.; Zust, R.; Weber, F.; Spiegel, M.; Lang, K.S.; Akira, S.; Thiel, V.; Ludewig, B. Control of coronavirus infection through plasmacytoid dendritic-cell-derived type I interferon. Blood 2007, 109, 1131–1137. [Google Scholar] [CrossRef] [Green Version]
- Zhang, Q.; Raoof, M.; Chen, Y.; Sumi, Y.; Sursal, T.; Junger, W.; Brohi, K.; Itagaki, K.; Hauser, C.J. Circulating mitochondrial DAMPs cause inflammatory responses to injury. Nature 2010, 464, 104–107. [Google Scholar] [CrossRef] [Green Version]
- Digard, P.; Lee, H.M.; Sharp, C.; Grey, F.; Gaunt, E. Intra-genome variability in the dinucleotide composition of SARS-CoV-2. Virus Evol. 2020, 6, veaa057. [Google Scholar] [CrossRef]
- Smith, J.A. STING, the Endoplasmic Reticulum, and Mitochondria: Is Three a Crowd or a Conversation? Front. Immunol. 2020, 11, 611347. [Google Scholar] [CrossRef]
- Zhong, B.; Zhang, L.; Lei, C.; Li, Y.; Mao, A.P.; Yang, Y.; Wang, Y.Y.; Zhang, X.L.; Shu, H.B. The ubiquitin ligase RNF5 regulates antiviral responses by mediating degradation of the adaptor protein MITA. Immunity 2009, 30, 397–407. [Google Scholar] [CrossRef] [Green Version]
- Wei, X.; Du, M.; Xie, J.; Luo, T.; Zhou, Y.; Zhang, K.; Li, J.; Chen, D.; Xu, P.; Jia, M.; et al. Mutations in TOMM70 lead to multi-OXPHOS deficiencies and cause severe anemia, lactic acidosis, and developmental delay. J. Hum. Genet. 2020, 65, 231–240. [Google Scholar] [CrossRef] [Green Version]
- Liu, X.Y.; Wei, B.; Shi, H.X.; Shan, Y.F.; Wang, C. Tom70 mediates activation of interferon regulatory factor 3 on mitochondria. Cell Res. 2010, 20, 994–1011. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Elesela, S.; Lukacs, N.W. Role of Mitochondria in Viral Infections. Life 2021, 11, 232. [Google Scholar] [CrossRef] [PubMed]
- Fu, Y.Z.; Wang, S.Y.; Zheng, Z.Q.; Yi, H.; Li, W.W.; Xu, Z.S.; Wang, Y.Y. SARS-CoV-2 membrane glycoprotein M antagonizes the MAVS-mediated innate antiviral response. Cell. Mol. Immunol. 2021, 18, 613–620. [Google Scholar] [CrossRef] [PubMed]
- Han, L.; Zhuang, M.W.; Deng, J.; Zheng, Y.; Zhang, J.; Nan, M.L.; Zhang, X.J.; Gao, C.; Wang, P.H. SARS-CoV-2 ORF9b antagonizes type I and III interferons by targeting multiple components of the RIG-I/MDA-5-MAVS, TLR3-TRIF, and cGAS-STING signaling pathways. J. Med. Virol. 2021, 93, 5376–5389. [Google Scholar] [CrossRef]
- Yuen, C.K.; Lam, J.Y.; Wong, W.M.; Mak, L.F.; Wang, X.; Chu, H.; Cai, J.P.; Jin, D.Y.; To, K.K.; Chan, J.F.; et al. SARS-CoV-2 nsp13, nsp14, nsp15 and orf6 function as potent interferon antagonists. Emerg. Microbes Infect. 2020, 9, 1418–1428. [Google Scholar] [CrossRef]
- Minakshi, R.; Padhan, K.; Rani, M.; Khan, N.; Ahmad, F.; Jameel, S. The SARS Coronavirus 3a protein causes endoplasmic reticulum stress and induces ligand-independent downregulation of the type 1 interferon receptor. PLoS ONE 2009, 4, e8342. [Google Scholar] [CrossRef] [Green Version]
- Kokic, G.; Hillen, H.S.; Tegunov, D.; Dienemann, C.; Seitz, F.; Schmitzova, J.; Farnung, L.; Siewert, A.; Hobartner, C.; Cramer, P. Mechanism of SARS-CoV-2 polymerase stalling by remdesivir. Nat. Commun. 2021, 12, 279. [Google Scholar] [CrossRef]
- Nevalainen, O.P.O.; Horstia, S.; Laakkonen, S.; Rutanen, J.; Mustonen, J.M.J.; Kalliala, I.E.J.; Ansakorpi, H.; Kreivi, H.R.; Kuutti, P.; Paajanen, J.; et al. Effect of remdesivir post hospitalization for COVID-19 infection from the randomized SOLIDARITY Finland trial. Nat. Commun. 2022, 13, 6152. [Google Scholar] [CrossRef]
- Boglione, L.; Meli, G.; Poletti, F.; Rostagno, R.; Moglia, R.; Cantone, M.; Esposito, M.; Scianguetta, C.; Domenicale, B.; Di Pasquale, F.; et al. Risk factors and incidence of long-COVID syndrome in hospitalized patients: Does remdesivir have a protective effect? QJM Int. J. Med. 2022, 114, 865–871. [Google Scholar] [CrossRef]
- Geiler, J.; Michaelis, M.; Naczk, P.; Leutz, A.; Langer, K.; Doerr, H.W.; Cinatl, J., Jr. N-acetyl-L-cysteine (NAC) inhibits virus replication and expression of pro-inflammatory molecules in A549 cells infected with highly pathogenic H5N1 influenza A virus. Biochem. Pharmacol. 2010, 79, 413–420. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cai, J.; Chen, Y.; Seth, S.; Furukawa, S.; Compans, R.W.; Jones, D.P. Inhibition of influenza infection by glutathione. Free Radic. Biol. Med. 2003, 34, 928–936. [Google Scholar] [CrossRef] [PubMed]
- Shi, X.; Shi, Z.; Huang, H.; Zhu, H.; Zhou, P.; Zhu, H.; Ju, D. Ability of recombinant human catalase to suppress inflammation of the murine lung induced by influenza A. Inflammation 2014, 37, 809–817. [Google Scholar] [CrossRef] [PubMed]
- Soto, M.E.; Guarner-Lans, V.; Soria-Castro, E.; Manzano Pech, L.; Perez-Torres, I. Is Antioxidant Therapy a Useful Complementary Measure for Covid-19 Treatment? An Algorithm for Its Application. Medicina 2020, 56, 386. [Google Scholar] [CrossRef] [PubMed]
- Melov, S.; Schneider, J.A.; Day, B.J.; Hinerfeld, D.; Coskun, P.; Mirra, S.S.; Crapo, J.D.; Wallace, D.C. A novel neurological phenotype in mice lacking mitochondrial manganese superoxide dismutase. Nat. Genet. 1998, 18, 159–163. [Google Scholar] [CrossRef]
- Melov, S.; Doctrow, S.R.; Schneider, J.A.; Haberson, J.; Patel, M.; Coskun, P.E.; Huffman, K.; Wallace, D.C.; Malfroy, B. Lifespan extension and rescue of spongiform encephalopathy in superoxide dismutase 2 nullizygous mice treated with superoxide dismutase-catalase mimetics. J. Neurosci. 2001, 21, 8348–8353. [Google Scholar] [CrossRef] [Green Version]
- De, R.; Mazumder, S.; Sarkar, S.; Debsharma, S.; Siddiqui, A.A.; Saha, S.J.; Banerjee, C.; Nag, S.; Saha, D.; Bandyopadhyay, U. Acute mental stress induces mitochondrial bioenergetic crisis and hyper-fission along with aberrant mitophagy in the gut mucosa in rodent model of stress-related mucosal disease. Free Radic. Biol. Med. 2017, 113, 424–438. [Google Scholar] [CrossRef]
- Petcherski, A.; Sharma, M.; Satta, S.; Daskou, M.; Vasilopoulos, H.; Hugo, C.; Ritou, E.; Dillon, B.J.; Fung, E.; Garcia, G.; et al. Mitoquinone mesylate targets SARS-CoV-2 infection in preclinical models. bioRxiv 2022. [Google Scholar] [CrossRef]
- Dittmar, M.; Lee, J.S.; Whig, K.; Segrist, E.; Li, M.; Kamalia, B.; Castellana, L.; Ayyanathan, K.; Cardenas-Diaz, F.L.; Morrisey, E.E.; et al. Drug repurposing screens reveal cell-type-specific entry pathways and FDA-approved drugs active against SARS-Cov-2. Cell Rep. 2021, 35, 108959. [Google Scholar] [CrossRef]
- Toth, E.; Maleth, J.; Zavogyan, N.; Fanczal, J.; Grassalkovich, A.; Erdos, R.; Pallagi, P.; Horvath, G.; Tretter, L.; Balint, E.R.; et al. Novel mitochondrial transition pore inhibitor N-methyl-4-isoleucine cyclosporin is a new therapeutic option in acute pancreatitis. J. Physiol. 2019, 597, 5879–5898. [Google Scholar] [CrossRef] [Green Version]
- Shakoory, B.; Carcillo, J.A.; Chatham, W.W.; Amdur, R.L.; Zhao, H.; Dinarello, C.A.; Cron, R.Q.; Opal, S.M. Interleukin-1 Receptor Blockade Is Associated with Reduced Mortality in Sepsis Patients With Features of Macrophage Activation Syndrome: Reanalysis of a Prior Phase III Trial. Crit. Care Med. 2016, 44, 275–281. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Norelli, M.; Camisa, B.; Barbiera, G.; Falcone, L.; Purevdorj, A.; Genua, M.; Sanvito, F.; Ponzoni, M.; Doglioni, C.; Cristofori, P.; et al. Monocyte-derived IL-1 and IL-6 are differentially required for cytokine-release syndrome and neurotoxicity due to CAR T cells. Nat. Med. 2018, 24, 739–748. [Google Scholar] [CrossRef] [PubMed]
- Le, R.Q.; Li, L.; Yuan, W.; Shord, S.S.; Nie, L.; Habtemariam, B.A.; Przepiorka, D.; Farrell, A.T.; Pazdur, R. FDA Approval Summary: Tocilizumab for Treatment of Chimeric Antigen Receptor T Cell-Induced Severe or Life-Threatening Cytokine Release Syndrome. Oncologist 2018, 23, 943–947. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kotch, C.; Barrett, D.; Teachey, D.T. Tocilizumab for the treatment of chimeric antigen receptor T cell-induced cytokine release syndrome. Expert Rev. Clin. Immunol. 2019, 15, 813–822. [Google Scholar] [CrossRef] [PubMed]
- Mehta, P.; McAuley, D.F.; Brown, M.; Sanchez, E.; Tattersall, R.S.; Manson, J.J.; Hlh Across Speciality Collaboration, U.K. COVID-19: Consider cytokine storm syndromes and immunosuppression. Lancet 2020, 395, 1033–1034. [Google Scholar] [CrossRef] [PubMed]
- Giavridis, T.; van der Stegen, S.J.C.; Eyquem, J.; Hamieh, M.; Piersigilli, A.; Sadelain, M. CAR T cell-induced cytokine release syndrome is mediated by macrophages and abated by IL-1 blockade. Nat. Med. 2018, 24, 731–738. [Google Scholar] [CrossRef] [PubMed]
- Vijay, R.; Fehr, A.R.; Janowski, A.M.; Athmer, J.; Wheeler, D.L.; Grunewald, M.; Sompallae, R.; Kurup, S.P.; Meyerholz, D.K.; Sutterwala, F.S.; et al. Virus-induced inflammasome activation is suppressed by prostaglandin D(2)/DP1 signaling. Proc. Natl. Acad. Sci. USA 2017, 114, E5444–E5453. [Google Scholar] [CrossRef] [Green Version]
- Cavalli, G.; De Luca, G.; Campochiaro, C.; Della-Torre, E.; Ripa, M.; Canetti, D.; Oltolini, C.; Castiglioni, B.; Tassan Din, C.; Boffini, N.; et al. Interleukin-1 blockade with high-dose anakinra in patients with COVID-19, acute respiratory distress syndrome, and hyperinflammation: A retrospective cohort study. Lancet Rheumatol. 2020, 2, e325–e331. [Google Scholar] [CrossRef]
- Piccoli, C.; Scrima, R.; Quarato, G.; D’Aprile, A.; Ripoli, M.; Lecce, L.; Boffoli, D.; Moradpour, D.; Capitanio, N. Hepatitis C virus protein expression causes calcium-mediated mitochondrial bioenergetic dysfunction and nitro-oxidative stress. Hepatology 2007, 46, 58–65. [Google Scholar] [CrossRef]
- Woods, J.J.; Wilson, J.J. Inhibitors of the mitochondrial calcium uniporter for the treatment of disease. Curr. Opin. Chem. Biol. 2020, 55, 9–18. [Google Scholar] [CrossRef]

| Proteins | Length (AA) 1 | Function, Key Features |
|---|---|---|
| NSP1 | 180 | Suppresses host genes and enhances viral RNA expression |
| NSP2 | 638 | For SARS-CoV replication |
| NSP3 | 1945 | Component of the replication/transcription complex |
| NSP4 | 500 | Assembly of cytoplasmic double-membrane vesicles required for viral replication |
| NSP5 | 306 | Cleavage of viral polyproteins |
| NSP6 | 290 | Inhibits the lysosomal autophagy system and stimulates the NLRP3 inflammasome-dependent pyroptotic pathway |
| NSP7 | 83 | Involved in the replication and transcription of SARS |
| NSP8 | 198 | Viral RNA primers co-localize with RdRP to replicate SARS-CoV |
| NSP9 | 113 | Interact with the replication complex |
| NSP10 | 139 | Interact with NSP14 and NSP16 |
| NSP11 | 13 | Unknown |
| NSP12 | 932 | RNA-dependent RNA polymerase |
| NSP13 | 601 | Zinc-binding domain, NTPase, dNTPase. 5′-to-3′ RNA and DNA helicase, RNA 5′-triphosphate |
| NSP14 | 527 | 3′-to-5′ exoribonuclease, zinc-binding domain, and N7-methyltransferase |
| NSP15 | 346 | Uridylate-specific endoribonuclease, homohexamer |
| NSP16 | 298 | Putative ribose-2′-O-methyltransferase |
| Proteins | Length (AA) | Function (Ref) |
|---|---|---|
| ORF3a | 275 | Ion channel [25] Replication and virulence [26] Activating pro-IL-1β gene expression and IL-1β secretion [27] Ultimately activating NFκB signaling and NLRP3 inflammasomes [27] Promoting the cytokine storm [27] Necrotic cell death and lysosomal damage [28] |
| ORF3b | 22 | IFN antagonism [29] |
| ORF3c | 41 | Unknown [30] |
| ORF3d | 57 | Interaction with STOML2 [31] With ORF8, elicits the strongest antibody responses [32] |
| ORF6 | 61 | Localized to the ER, autophagosomes, and lysosomes [33] Blocks STAT transportation from the cytoplasm to the nucleus [34] IFN antagonist [34,35] Inhibits STAT1 nuclear transportation [36] Accumulation of hnRNPA1 in the nucleus [37] |
| ORF7a | 121 | Antagonizes the IFN-I response [38] Hijacks the host ubiquitin system to enhance and antagonize IFN-I responses [39] Decreases antigen-presenting ability and induces expression of pro-inflammatory cytokines [40] |
| ORF7b | 43 | Transmembrane protein localized in the Golgi [41] |
| ORF8 | 121 | Apoptosis [42] Antagonizing the IFN signaling pathway [43] Interacts with MHC-I [24] IFN I antagonist [44] Activates IL-17 signaling pathway and promote proinflammatory factors [45] |
| ORF9b | 97 | Induces autophagy in host cells mediated by ATG5 [46] Tom70 forms a complex with ORF9b to modulate the host immune response by compromising type I IFN synthesis [47] Activation of inflammasome [48] Promotes proteasomal degradation of Drp1 [46] |
| ORF9c | 73 | Protein interaction between ORF9c protein and NFκB-related molecules [31] Interactions with NFκB-related molecules [49] Interferes with IFN signaling [31,50,51] |
| ORF 10 | 38 | Unknown [52] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chen, T.-H.; Chang, C.-J.; Hung, P.-H. Possible Pathogenesis and Prevention of Long COVID: SARS-CoV-2-Induced Mitochondrial Disorder. Int. J. Mol. Sci. 2023, 24, 8034. https://doi.org/10.3390/ijms24098034
Chen T-H, Chang C-J, Hung P-H. Possible Pathogenesis and Prevention of Long COVID: SARS-CoV-2-Induced Mitochondrial Disorder. International Journal of Molecular Sciences. 2023; 24(9):8034. https://doi.org/10.3390/ijms24098034
Chicago/Turabian StyleChen, Tsung-Hsien, Chia-Jung Chang, and Peir-Haur Hung. 2023. "Possible Pathogenesis and Prevention of Long COVID: SARS-CoV-2-Induced Mitochondrial Disorder" International Journal of Molecular Sciences 24, no. 9: 8034. https://doi.org/10.3390/ijms24098034
APA StyleChen, T. -H., Chang, C. -J., & Hung, P. -H. (2023). Possible Pathogenesis and Prevention of Long COVID: SARS-CoV-2-Induced Mitochondrial Disorder. International Journal of Molecular Sciences, 24(9), 8034. https://doi.org/10.3390/ijms24098034





